Abstract
We examined transcriptional and epigenetic mechanism(s) behind diminished skeletal muscle GLUT4 mRNA in intrauterine growth-restricted (IUGR) female rat offspring. An increase in MEF2D (inhibitor) with a decline in MEF2A (activator) and MyoD (co-activator) binding to the glut4 promoter in IUGR versus control was observed. The functional role of MEF2/MyoD-binding sites and neighboring three CpG clusters in glut4 gene transcription was confirmed in C2C12 muscle cells. No differential methylation of these three and other CpG clusters in the glut4 promoter occurred. DNA methyltransferase 1 (DNMT1) in postnatal, DNMT3a, and DNMT3b in adult was differentially recruited with increased MeCP2 (methyl CpG-binding protein) concentrations to bind the IUGR glut4 gene. Covalent modifications of the histone (H) code consisted of H3.K14 de-acetylation by recruitment of histone deacetylase (HDAC) 1 and enhanced association of HDAC4 enzymes. This set the stage for Suv39H1 methylase-mediated di-methylation of H3.K9 and increased recruitment of heterochromatin protein 1α, which partially inactivates postnatal and adult IUGR glut4 gene transcription. Further increased interactions in the adult IUGR between DNMT3a/DNMT3b and HDAC1 and MEF2D and HDAC1/HDAC4 and decreased association between MyoD and MEF2A existed. We conclude that epigenetic mechanisms consisting of histone code modifications repress skeletal muscle glut4 transcription in the postnatal period and persist in the adult female IUGR offspring.
Pre- and postnatal nutritional deficiency culminating in intrauterine and postnatal growth restriction (IUGR)2 leads to insulin resistance, a forerunner of gestational and type 2 diabetes mellitus (1–3). The predominant adaptive mechanism underlying this adult phenotypic presentation consists of aberrant glucose transport into insulin-sensitive tissues (4–6). Glucose transport, a rate-limiting step in glucose utilization under normal physiological circumstances, occurs by facilitated diffusion (7). This process is mediated by a family of structurally related membrane-spanning glycoproteins, termed the facilitative glucose transporters (GLUT; Slc2 family of transport proteins) (8, 9). Of the isoforms cloned to date, GLUT4 is the major insulin-responsive isoform expressed in insulin-sensitive tissues such as skeletal muscle, adipose tissue and cardiac muscle (9, 10). In addition, GLUT4 is expressed in skeletal muscle cell lines during myoblast differentiation to insulin-sensitive myotubes (11, 12). Targeted disruption of skeletal muscle GLUT4 resulting in severe insulin resistance and glucose intolerance confirmed the vital role of this isoform in mediating insulin sensitivity (13).
We and others have observed that a central basis for insulin resistance in the IUGR offspring stems from key changes in skeletal muscle GLUT4. These changes include the following: 1) decline in skeletal muscle GLUT4 mRNA and protein concentrations in rats (4, 6, 14) and human (6), suggesting aberrations in transcriptional control; and 2) insulin resistance of post-translational translocation of GLUT4 from intracellular vesicles to plasma membrane (4). Although we have previously determined the molecular basis for the latter (15), the molecular mechanisms responsible for the former remain to be investigated. Transgenic investigations established the crucial in vivo role for conserved glut4 promoter regions in skeletal muscle gene expression. Disruption of the myocyte enhancer factor 2 (MEF2)-binding site (–473 to –464 bp) ablated tissue-specific glut4 expression in transgenic mice (16, 17). MyoD on the other hand is responsible for glut4 expression in vitro during myoblast to myocyte differentiation (18). MyoD binding with that of MEF2 and TRα1 spans the –502- to –420-bp region of the glut4 gene in skeletal muscle (19). MyoD directly interacts with MEF2 synergistically driving gene expression necessary for myogenesis (19–21), including skeletal muscle glut4 transcription and gene expression (18, 19, 22).
