Abstract
Mammalian target of rapamycin (mTOR) is a key regulator for cell growth through modulating components of the translation machinery. Previously, numerous pharmacological studies using rapamycin suggested that mTOR has an important role in regulating cardiac hypertrophic growth. To further investigate this assumption, we have generated two lines of cardiac specific mTOR transgenic mice, kinase-dead (kd) mTOR and constitutively active (ca) mTOR, using α-myosin heavy chain promoter. α-Myosin heavy chain (αMHC)-mTORkd mice had a near complete inhibition of p70 S6k and 4E-BP1 phosphorylation, whereas αMHC-mTORca had a significant increase in p70 S6k and 4E-BP1 phosphorylation. Although the cardiac function of αMHC-mTORkd mice was significantly altered, the cardiac morphology of these transgenic mice was normal. The cardiac hypertrophic growth in response to physiological and pathological stimuli was not different in αMHC-mTORkd and αMHC-mTORca transgenic mice when compared with that of nontransgenic littermates. These findings suggest that the mTOR-mediated signaling pathway is not essential to cardiac hypertrophic growth but is involved in regulating cardiac function. Additional analysis of cardiac responses to fasting-refeeding or acute insulin administration indicated that αMHC-mTORkd mice had a largely impaired physiological response to nutrient energy supply and insulin stimulation.
The generation of a normal sized organ is achieved by the control of cell number and cell size. Similarly, the size of the heart is regulated by hyperplastic growth (increase of cell number) and hypertrophic growth (increase of cell size). Normal cardiomyocyte proliferation and hypertrophic growth have a critical role in postnatal cardiac functions (1, 2). The molecular mechanism of regulating the size of cardiomyocytes remains as one of the major research interests in molecular cardiology.
Mammalian target of rapamycin (mTOR)4 is a large (289 kDa) and evolutionarily conserved member of phosphatidylinositol kinase-related kinase family. mTOR appears to have multiple biological functions. The most well characterized area is its control of cellular growth and proliferation via the regulation of protein translational machinery. Under most circumstances, protein synthesis is regulated by translation initiation, a complex process in which mRNA template, the ribosome, and aminoacylated tRNA are assembled in a correct configuration. The assembly of this translational machinery requires a group of proteins called eukaryotic initiation factors (eIFs). An important finding over the past decade is that mTOR and its mediated signaling pathway regulate the assembly of the translational machinery. In mammalian cells, mTOR is associated with two additional proteins, Raptor and mLST8/GβL, to form complex 1 (mTORC1). Raptor and mLST8/GβL are homologues of the yeast KOG1p and LST8p, respectively (3). Via Raptor, mTOR interacts with and phosphorylates p70 S6k and eIF4E-binding proteins (4E-BPs), including 4E-BP1, 4E-BP2, and 4E-BP3, and thereby regulating protein synthesis and cell growth (4, 5). p70 S6k directly phosphorylates the 40 S ribosomal protein S6. Increased S6 phosphorylation enhances the translation of transcripts with 5′-terminal oligopyrimidine sequences (6). 4E-BP1 can be phosphorylated at multiple sites after stimulation, and phosphorylated 4E-BP1 results in its dissociation from eIF4E, thereby allowing eIF4E to assemble with eIF4G, facilitating the recruitment of other translation initiation factors to form the eIF4F complex and initiate cap-dependent translation (5). In addition to the mediation of mitogenic signals (7–9), mTOR also likely functions as a nutritional checkpoint, as its downstream effectors are sensitive to both amino acid concentrations (10) and energy status (11).
Recent studies show that mTOR also exists in a second signaling complex with mLST8/GβL and a protein termed Rictor, rather than Raptor. This second mTOR complex (mTORC2), unlike mTORC1, is rapamycin-resistant and functions as an Akt kinase (12, 13). It has been shown that the function of mTORC2 is regulated by Sin1 (14). Sin1 appeared to be required for mTORC2 kinase activity in phosphorylating Akt on Ser-473 and to facilitate the interaction between Rictor and mTOR.
A key feature for cardiomyocyte hypertrophic growth is the significant increase of protein synthesis. Previous pharmacological studies using rapamycin demonstrated a potential use of rapamycin in inhibiting the cardiomyocyte hypertrophic response and the activation of protein synthesis induced by phenylephrine (PE) (15), endothelin (ET-1) (16), angiotensin II (17), or insulin (18) in cultured cardiomyocytes. However, the results derived from in vivo experiments were inconsistent. Several studies demonstrated that rapamycin could attenuate or regress pressure overload- and constitutively active Akt-induced cardiac hypertrophy (16, 19, 20), suggesting that mTOR is an important regulator for cardiac hypertrophy. However, a number of other lines of evidence argued against the importance of mTOR in cardiac hypertrophic growth. Treatment of mice from 3 to 4 weeks of age with rapamycin (4 mg/kg/day) significantly decreased p70 S6k kinase activity, but the treatment did not suppress postnatal cardiac growth when compared with mice treated with vehicle (19). The ablation of p70 S6k (S6k1) and p54 S6k (S6k2), which was believed to be regulated by mTOR, had no impact on the development of pathological, physiological, insulin-like growth factor 1 receptor-mediated, and PI3K-mediated cardiac hypertrophy (21). In addition, transgenic mice with cardiac specific overexpression of kinase-dead Akt or knock-out mice deficient in p70 S6k gave rise to hearts with normal sizes (19, 22).