The insulin-resistant phenotype of the IUGR adult female offspring has been trans-generationally transmitted (23–25). These observations support an epigenetic basis for the propagation of insulin resistance. DNA methylation of CpG islands in a gene promoter, mediated by DNA methyltransferase (DNMT) enzymes, notably silences gene expression (26, 27). Alternatively, DNMT enzymes directly complex with histone-modifying enzymes and perturb the chromatin structure (28). Changes in chromatin structure and locus accessibility predetermine the epigenetic imprint on gene expression (29, 30). DNA methylation may permanently silence a gene throughout development (31). In contrast, the dynamic flexibility of histone post-translational modifications, such as de-acetylation, de-phosphorylation, and methylation of specific amino acid residues in the N-terminal tails, exerts diversified effects on gene transcriptional regulation (30). These effects may decrease gene expression rather than complete silencing. An example is the association of MEF2 with class II histone deacetylating enzymes suppressing MEF2-mediated downstream gene expression (32).
Because DNA methylation and concerted changes in the combinatorial histone code regulate tissue-specific gene expression (26–31), we hypothesized that epigenetic phenomena may underlie aberrations in MEF2- and/or MyoD-mediated transcriptional induction of skeletal muscle glut4 expression in the IUGR offspring. We tested this hypothesis in a well characterized rat model, where the offspring was exposed to prenatal and postnatal nutrient restriction causing intrauterine and postnatal growth restriction (4).
EXPERIMENTAL PROCEDURES
Materials
Oligonucleotides—Synthetic oligonucleotides (Retrogen Inc., Carlsbad, CA; Integrated DNA Technologies, San Diego) were used in these experiments. Double-stranded oligonucleotides were generated by annealing synthetic oligonucleotides with respective complementary sequences under standard conditions.
Antibodies—Rabbit polyclonal anti-MyoD, anti-MEF2A, anti-MEF2C, anti-MEF2D, anti-HDAC1, anti-HDAC4, anti-DNMT1, anti-DNMT3a, and anti-DNMT3b were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-MEF2D used for Western blot analysis was purchased from BD Biosciences. Anti-acetyl-histone H3, anti-acetyl-histone H4, anti-acetyl-histone H3 (Lys-9), anti-acetyl-histone H3 (Lys-14), anti-acetyl-histone H3 (Lys-27), anti-SUV39H1 clone MG44, anti-HP1α, clone 15.19s2, and anti-MeCP2 were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). Anti-dimethyl-histone H3 (Lys-9) was purchased from Abcam Inc. (Cambridge, MA), and anti-polymerase II antibody was from Active Motif (Carlsbad, CA). Horseradish peroxidase-linked anti-rabbit and anti-mouse IgGs were from Amersham Biosciences. For gel shift-supershift experiments the more concentrated (2 μg/μl) form (Santa Cruz Biotechnology) of anti-MyoD, anti-MEF2A, anti-MEF2C, and anti-MEF2D antibodies was used.
Methods
Cells—C2C12 murine cells (American Type Culture Collection, Manassas, VA) were grown at 37 °C with 95% air, 5% CO2 in poly-l-lysine-coated culture flasks and maintained in Dulbecco's modified Eagle's medium supplemented with 2 mm glutamine, sodium pyruvate (110 mg/ml), penicillin (100 units/ml), streptomycin (100 units/ml), 4.5% glucose, and 10% fetal bovine serum.
DNA Cloning and Site-directed Mutagenesis—Standard recombinant molecular biology techniques were used in cloning ∼1-kb rat glut4 upstream DNA sequences obtained by PCR amplification of genomic DNA from 450-day female (control) rat skeletal muscle. Bidirectional cloning of the PCR amplification product obtained by using the Herculase hot start enzyme (Stratagene Inc., La Jolla, CA) was accomplished by ligating the KpnI- and HindIII-restricted glut4 DNA fragment to the pGL3 vector proximal to the contained firefly luciferase reporter gene (Promega Inc., Madison, WI). DNA sequences of the glut4-amplified product and its distal luciferase reporter gene were confirmed by DNA sequencing using specific primers. These sequences were aligned and compared with the rat, mouse, and human glut4 upstream sequences in GenBank™ using the ClustalW alignment search software (33). Next, a search for transcription factor-binding sites that demonstrated high fidelity was performed using TRANSFAC 4.0 data base search (IMD = version 1.1, CBIL/GibbsMat = version 1.1, available on line) (34). In particular, the highly conserved MEF2- and MyoD-binding sites were delineated within the rat glut4 upstream sequences. In addition, using the CpG islander searcher software (35), we identified multiple highly stringent CpG dinucleotide-enriched sequences in and around the proximal region of the glut4 promoter. These CpG dinucleotides in close proximity to the MEF2/MyoD-binding site were artificially grouped into three clusters, CpG-I, CpG-II, and CpG-III to facilitate analysis without disrupting the transcriptional binding sites. An additional eight CpG islands (numbered 21–28) 3′ to the MEF2/MyoD-binding site but 5′ to the transcription start site were also identified for analysis. Both primer-specific PCR-mediated (QuikChange mutagenesis kits, Stratagene) deletions and/or substitutions of target sequences were performed and confirmed by DNA sequence analyses. The primer sequences employed for DNA cloning and mutational analysis are shown in supplemental Table 1.
Transient Transfection and Reporter Activity Assays—Transient transfection of cultured C2C12 cells was achieved by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Briefly, 2–4 μg of glut4-luciferase DNA constructs along with 0.5 μg of the β-galactosidase expressing plasmid were incubated with Lipofectamine 2000 at room temperature, in reduced serum containing Opti-MEM medium to facilitate DNA-liposome complex formation. After a 6–8-h incubation of the washed adherent cells with the DNA-liposome complex in either 6-well or 60-mm plates, fresh medium was added and incubation continued for varying durations. Following cell lysis, luciferase reporter activity was assessed in 20 μl of the cell extract that was mixed with 100 μl of the luciferase assay reagent, and firefly luciferase activity was measured as light output (10 s) by a standard luminometer (Monolight 2010, Analytical Luminescence Laboratory, San Diego). To determine the transfection efficiency, β-galactosidase activity was also assayed by the luminometer according to the manufacturer's protocol (Promega, Madison, WI). The glut4 promoter-driven luciferase enzyme activity was expressed as a ratio to the corresponding β-galactosidase activity per cellular protein concentrations (36).
Effect of in Vitro Manipulations on Reporter Gene Activity—To methylate DNA in vitro, 10–20 μg of pglut4-Luc containing pGL3 plasmid DNA was treated with 10–20 units of SssI DNA methylase enzyme (New England Biolab, Ipswich, MA) for 6 h at 37 °C in a 20–50-μl volume and then purified in a quick column (Qiagen Inc., Valencia, CA) after deactivation. The CpG-methylated reporter DNA was digested by HpaII and BstU1 and compared with the corresponding wild type DNA on agarose gel electrophoresis. To assess the effect of DNA methylation on glut4 promoter activity, the WT and modified glut4-luciferase DNA were transiently transfected into C2C12 cells, and luciferase reporter activity was assessed after 48 h (36).
To inhibit histone deacetylase (HDAC) enzymes in general, 0, 0.3, and 0.6 μm trichostatin A (Upstate Inc., Lake Placid, NY) was added to cultured cells for 12 h prior to assessing reporter luciferase enzyme activity. In addition, the effect of glucose and amino acid deprivation was examined in vitro by culturing C2C12 cells in starvation medium (Hanks' balanced salt solution with calcium chloride, magnesium chloride, sodium pyruvate and dialyzed 10% fetal bovine serum) (Sigma). The cells were initially maintained in complete Dulbecco's modified Eagle's medium with high glucose (4.5 g/liter), l-glutamine, and sodium pyruvate along with 10% fetal bovine serum. Following overnight transfection in Opti-MEM media of the glut4 promoter-luciferase DNA construct, 50% glucose and amino acid content in culturing media was achieved for 24 h. The reporter luciferase gene activity was measured after cell lysis according to the manufacturer's protocol.