To further examine the role of mTOR in cardiac hypertrophic growth, we generated two lines of transgenic mice expressing constitutively active (ca) or kinase-dead (kd) mutant mTOR exclusively in myocardium. Our results showed that cardiac specific overexpression of αMHC-mTORca or αMHC-mTORkd mutant proteins by 4–6-fold led to significant alterations in mTOR-mediated signaling. αMHC-mTORkd mice had a near complete inhibition of p70 S6k and 4E-BP1 phosphorylation, whereas αMHC-mTORca had a significant increase in p70 S6k and 4EBP1 phosphorylation. Interestingly, the cardiac morphology of these transgenic mice was normal. The cardiac hypertrophic growth of αMHC-mTORkd transgenic mice to physiological and pathological stimuli was not different from that of nontransgenic littermates. However, the cardiac function of αMHC-mTORkd mice was significantly altered. These findings suggest that mTOR-mediated signaling pathway is not essential to cardiac hypertrophic growth but is involved in regulating cardiac function. Additional analysis of cardiac responses to feeding or acute insulin administration indicated that αMHC-mTORkd mice had a largely impaired physiological response to nutrient energy supply and insulin stimulation, which might explain altered cardiac functions in αMHC-mTORkd mice.
MATERIALS AND METHODS
mTOR Mutants—pcDNA3/mTOR eukaryotic expression plasmids encoding kinase-dead (D2338A) or constitutively active (deletion of amino acid residues 2430–2450) rat mTOR were generated previously (9). The expression of the mutant proteins was confirmed by transient transfection into C2C12 cells, followed by Western blot analysis.
Generation of Transgenic Mice—The αMHC promoter drives transgenes to express exclusively in cardiac myocytes and has been used extensively in transgenic studies. To generate mTORkd and mTORca cardiac specific transgenic mice, we placed an αMHC promoter at the 5′ end of mTOR mutant cDNAs. To distinguish transgenic mTOR from endogenous mTOR, AU1 tag was placed at the 5′ end of mTOR cDNA (Fig. 1A). Transgenic DNAs were microinjected into zygotes by standard methodology as described previously (23). Eight independently derived founders for αMHC-mTORkd and six founders for αMHC-mTORca were produced. To minimize gene silencing because of genomic DNA methylation, transgenic mouse lines were maintained in DBA/2J background (back-crossed for at least 7 generations). All animal experiments were in strict compliance with the National Institutes of Health guidelines within an American Association for Laboratory Animal Care certified facility, and the study protocols were approved by the Institutional Animal Care and Use Committee.
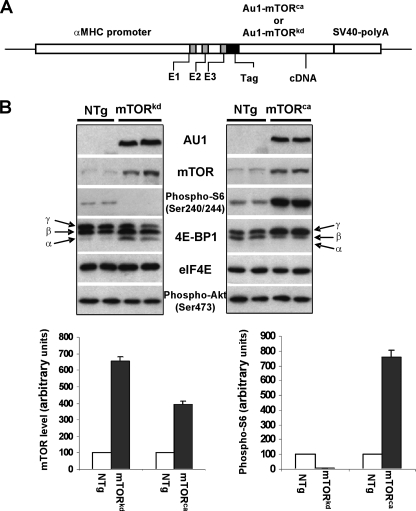
FIGURE 1.
Generation of αMHC-mTORkd and αMHC-mTORkd transgenic mice. A, schematic diagram of constructs. B, Western blot analysis of the level of transgenic mTOR expression, total mTOR, phospho-S6, phospho-Akt, and different phosphorylated 4E-BP1 isoforms in αMHC-mTORkd and αMHC-mTORca transgenic mice and littermate controls. Cardiac protein extracts were probed with antibodies against AU1 tag, mTOR, phospho-S6, 4E-BP1, and eIF4E (loading control). 4E-BP1 phosphorylation was demonstrated by changes in the migration rate of the protein on 15% SDS-PAGE, with increased phosphorylation (γ) corresponding to decreased electrophoretic mobility. NTg, nontransgenic.
Histological Analysis—Heart samples were harvested, fixed, sectioned, stained with hematoxylin and eosin for cardiac histology and Masson's trichrome for potential cardiac fibrosis, and subjected to in situ hybridization and immunohistochemical analyses as described previously (24).
Morphometric Measurement of Isolated Cardiomyocytes— Cardiomyocytes were enzymatically dissociated from the heart with 0.17% type I collagenase (Worthington) and stained with Hoechst to demonstrate nuclei. Only typical rod-shaped cardiomyocytes were included in the morphometric measurement. Using ImagePro plus 5.1 software, the long axis, short axis, and cell area were measured and compared between transgenic and control mice. To count the total number of cardiomyocytes in adult hearts, formalin-fixed hearts (the atria were removed) were digested with 50% KOH for 24 h (25). After careful washing with phosphate-buffered saline, rod-shaped cells were counted using a hemocytometer.
Echocardiogram and Electrocardiogram (ECG) Recording— Transthoracic echocardiograms were performed as described previously (26) under 1.5–2.0% isoflurane. Two-dimensional short-axis images were obtained using a high-resolution Vevo 770 Imaging System (Visualsonics Inc., Toronto, Canada) equipped with a 35-MHz scan probe. Left ventricular chamber dimension and wall thickness at systolic and diastolic phases were measured from M-mode recording. Heart rate was calculated from simultaneous electrocardiogram recording. Left ventricular volume, fractional shortening, and ejection fraction were calculated using the Vevo Analysis program as described (26).
For ECG recording, mice were anesthetized with 4% isoflurane before placing the electrodes. With continuous 2% isoflurane through a face mask, three subdermal needle electrodes (model F-E2, Grass Technologies, West Warwick, RI) were impaled in both the front legs and the left rear leg, where the left front served as the positive input, the right front as the negative input, and the left rear leg as the reference to the isolated biological amplifier (ISO-Dam, World Precision Instruments Inc, Sarasota, FL). The gain was set at 1000, and the filters were set at 0.1 Hz for the low filter and 1 kHz for the high filter, respectively. The base line was adjusted to zero. The ECG was then sampled at 2 kHz and digitized using a 16-bit data acquisition card (DAQCard-AI-16XE-50 National Instruments, Austin, TX) under the control of a custom-written program in Lab-Windows/CVI (National Instruments, Austin, TX). The ECG was displayed in real time and was saved for analysis. ECG traces were further filtered with an 8th order Butterworth filter (0.5–300 Hz) and plotted in Matlab (The MathWorks, Inc., Natick, MA).