Animals—Sprague-Dawley rats (Charles River Breeding Laboratories, Hollister, CA) were housed in individual cages, exposed to 12:12-h light-dark cycles at 21–23 °C, and allowed ad libitum access to standard rat chow. Animal care and use were approved by the Animal Research Committee at UCLA in accordance with the guidelines from the National Institutes of Health.
Prenatal Semi-nutrient Restriction Model—Pregnant rats received 50% of their daily food intake (11 g/day) beginning from day 11 through day 21 of gestation, which constitutes mid- to late gestation, as compared with their control counterparts that received ad libitum access to rat chow (∼22g/day). Both groups had ad libitum access to drinking water (3, 4). At birth, the litter size was culled to six. The newborn rats born to semi-nutrient-restricted mothers were reared by the same mother that continued to be semi-nutrient restricted by receiving 20g/day food intake through lactation (IUGR) (3). Similarly, newborn pups born to control mothers were reared by the control mother with ad libitum access to rat chow (∼40g/day) (CON). This food restriction scheme ensured that the semi-nutrient restricted maternal rats received about ∼50% of the ad libitum food intake through mid- to late pregnancy and lactation (3). At day 21, in both experimental groups, the pups were weaned from the mother and maintained in individual cages with ad libitum access to a similar diet of standard rat chow (3, 4).
Skeletal Muscle Preparation—Hind limb skeletal muscle was rapidly separated from surrounding tissues, quickly snap-frozen in liquid nitrogen, and stored at –70 °C as described previously (4). Skeletal muscle was powdered under liquid nitrogen prior to use for varying extractions.
Northern Blot Analysis—Total RNA was isolated using the TRIzol reagent (Invitrogen). The extracted RNA (15 μg/lane) was subjected to Northern blot analysis as described previously (4). A 32P-labeled 439-bp fragment of the rat glut4 cDNA served as the probe. The probe was prepared by amplifying rat glut4 cDNA containing the coding region spanning exons 4–7, with the forward primer 5′-ccggaattcctatgctggccaacaatgtc-3′ that primed rat glut4 cDNA at the beginning of exon 4 and the reverse primer 5′-cacacaagcttagtgcatcagacacatcagc-3′ that primed rat glut4 cDNA at the beginning of exon 7. PCR parameters for the glut4 cDNA amplification over 30 cycles consisted of 95 °C for 2 min initially, followed by denaturation at 95 °C for 30 s, annealing at 60 °C for 60 s, extension at 72 °C for 30 s, and the last step consisting of 72 °C for 5 min. Inter-lane loading variability of Northern blots was standardized by re-hybridization of stripped filters with a 32P-labeled rat 589-bp fragment of the β-actin cDNA probe (37). The rat β-actin probe was amplified with a forward primer 5′-acctgacagactacctcatg-3′ and a reverse primer 5′-taacagtccgcctagaagca-3′ with similar PCR conditions except for annealing at 55 °C for 30 s and extension at 72 °C for 90 s. In the case of insulin, pancreatic RNA was extracted and Northern blot analysis performed using similar conditions with a PCR-amplified rat I insulin probe (38) using an internal control of rat β-actin (37). The rat insulin I probe was amplified using the following primers: forward primer 5′-atagaccatcagcagcaagcagg-3′ and reverse primer 5′-tccagttgtggcacttgcg-3′ (GenBank™ accession number V01242) in PCR beginning at 94 °C for 5 min followed by denaturation at 94 °C for 30 s, annealing at 58 °C for 1 min, and extension at 72 °C for 1 min over 35 cycles followed by 72 °C for 5 min. The insulin I probe was digoxigenin-labeled (Roche Diagnostics) and used with 15 μg of RNA/lane loading. The signals on blots were quantified with a Variable Mode Imager (Typhoon 9410, Amersham Biosciences). The results were expressed as a ratio between glut4 and β-actin mRNA or insulin and β-actin PhosphorImager values.