Swimming Training—The protocol was as described previously (2). Briefly, 8-week-old male mice weighing 15–20 g were made to swim in tanks with a surface area of 225 cm2 and a depth of 15 cm and water temperature of 30–32 °C. Animals were initially exercised for 20 min, twice daily, and the duration of exercise was increased in 10-min increments daily, reaching 90 min, twice daily, by the end of the 2nd week. The duration of exercise was maintained until the end of the study (4 weeks). At the end of the study, the animals were sacrificed, and their heart weights and body weights were determined.
Isoproterenol Treatment Using Osmotic Pumps—Three-month-old male αMHC-mTORkd mice and their nontransgenic siblings were treated with the β-adrenergic agonist, isoproterenol, using Alzet osmotic mini-pumps (7-day infusion, model 2001, flow rate of 1 μl/h, 0.028 g/ml isoproterenol dissolved in saline) as described previously (28).
Insulin, Fasting, and Refeeding Study—Eight-week-old male mice were used in these studies. The animals were first fasted for 18 h, followed by refeeding (simply reintroducing chow to the cage), or a single intraperitoneal injection of insulin (2 IU/kg) (pork insulin, Lilly). Sixty minutes after insulin injection, the animals were sacrificed, and heart and skeletal muscle samples were harvested. The tissue samples were briefly rinsed with ice-cold phosphate-buffered saline and immediately frozen in liquid nitrogen.
Western Blotting—The heart and skeletal muscle samples were homogenized, and Western blotting was performed as described (29). Phosphorylation of 4E-BP1, Akt, eIF4G, p70 S6k, and S6 at designated sites was measured similarly except that phospho-specific antibodies (all from Cell Signaling Technology, Inc., Beverly, MA) were used. Phosphorylation of 4E-BP1 and p70 S6k was also determined by viewing the changes of the migration rate of the proteins on SDS-PAGE. Phosphorylation of 4E-BP1 and p70 S6k retarded the protein migration rate on SDS-polyacrylamide gel. Consequently, when protein extract is subjected to SDS-PAGE, multiple electrophoretic forms are resolved, with increased phosphorylation corresponding to decreased electrophoretic mobility. In addition, association of 4E-BP1 with eIF4E was assessed by measuring the amount of 4E-BP1 recovered from eIF4E extraction as described (29).
Statistics—Results are presented as the mean ± S.E. Densitometry data were normalized to nontransgenic littermate controls on the same blot. Differences between the groups were compared by using the Student's t test. p < 0.05 was considered significant.
RESULTS
Generation of αMHC-mTORkd and αMHC-mTORca Transgenic Mice—To study the impact of mTORkd and mTORca on postnatal cardiac growth and function, we generated αMHC-mTORkd and αMHC-mTORca transgenic mice (Fig. 1A). The cardiomyocyte specific αMHC promoter has a transient burst of activity in embryonic heart between E9.5 and 10.5 and is reactivated during early postnatal life and remains persistently high into adulthood (30). Eight transgenic founders carrying the αMHC-mTORkd transgene and six transgenic founders carrying the αMHC-mTORca transgene were generated. Reverse transcription-PCR and Western blotting analysis confirmed cardiac overexpression of mTOR transgenes. These transgenic mice appear to have normal embryonic cardiac development and normal life span (survive over 12 months of age). Detailed characterization of αMHC-mTORkd and αMHC-mTORca transgenic mice was performed on the transgenic lines with highest transgenic expression level. Fig. 1B represents the use of anti-AU1 and anti-mTOR antibodies in determining the level of expression in αMHC-mTORkd and αMHC-mTORca mice. Western blots showed that these transgenic mTORs (mTORkd and mTORca) were expressed 4–6-fold higher in transgenic mouse hearts (Fig. 1B).
Altered mTOR-Mediated Signaling in αMHC-mTORkd and αMHC-mTORca Transgenic Hearts—It has been demonstrated previously in cultured cells that a point mutation at aspartic acid residue 2338 (D2338A) prevented mTOR autophosphorylation and mTOR kinase activity (31), and that an mTOR mutant lacking amino acid residues 2430–2450 constantly displayed an elevated level of protein kinase activity relative to full-length mTOR wild type (9). To confirm this in transgenic hearts, we measured the phosphorylation levels of the ribosomal proteins S6 and 4E-BP1, two of the well established mTORC1-mediated downstream targets, in the hearts of αMHC-mTORkd and αMHC-mTORca transgenic mice. As shown in Fig. 1B, the level of S6 phosphorylation decreased to almost an undetectable level in αMHC-mTORkd heart but increased by 8-fold in αMHC-mTORca heart. Similarly, the phosphorylation of 4E-BP1 was dramatically reduced in αMHC-mTORkd heart, but was significantly increased in αMHC-mTORca heart as compared with nontransgenic littermate control hearts. In nontransgenic mice, 4E-BP1 phosphorylation, expressed as percentage of the γ form, was about 50%. This figure dropped to 20% in the heart of αMHC-mTORkd hearts and increased to 85% in the αMHC-mTORca hearts. These experiments indicate that mTOR-mediated signaling via mTORC1 is altered in the heart of αMHC-mTORkd and αMHC-mTORca transgenic mice. To determine whether overexpression of mTORkd and mTORca would also have impact on Akt phosphorylation, an event could be potentially mediated through mTORC2 (13), we performed Western blot analysis using specific antibody against pAkt-Ser-473. Our results showed that the level of Akt phosphorylation was not altered in the hearts of either αMHC-mTORkd or αMHC-mTORca mice (Fig. 1B). Taken together, our data suggested that overexpression of mTORkd or mTORca in the heart affected the mTORC1-mediated pathway but not the level of Akt phosphorylation.
Altered Cardiac Function but No Morphological Defect in αMHC-mTORkd Mice—Despite clear alterations in mTOR-mediated signaling in αMHC-mTORkd and αMHC-mTORca transgenic hearts, we failed to find any visible abnormal morphological and histological alteration in the size of the hearts. First, we compared heart weight, body weight, and the ratio of heart weight to body weight in αMHC-mTORkd, αMHC-mTORca, and nontransgenic littermate controls at 1, 3, and 5 months of age. There was no evidence of either an increase or decrease in heart weight and size in αMHC-mTORkd and αMHC-mTORca transgenic mice (Fig. 2A). Histological analysis did not demonstrate that αMHC-mTORkd and αMHC-mTORca transgenic mice developed any significant cardiac hypertrophy, atrophy, cardiac fibrosis, or myocyte disarray, indicating that overexpression of either mTORkd or mTORca did not assert a large impact on cardiac growth under normal physiological conditions (Fig. 2A). Furthermore, we also used [3H]thymidine labeling to analyze the cell cycle activity of the transgenic heart. There was no abnormal cell cycle activity observed in αMHC-mTORkd and αMHC-mTORca transgenic hearts (data not shown). In addition, we measured cell size of isolated cardiomyocytes from αMHC-mTORkd and αMHC-mTORca transgenic and nontransgenic control mice and confirmed at cellular level that there was no alteration in cell size in both transgenic hearts (Fig. 3A).
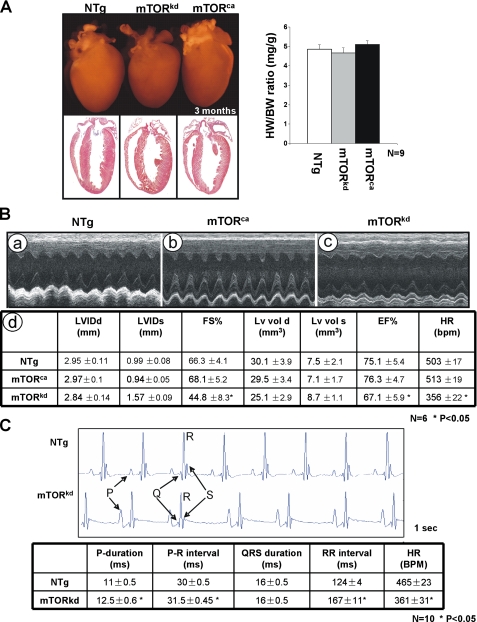
FIGURE 2.
Characterization of αMHC-mTORkd and αMHC-mTORkd transgenic mice. A, left panel, comparison of the gross morphology and histology of αMHC-mTORkd and αMHC-mTORca transgenic and littermate nontransgenic (NTg) control heart. There was no obvious morphological defect in both αMHC-mTORkd and αMHC-mTORca transgenic hearts when compared with littermate controls. Right panel, quantitative comparison of the ratio of heart weight versus body weight of αMHC-mTORkd and αMHC-mTORca transgenic hearts and nontransgenic littermate control. B, echocardiograph analysis of αMHC-mTORkd and αMHC-mTORca transgenic mice and littermate controls (3-month-old male). Representative M-Mode images are shown in panels a–c. Measurements of various parameters and statistics analysis are summarized in panel d. C, ECG recording of the αMHC-mTORkd and littermate control mice (3-month-old male). There was a difference in the function of atria and sinoatrial node in αMHC-mTORkd hearts compared with littermate controls.
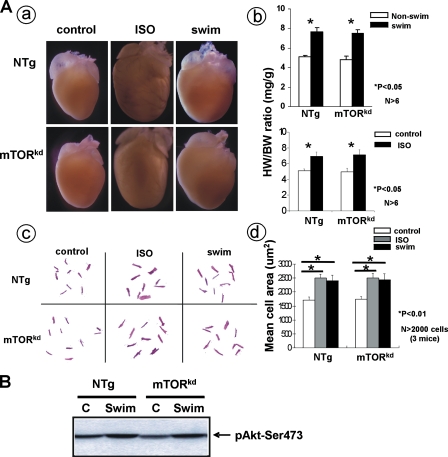
FIGURE 3.
Testing cardiac response of αMHC-mTORkd transgenic mice to hypertrophic stimulation. A, comparison ofαMHC-mTORkd transgenic and littermate control hearts (3-month-old male) in response to isoproterenol and swimming stimulation. A, panel a, gross morphology of the hearts; panel b, statistic comparison of the ratio of heart weight versus body weight without and with swimming exercise (swim) or isoproterenol treatment (iso). There is no significant difference in change of heart weight after swimming exercise or isoproterenol treatment among these mice. Panel c, morphological comparison of the size of dispersed cardiomyocytes of nontreated control, swimming exercised, and isoproterenol-treated mice. Panel d, statistic comparison of the cell size. Similar degree of hypertrophic response was observed in nontransgenic (NTg) and αMHC-mTORkd transgenic cardiomyocytes. B, Western blot analysis of activated Akt level in basal (nonexercised) and exercised hearts. Akt activation is not altered in αMHC-mTORkd mice. C, control.
To determine the functional aspect of αMHC-mTORkd and αMHC-mTORca transgenic hearts, we have used an echocardiograph to analyze the cardiac structure and function. Surprisingly, contrary to an overall normal cardiac histology, αMHC-mTORkd mice displayed slower heart rates and significantly altered diastolic and systolic ventricular chamber volumes associated with reduced cardiac function. Both ejection fraction and fraction shortening were reduced (Fig. 2B). However, αMHC-mTORca mice have normal cardiac function (Fig. 2B). The slower heart rate was further confirmed by ECG analysis (Fig. 2C). Although the QRS duration was not altered in αMHC-mTORkd mice, the P wave duration and the RR interval were significantly prolonged in αMHC-mTORkd mice. In addition, beat-to-beat RR interval variation was also evident in all αMHC-mTORkd mice, indicating altered pacemaker function of the sinoatrial node. This observation suggests that the conduction velocity of atria, but not ventricle, is affected in αMHC-mTORkd transgenic heart. These data suggest that mTOR kinase activity is not critical to normal cardiac hypertrophic growth but is more relevant in regulating cardiac function.