Electromobility Shift Assay (EMSA)—The 1-kb upstream glut4 wild type and mutant clones carrying deletions of MyoD-I, MyoD-II, and MEF2 binding domains were amplified by PCR. Primers encompassing MyoD-I-, MyoD-II-, and MEF2-binding sites individually were employed to produce ∼100-bp size DNA fragments carrying either only one binding site each or a deleted binding site that was then end-labeled with [γ-32P]ATP (Perkin Elmer Life Sciences) and T4 polynucleotide kinase. The primers used for amplifying the wild type and mutated probes are listed in supplemental Table 2. Approximately 6 fmol (specific activity = 3000 Ci/mmol) of the labeled DNA probe was added to 5 μg of skeletal muscle nuclear extract in a final volume of 20 μl containing 1 μg of poly(dI-dC), 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.5 mm EDTA, 1 mm MgCl2, 4% glycerol, 1 mm dithiothreitol and incubated for 15 min at room temperature. Subsequently, the DNA-protein complexes were separated from unbound DNA by electrophoresis through a 5% nondenaturing polyacrylamide gel in a 90 mm Tris borate, 2 mm EDTA buffer. The gels were dried and subjected to autoradiography (39). Competition occurred in the presence of 10–100-fold excess of unlabeled DNA oligonucleotides. In electrophoretic mobility super-shift assays, 2 μg of the respective antibody (IgG) was included in the reaction mix and incubated for 30 min. DNA target sequences (oligonucleotides) used in gel shift reactions are listed in supplemental Table 3.
DNA Bisulfite Modification Assay—Genomic DNA was extracted from 450-day female CON and IUGR and 2-day male and female CON and IUGR skeletal muscle using the DNeasy tissue kit (Qiagen Inc., Valencia, CA). The extracted DNA was subjected to bisulfite modification using the CpGenome fast DNA modification kit (Chemicon International Inc., Temecula, CA). The bisulfite-modified naked DNA served as the template in a PCR where specific regions containing the CpG-I, CpG-II, and CpG-III islands and other 3′-CpG-rich regions of the upstream glut4 promoter 5′ to the transcription start site were amplified with primers created by the MethPrimer software (40). The PCR-amplified DNA was cloned in a TOPO-TA vector (Invitrogen) after the addition of a 3′-A. These clones were then sequenced to detect either unmodified CpG islands protected by a methyl group or modified TpG islands because of de-amination and sulfonation of the unmethylated cytosines (40).
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assays were performed as described previously (41) with some modifications. Powdered skeletal muscle was fixed in 1% formaldehyde for 45 min at room temperature. The tissue pellet was resuspended in cell lysis buffer (5 mm Pipes (KOH), pH 8.0, 85 mm KCl, 0.5% Nonidet P-40) containing protease inhibitors (100 mm phenylmethylsulfonyl fluoride at 1:100 dilution in ethanol, aprotinin at 10 mg/ml concentration diluted to 1:1000 in 0.01 m HEPES, pH 8.0, and leupeptin at 10 mg/ml diluted to 1:1000 in water) and homogenized with an Omni tissue homogenizer, 115 V (Omni International, Inc., Marietta, GA). The separated nuclei were lysed in nuclear lysis buffer (50 mm Tris, pH 8.1, 10 mm EDTA, 1% SDS) containing protease inhibitors. The resultant chromatin was sonicated (Fisher model 100 Sonic Dismembrator) on ice with 20 pulses for 15 s each at setting 4 with a 1-min rest interval between pulses. The average length of sonicated chromatin was determined by resolving on a 1.5% agarose gel and found to be ∼500 bp. The sample was then centrifuged at 4 °C (10 min at 14,000 rpm) to remove cell debris from the crude chromatin lysate. Ten percent of the lysate was used as the input control for PCR. Fifty μg (at A260) of sheared chromatin was added to a final volume of 500 μl of the immunoprecipitation (IP) dilution buffer and pre-cleared with 25 μl of salmon sperm DNA/protein A-agarose slurry (Upstate Biotechnology, Inc., Lake Placid, NY) at 4 °C for 30 min. The precleared chromatin was subsequently incubated overnight on a nutator with 2 μg of a primary polyclonal antibody at 4 °C. Fifty μl of pre-blocked protein A-agarose beads were then added to the pre-cleared chromatin for 2 h, followed by centrifugation at 14,000 rpm for 3 min at room temperature. The pellet was washed twice with 0.4 ml of wash buffer A (50 mm HEPES, pH 8.0, 0.1% SDS, 1% Triton X-100, 0.1% deoxycholate, 1 mm EDTA, 140 mm NaCl) followed by two washes with 0.4 ml each of wash buffer B (50 mm HEPES, pH 8.0, 0.1% SDS, 1% Triton X-100, 0.5% deoxycholate, 1 mm EDTA, 500 mm NaCl), then LiCl-containing buffer (20 mm Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 0.5% deoxycholate, 1 mm EDTA, 250 mm LiCl, and TE (10 mm Tris-HCl, pH 8.0, 1 mm EDTA)). Antibody-protein-DNA complexes were eluted from protein A-agarose beads by adding 100 μl of the elution buffer (50 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 1% SDS) followed by 150 μl of TE buffer with 0.67% SDS at 65 °C for 10 min each. The combined supernatants were incubated overnight at 65 °C after addition of 1 μl of RNase A (10 mg/ml) to reverse the formaldehyde cross-links. The complex was then treated with proteinase K at 55 °C for 2 h, extracted once with phenol/chloroform/isoamyl alcohol and once with chloroform/isoamyl alcohol, and subsequently precipitated overnight at –20 °C with ethanol in the presence of 5 μg of tRNA and 5 μg of glycogen. The DNA concentration in the complex was determined by a Dip Stick kit (Invitrogen), and ∼2–4 ng of immunoprecipitated DNA was used as a template in each PCR. The PCR consisted of 25 μl of the reaction mix containing 8 μl of the DNA template, 0.75 μm of forward primer, 0.25 μm of reverse primer, 10× PCR buffer with 1.5 mm MgCl2, 200 μm dNTPs, and 0.25 μl of Taq polymerase (5 units/μl). The PCR amplification in a T3 thermocycler (Biometra, Goettingen, Germany) for the glut4 promoter region from –836 to –452 bp was performed initially at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and then at 72 °C for 4 min. The PCR employed for GAPDH (amplification product targeted at the translational start site 1–231 bp spans exons 1–4) consisted of 25 μl of the reaction mix containing 2 μl of the DNA template, 0.5 μm forward primer, 0.5 μm reverse primer, 10× PCR buffer with 1.5 mm MgCl2, 200 μm dNTPs, and 0.25 μl of Taq polymerase (5 units/μl), which was subjected to amplification in a T3 thermocycler. The PCR was performed as described above except for an annealing temperature of 60 °C over 60 s (38, 39). The primers used in these PCRs are listed in supplemental Table 4. Other controls consisted of a 248-bp amplification product ∼1 kb upstream from MyoD-MEF2 binding region of rat glut4 (forward primer spanning –2172 bp to 2149 bp, 5′-accaagtgtaatcccaggcctcat-3′, and reverse primer spanning –1947 bp to –1924 bp, 5′-ttgttgccctgtggtcaagtttgg-3′) in the presence or absence of anti-RNA polymerase II IgG and a nonspecific IgG.
Western Blot Analysis—Fifty μg of the extracted nuclear protein fraction (per the nuclear extraction kit manufacturer's instructions; Active Motif, Carlsbad, CA) was solubilized in 50 mm Tris, pH 6.8, containing 2% SDS and the protein concentration determined by the Bio-Rad dye binding assay (42) prior to undertaking Western blots as described previously (39).