Cardiac Hypertrophic Response in αMHC-mTORkd Transgenic Mice—A previous study indicated that rapamycin could attenuate the development of hypertrophy induced by physiological stimuli, such as swimming exercise (19). We initially hypothesized that mTOR might be an important player in cardiac physiological hypertrophic response. To test this, a swimming exercise test was performed on 3-month-old αMHC-mTORkd transgenic male mice as described previously (2). Sex-matched nontransgenic littermates were used as controls. Surprisingly, the cardiac mass in αMHC-mTORkd mice subjected to 4 weeks of swimming exercise had a similar level of increase when compared with littermate controls (Fig. 3A). The percentage increase of the heart weight/body weight ratio (mg/g) in response to swim training was similar between nontransgenic control (59%) and αMHC-mTORkd mice (65%) (n = 6), suggesting that reduced mTOR kinase activity has no impact on the cardiomyocyte hypertrophic growth in response to physiological stimuli.
Akt activation was shown to be associated with cardiac hypertrophy induced by swimming exercise. Because Akt activity could also be mediated by mTOR via direct phosphorylation on Ser-473, we asked whether exercise-induced Akt signaling was altered in αMHC-mTORkd hearts subjected to swimming exercise. Using Western blot analysis, we measured the level of phosphorylated Akt on Ser-473 in the hearts of αMHC-mTORkd transgenic and nontransgenic littermate controls. Consistent with the normal cardiac phenotype, we found that Akt phosphorylation in response to swimming exercise was not altered in αMHC-mTORkd hearts (Fig. 3B), which further confirmed that overexpression of mTORkd had no impact on Akt signaling.
Previously, a series of pharmacological studies using rapamycin demonstrated a potential use of rapamycin in inhibiting the hypertrophic response by certain pathological stimuli, such as PE (15), ET-1 (16), and angiotensin II (17) in cultured cardiomyocytes. However, this hypertrophic inhibiting activity has not been assessed in vivo. To determine whether cardiac overexpression of mTORkd would alter the ability to respond to β-adrenergic agonist-induced cardiac hypertrophy, 3-month-old αMHC-mTORkd mice and their nontransgenic littermates were treated with isoproterenol as described previously (28). This treatment typically results in a 30–40% increase in heart weight/body weight ratio, which is reflected by uniform hypertrophic cardiomyocyte growth (28). Control mice were treated with saline-filled minipumps. Similar to the response from the swimming exercise, a marked and similar increase in cardiac mass and cardiomyocyte size were apparent in both the αMHC-mTORkd and nontransgenic mice treated with isoproterenol (Fig. 3A), indicating that disrupted mTOR kinase activity does not alter β-adrenergic signaling in postnatal myocardium in vivo. Once again, this result disagrees the previous pharmacological studies using rapamycin as an mTOR inhibitor in cultured neonatal cardiomyocytes in vitro (15, 16).
Impaired Response to Nutrient Energy Supply and Insulin Stimulation in αMHC-mTORkd Mice—Although we did not observe a significant defect in cardiac growth and hypertrophic response, the alteration in cardiac function in αMHC-mTORkd mice suggested that inhibition of mTORC1-mediated signaling could nevertheless impact cardiac physiology. Hormones (e.g. insulin) and nutrients (e.g. amino acid and glucose) have been shown to play an important role in both fetal and postnatal cardiac growth and function. Heart weight and myocyte size are reduced in starved subjects and in individuals on severe weight loss diets. Mice with cardiomyocyte-selective insulin receptor knock-out exhibit significant reduction in cardiac myocyte size and persistent expression of the fetal β-myosin heavy chain isoform (32). In contrast, insulin supplementation has been shown to promote cardiomyocyte hypertrophy in cells in culture (33). Because one of the major physiological functions for mTOR via mTORC1-mediated pathway is its involvement in cellular response to nutrient and energy supplies, we speculated that the abnormal cardiac function in αMHC-mTORkd transgenic mice was associated with altered responses to nutrient energy and insulin treatment. Therefore, we conducted a set of studies to investigate the response of mTOR-mediated signaling to fasting-refeeding and insulin stimulation in αMHC-mTORkd mice.
Our results showed that feeding-induced Akt phosphorylation in the hearts of αMHC-mTORkd mice was not altered when compared with nontransgenic mice, but the mTOR-mediated signaling (p70 S6k phosphorylation at Thr-389) was markedly inhibited in the hearts of αMHC-mTORkd mice, suggesting that feeding-induced activation of mTORC1-mediated signaling was largely impaired in the αMHC-mTORkd heart (Fig. 4A).
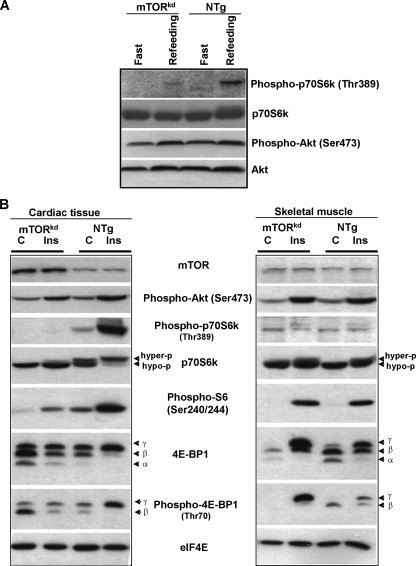
FIGURE 4.
Testing cardiac response of αMHC-mTORkd mice to nutrient energy supply and insulin stimulation. A, Western blot analysis of αMHC-mTORkd and littermate nontransgenic control mice subjected to fast-refeeding protocol. p70 S6k (Thr-389) was activated by refeeding in control heart, but severely inhibited in αMHC-mTORkd heart, whereas Akt activation is not affected in αMHC-mTORkd mice. B, Western blot analysis of αMHC-mTORkd and littermate nontransgenic control (C) mice subjected to insulin (Ins) stimulation. The levels of phosphorylation of p70 S6k and 4E-BP1 were significantly reduced in αMHC-mTORkd hearts when compared with littermate nontransgenic (NTg) control subjected with insulin administration. The skeletal muscle of αMHC-mTORkd mice was not affected when compared with controls.
Similar to the feeding experiments, Akt phosphorylation in αMHC-mTORkd hearts in response to insulin administration was normal; however, the phosphorylation of 4E-BP1, S6, and p70 S6k was greatly impaired in αMHC-mTORkd hearts when compared with control hearts (Fig. 4B, left panel). Using skeletal muscle samples as internal controls, a similar response of the mTOR-mediated signaling to insulin stimulation was seen in skeletal muscles isolated from identical αMHC-mTORkd transgenic mice and nontransgenic littermates (Fig. 4B, right panel).
The reduced response of the mTOR signaling to insulin stimulation in αMHC-mTORkd mice was also reflected by alterations in the amount of 4E-BP1 associated with eIF4E. Previous studies have demonstrated that insulin decreases eIF4E-4E-BP1 interaction because of enhanced 4E-BP1 phosphorylation. In this study, we determined the amount of 4E-BP1 in eIF4E-4E-BP1 complexes using the m7GTP-Sepharose 4B binding assay. Consistent with reduced 4E-BP1 phosphorylation level, the reduction in eIF4E-4E-BP1 interaction in response to insulin treatment was only observed in αMHC-mTORkd skeletal muscle but not in αMHC-mTORkd hearts (Fig. 5, A–C).
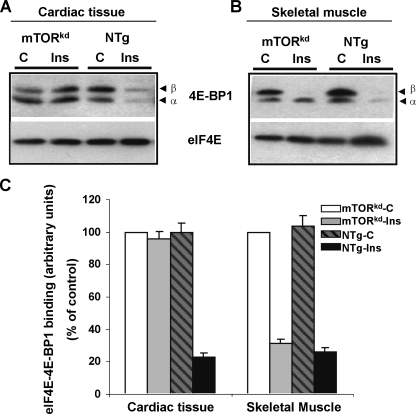
FIGURE 5.
Biochemical analysis of the level of 4E-BP1 bound to eIF4E in αMHC-mTORkd mice. A, in nontransgenic (NTg) mice, insulin (Ins) was able to reduce the amount of 4E-BP1 bound to eIF4E. In contrast, the amount of 4E-BP1 bound to eIF4E was not found reduced in αMHC-mTORkd heart. B, skeletal muscle sample from identical mice presented in A was used as internal control. C, quantitative and statistic comparison from independent experiments. Each bar represents the average of three experiments, and values are expressed as percentage versus control (set at 100%).
DISCUSSION
Our study was initially designed to address and clarify the role of mTOR-mediated signaling in cardiac hypertrophic growth. Using a cardiomyocyte-specific promoter, we were able to overexpress two forms of mutant mTOR (mTORkd and mTORca) in myocardium. The point mutation at aspartic acid residue 2338 led to a dominant negative mutant mTOR that was able to inhibit both S6 and 4E-BP1 phosphorylation; the deletion of amino acid residues 2430–2450 led to a constitutively active mTOR that was able to enhance both S6 and 4E-BP1 phosphorylations. Our findings are consistent with previous in vitro studies using cultured cells (9) and confirm that mTORkd functions in a dominant negative fashion, and mTORca acts as a constitutively active mutant in vivo. One interesting aspect is that these mutant mTORs apparently have a greater effect on mTORC1. Akt (Ser-473) phosphorylation, a potential target of mTORC2, is not altered in these transgenic mice. The underlying mechanism is not clear. Nevertheless, our data provide us a unique assessment of the role of kinase activity of mTOR in regulating cardiac growth and function.
It is likely that mTORkd overexpression (about 6-fold higher than endogenous mTOR level) competes with endogenous mTOR for partner proteins. The observation that the phosphorylation of p70 S6k at Thr-389 and the phosphorylation of the S6 protein at Ser-240/244 were near undetectable in the hearts of αMHC-mTORkd mice suggested that the remaining activity of endogenous mTOR was effectively diminished, although we cannot rule out some mTOR activity might still remain in αMHC-mTORkd transgenic hearts as this was not a complete loss of function approach such as genetic targeted deletion, and the remaining mTOR activity could well be responsible for the normal cardiac growth phenotype. Initially we were concerned whether this potentially remaining endogenous mTOR activity would contribute to the mild abnormal phenotype in αMHC-mTORkd transgenic hearts. However, the lack of an obvious defect in cardiac hypertrophy growth in αMHC-mTORkd mice is in strong agreement with a number of recent reports using genetic modified mouse models. Cardiac specific overexpression of kinase-dead mutants of p70 S6k and p54 S6k did not give rise to defects in cardiac hypertrophic growth (21). Furthermore, ablation of both p70 S6k (S6K1) and p54 S6k (S6K2) did not attenuate pathological, physiological, insulin-like growth factor 1 receptor-induced, or PI3K-induced cardiac hypertrophy (21). Interestingly, overexpressing a kinase-dead mutant of Akt in myocardium led only to a defect in physiological hypertrophic growth (exercise-induced), but not to the normal cardiac growth, despite significant reduction in activation of p70 S6k activity (presumably mediated by mTOR) in these transgenic mice (19). In addition, αMHC-mTORca mice, a gain of function model, also fail to develop hypertrophic hearts. Taken together, we believe that mTOR alone is not an essential element in regulating cardiac hypertrophic growth.