Co-immunoprecipitation (Co-IP)—Hundred μg of nuclear protein was pre-cleared with 40 μl of 50% protein A-agarose slurry (1:1 dilution) (Pierce) in the lysis buffer consisting of 25 mm Tris, pH 8.0, 100 mm NaCl, 10% glycerol, Nonidet P-40, 1.5 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, and 1 μg/ml pepstatin A at 4 °C (all procedures were carried out in the cold unless stated otherwise) by gentle agitation for 30 min. The samples were centrifuged (2000 rpm, 2 min) and 10 μl of anti-HDAC1, anti-MEF2A, anti-MEF2D, anti-HDAC4, or anti-MyoD was added, and incubation was carried out overnight with gentle agitation. Fifty μl of the protein A-agarose slurry (1:1 dilution) was added and incubation was continued for 2 h. Protein A-agarose beads were washed twice for 5 min each in 500 μl of the lysis buffer and then boiled for 5 min in SDS sample buffer with 100 mm dithiothreitol. The proteins were separated on 4–15% SDS-PAGE and then processed for Western blotting (39). The membranes were probed with 1:50 dilution of the anti-HDAC1, anti-HDAC4, anti-MEF2A, anti-MEF2D, anti-DNMT3a, anti-DNMT3b, or anti-MyoD (1:1000 dilution) antibodies.
Data Analysis—All data are depicted as the means ± S.E. Differences between two groups were validated by the Student's t test. Significance was assigned when the p value was ≤0.05.
RESULTS
Cis-elements of the Rat glut4 Gene Promoter
Data base (CpG Island Searcher) (35) search demonstrated highly stringent ∼58–59% GC-rich area (observed CpG/expected CpG is 0.65) in a length of 407 bp spanning from –748 to –342 bp in the rat glut4 gene promoter (GenBank™ accession number L36125). These CpG islands consisting of CpG-I (7 CpGs in 114-bp region), CpG-II (7 CpGs in 98 bp), and CpG-III (2 CpGs in 30 bp) are in close proximity and intertwined by one MyoD (-CACCTG; –739 to –734 bp) and one myocyte enhancer factor 2 (MEF2) transcription factor binding domains (-CTAAAAATAG-) (–466 to –457 bp). We found another conserved MyoD binding sequence region (CAGCTG) in the glut4 5′-flanking upstream DNA to encompass the MEF2 binding region. The proximal MyoD-II (–152 to –147 bp) sequence (-cagctg-) (TRANSFAC, 4.0, Job W0854015084) (34) is not identical to that of the distal MyoD-I (–739 to –734 bp) (-cacctg-) sequence (Fig. 1A).
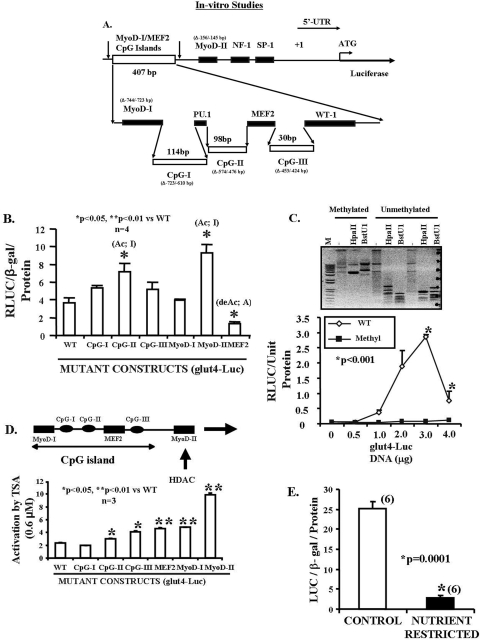
FIGURE 1.