Our results are seemingly contradictory to previous other reports based on the use of rapamycin. Several studies pointed to a role of mTOR in the development of physiological and pathological cardiac hypertrophy under certain circumstances. Rapamycin blocks the activation of protein synthesis induced by PE or ET-1 (15, 16). Rapamycin also blocks the increase in heart weight and/or cardiomyocyte size induced by overload, PE, angiotensin II, or overexpression of constitutively active Akt (15, 17, 19, 20, 34). It is interesting to note that almost all pharmacological studies using rapamycin support the idea that mTOR may play a role in the cardiac hypertrophic growth, whereas genetic interference of the PI3K-mTOR-p70 S6k pathways usually does not lead to measurable phenotypes. The one exception demonstrated that αMHC-dnPI3K mice did develop smaller hearts (35). However, PI3K is an upstream kinase that impacts on multiple pathways. Initially, it was speculated that the smaller hearts in αMHC-dnPI3K mice were because of the reduction of p70 S6k activity. An attempt to rescue this small heart phenotype via the genetic cross of αMHC-dnPI3K mice and p70 S6k transgenic mice was not successful (21), suggesting that the small heart phenotype observed in αMHC-dnPI3K transgenic mice was not because of the reduced mTOR-mediated signaling in these mice. It should be noted that rapamycin does not alter mTOR kinase activity (27). Exactly how the rapamycin impacts on the function of mTOR is still an open question. It is also not entirely clear whether rapamycin has an uncharacterized effect on other signaling pathways. We could not rule out the possibility that some inhibitory effect of rapamycin to cardiac hypertrophic growth could be mediated via an unknown mechanism other than inhibiting the mTOR signaling. This apparent discrepancy will not be resolved until these questions are appropriately addressed in the future.
Despite the fact that we did not observe clear defects in cardiac hypertrophic growth in αMHC-mTORkd and αMHC-mTORca mice, we did find significantly altered cardiac function in αMHC-mTORkd mice, including defects in sinoatrial node pacemaker function, slower atrial conduction velocity, and decreased ventricular contractile function. This abnormal cardiac rhythm could be responsible for the altered cardiac contractile function in αMHC-mTORkd mice as revealed by echocardiograph. This finding suggests that mTOR has a unique role in regulating cardiac physiological function. Future studies will aim to determine the molecular mechanism that leads to this altered cardiac function in αMHC-mTORkd mice. Our data further demonstrate that αMHC-mTORkd heart is associated with reduced response to nutrient energy supply and insulin stimulation. This finding is not surprising considering the well characterized role of mTOR in nutrient signaling, energy signaling, and insulin signaling. The abnormal response to nutrient energy supply and insulin stimulation may explain the altered cardiac function in αMHC-mTORkd mice, as hormones/growth factors (e.g. insulin and insulin-like growth factor 1) and nutrients (e.g. amino acid and glucose) have been shown to play a significant role in both fetal and postnatal cardiac growth and function (32, 33). How mTOR regulates cardiac function via mediating nutrient and insulin signaling remains to be further investigated.
In summary, using transgenic approach, we have assessed the kinase activity of mTOR in cardiac hypertrophic growth and function and cardiac response to nutrient energy supply and insulin stimulation. We conclude that mTOR kinase activity is not essential to cardiac hypertrophic growth but is involved in regulating cardiac function and the physiological response of the cardiomyocytes to nutrient energy and insulin stimulation.
Acknowledgments
We thank Dr. Shaolian Jing and William Carter of Indiana University Mouse Core for their superb assistance in generating mTOR transgenic mice.
This work was supported, in whole or in part, by National Institute of Health grants (to W. S. and L. J. F.). This work was also supported by the Riley Children's Foundation (to W. S., L. J. F., D. W. B., and E. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: mTOR, mammalian target of rapamycin; eIF, eukaryotic initiation factor; αMHC, α-myosin heavy chain; PI3K, phosphoinositide 3-kinase; ca, constitutively active; kd, kinase-dead; ECG, electrocardiogram; PE, phenylephrine; ET-1, endothelin; 4E-BP, eIF4E-binding protein.
References
- 1.Soonpaa, M. H., and Field, L. J. (1998) Circ. Res. 83 15-26 [DOI] [PubMed] [Google Scholar]
- 2.Chen, H., Yong, W., Ren, S., Shen, W., He, Y., Cox, K. A., Zhu, W., Li, W., Soonpaa, M., Payne, R. M., Franco, D., Field, L. J., Rosen, V., Wang, Y., and Shou, W. (2006) J. Biol. Chem. 281 27481-27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., Tokunaga, C., Avruch, J., and Yonezawa, K. (2002) Cell 110 177-189 [DOI] [PubMed] [Google Scholar]
- 4.Dufner, A., and Thomas, G. (1999) Exp. Cell Res. 253 100-109 [DOI] [PubMed] [Google Scholar]
- 5.Gingras, A. C., Raught, B., and Sonenberg, N. (2001) Genes Dev. 15 807-826 [DOI] [PubMed] [Google Scholar]
- 6.Jefferies, H. B., Fumagalli, S., Dennis, P. B., Reinhard, C., Pearson, R. B., and Thomas, G. (1997) EMBO J. 16 3693-3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, Y., Vilella-Bach, M., Bachmann, R., Flanigan, A., and Chen, J. (2001) Science 294 1942-1945 [DOI] [PubMed] [Google Scholar]
- 8.Scott, P. H., Brunn, G. J., Kohn, A. D., Roth, R. A., and Lawrence, J. C., Jr. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7772-7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekulic, A., Hudson, C. C., Homme, J. L., Yin, P., Otterness, D. M., Karnitz, L. M., and Abraham, R. T. (2000) Cancer Res. 60 3504-3513 [PubMed] [Google Scholar]
- 10.Rohde, J., Heitman, J., and Cardenas, M. E. (2001) J. Biol. Chem. 276 9583-9586 [DOI] [PubMed] [Google Scholar]
- 11.Dennis, P. B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S. C., and Thomas, G. (2001) Science 294 1102-1105 [DOI] [PubMed] [Google Scholar]
- 12.Hresko, R. C., and Mueckler, M. (2005) J. Biol. Chem. 280 40406-40416 [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098-1101 [DOI] [PubMed] [Google Scholar]
- 14.Yang, Q., Inoki, K., Ikenoue, T., and Guan, K. L. (2006) Genes Dev. 20 2820-2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boluyt, M. O., Zheng, J. S., Younes, A., Long, X., O'Neill, L., Silverman, H., Lakatta, E. G., and Crow, M. T. (1997) Circ. Res. 81 176-186 [DOI] [PubMed] [Google Scholar]
- 16.Wang, L., and Proud, C. G. (2002) Circ. Res. 91 821-829 [DOI] [PubMed] [Google Scholar]
- 17.Sadoshima, J., and Izumo, S. (1995) Circ. Res. 77 1040-1052 [DOI] [PubMed] [Google Scholar]
- 18.Wang, L., Wang, X., and Proud, C. G. (2000) Am. J. Physiol. 278 H1056-H1068 [DOI] [PubMed] [Google Scholar]
- 19.Shioi, T., McMullen, J. R., Kang, P. M., Douglas, P. S., Obata, T., Franke, T. F., Cantley, L. C., and Izumo, S. (2002) Mol. Cell. Biol. 22 2799-2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shioi, T., McMullen, J. R., Tarnavski, O., Converso, K., Sherwood, M. C., Manning, W. J., and Izumo, S. (2003) Circulation 107 1664-1670 [DOI] [PubMed] [Google Scholar]
- 21.McMullen, J. R., Shioi, T., Zhang, L., Tarnavski, O., Sherwood, M. C., Dorfman, A. L., Longnus, S., Pende, M., Martin, K. A., Blenis, J., Thomas, G., and Izumo, S. (2004) Mol. Cell. Biol. 24 6231-6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pende, M., Um, S. H., Mieulet, V., Sticker, M., Goss, V. L., Mestan, J., Mueller, M., Fumagalli, S., Kozma, S. C., and Thomas, G. (2004) Mol. Cell. Biol. 24 3112-3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pashmforoush, M., Lu, J. T., Chen, H., Amand, T. S., Kondo, R., Pradervand, S., Evans, S. M., Clark, B., Feramisco, J. R., Giles, W., Ho, S. Y., Benson, D. W., Silberbach, M., Shou, W., and Chien, K. R. (2004) Cell 117 373-386 [DOI] [PubMed] [Google Scholar]
- 24.Chen, H., Shi, S., Acosta, L., Li, W., Lu, J., Bao, S., Chen, Z., Yang, Z., Schneider, M. D., Chien, K. R., Conway, S. J., Yoder, M. C., Haneline, L. S., Franco, D., and Shou, W. (2004) Development (Camb.) 131 2219-2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes, A. M., Onodera, T., Tamura, T., Said, S., Bohlmeyer, T. J., Abraham, W. T., and Bristow, M. R. (1998) J. Card. Fail. 4 343-348 [DOI] [PubMed] [Google Scholar]
- 26.Liao, P., Georgakopoulos, D., Kovacs, A., Zheng, M., Lerner, D., Pu, H., Saffitz, J., Chien, K., Xiao, R. P., Kass, D. A., and Wang, Y. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12283-12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, R. T., Beal, P. A., Comb, M. J., and Schreiber, S. L. (2000) J. Biol. Chem. 275 7416-7423 [DOI] [PubMed] [Google Scholar]
- 28.Soonpaa, M. H., and Field, L. J. (1994) Am. J. Physiol. 266 H1439-H1445 [DOI] [PubMed] [Google Scholar]
- 29.Shen, W. H., Boyle, D. W., Wisniowski, P., Bade, A., and Liechty, E. A. (2005) J. Endocrinol. 185 275-289 [DOI] [PubMed] [Google Scholar]
- 30.Gulick, J., Subramaniam, A., Neumann, J., and Robbins, J. (1991) J. Biol. Chem. 266 9180-9185 [PubMed] [Google Scholar]
- 31.Brunn, G. J., Hudson, C. C., Sekulic, A., Williams, J. M., Hosoi, H., Houghton, P. J., Lawrence, J. C., Jr., and Abraham, R. T. (1997) Science 277 99-101 [DOI] [PubMed] [Google Scholar]
- 32.Belke, D. D., Betuing, S., Tuttle, M. J., Graveleau, C., Young, M. E., Pham, M., Zhang, D., Cooksey, R. C., McClain, D. A., Litwin, S. E., Taegtmeyer, H., Severson, D., Kahn, C. R., and Abel, E. D. (2002) J. Clin. Investig. 109 629-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiojima, I., Yefremashvili, M., Luo, Z., Kureishi, Y., Takahashi, A., Tao, J., Rosenzweig, A., Kahn, C. R., Abel, E. D., and Walsh, K. (2002) J. Biol. Chem. 277 37670-37677 [DOI] [PubMed] [Google Scholar]
- 34.Ha, T., Li, Y., Gao, X., McMullen, J. R., Shioi, T., Izumo, S., Kelley, J. L., Zhao, A., Haddad, G. E., Williams, D. L., Browder, I. W., Kao, R. L., and Li, C. (2005) Free Radic. Biol. Med. 39 1570-1580 [DOI] [PubMed] [Google Scholar]
- 35.Shioi, T., Kang, P. M., Douglas, P. S., Hampe, J., Yballe, C. M., Lawitts, J., Cantley, L. C., and Izumo, S. (2000) EMBO J. 19 2537-2548 [DOI] [PMC free article] [PubMed] [Google Scholar]