In vitro studies. A, scheme of the rat glut4 upstream region. The rat glut4 promoter region (1 kb) contains the three CpG islands, MyoD-I, MyoD-II, and the MEF2 DNA binding consensus elements 5′ to the transcriptional (+1) and translational (ATG) start sites. In addition, NF-1, Sp-1, PU.1, and WT-1 consensus sequences are shown. The sequences deleted in mutant (Δ) constructs are depicted in parentheses. B, transient transfection of glut4-luciferase DNA constructs in C2C12 murine skeletal muscle cell line. Luciferase reporter activity of wild type (WT) and mutated glut4-luciferase DNA constructs is depicted as a ratio to that of the β-galactosidase gene. Differences between the wild type and various mutated constructs were established by analysis of variance (F = 11.6; p = 0.0001), and inter-group differences in comparison with the WT DNA construct were determined by the post-hoc Tukey's HSD test (* and **). Mutant CpG-II and MyoD-II activate (Ac) and MEF2 de-activates (de-Ac) luciferase gene transcription. C, DNA methylation. glut4-luciferase DNA construct was either pretreated or untreated with the SssI methylase enzyme. Top panel, demonstrates the methylated and unmethylated glut4 DNA that was restricted with methylation-sensitive HpaII and BstU1 enzymes, and the digested DNA products were separated by gel electrophoresis along with the unrestricted DNA. Although the unmethylated glut4 DNA was restricted yielding smaller products, the SssI methylase-treated methylated DNA was resistant to restriction enzyme digestion yielding larger DNA products. M = DNA markers. Lower panel demonstrates the luciferase activity of SssI methylase-treated and untreated glut4-luciferase (0–4 μg) transiently transfected in C2C12 cells. Differences between DNA concentration-matched methylated and unmethylated DNA-reporter activity was assessed by the Student's t test (*). D, histone de-acetylation. Transiently transfected C2C12 cells containing wild type (WT) and mutated glut4-luciferase DNA constructs were pretreated with trichostatin A (0.6 μm), a generic HDAC inhibitor. The mutant constructs were compared with the WT construct by analysis of variance (F = 96.1; p = 0.0001), and inter-group differences were established by the post-hoc Tukey's HSD test (*, **). E, in vitro nutrient restriction. The transiently transfected cells containing the wild type glut4-luciferase DNA construct were incubated in either a control or a nutrient-restricted (glucose and amino acid deprived) medium. Luciferase activity is depicted as a ratio to the β-galactosidase gene (internal control) per protein concentration. Student's t test demonstrated inter-group differences (*).
Inter-species comparison of the upstream region of the glut4 gene was performed by ClustalW program (33). Among rat, mouse, and human, the MEF2-, MyoD-I-, and MyoD-II-binding sites were highly conserved. Other binding sites of PU.1, WT-1, Sp1, and NF-1 were also present and conserved across species. Furthermore, random CpG sites (numbered 18–35) in the glut4 promoter were identified, and 9 of 18 dinucleotides (50%) and overall 17 of 35 CpGs (48.6%) that included the CpG-I, CpG-II, and CpG-III clusters were conserved across the three species. An additional six CpGs were also nearly conserved (numbered 2, 29, 32, 33, 34, and 35) across species suggesting a functional role in transcription. For our investigations, we subcloned the mutated CpG island region as three separate mutated DNA fragments and designated them as CpG-I (Δ-723 bp to –610 bp), CpG-II (Δ-574 bp to –476 bp), and CpG-III (Δ-453 to –424) retaining the intervening wild type transcription factor binding sequences intact (Fig. 1A).
In Vitro Studies, Cis-elements Regulate glut4 Gene Transcription
Transient transfection experiments in C2C12 murine myoblast cells using specific mutants in comparison with the wild type upstream glut4-luciferase construct revealed that CpG-II, MyoD-II (–156 to –145 bp), and MEF2 (–473 to –453 bp) deletion mutations, but not CpG-I, CpG-III, and MyoD-I (Δ-744 to –723 bp), significantly altered in vitro transcription in the cell culture reporter assay (Fig. 1B). These observations supported a role for MEF2 in activating, MyoD-II in inhibiting, and MyoD-I not affecting gene expression in vitro. In contrast, whereas CpG-I and CpG-III did not demonstrate an effect, CpG-II alone inhibits gene expression (Fig. 1B). These observations support the CpG-II region to be either methylated or differentially modified by repressor binding, because mutation of this site activated luciferase gene expression (Fig. 1B). We next SssI methylase-treated the glut4-luciferase DNA construct and noted a resistance to methylation-sensitive restriction enzyme(s) HpaII and BstU1 digestion as opposed to the unmethylated DNA (Fig. 1C, top panel). Furthermore, this methylase treatment led to complete suppression of reporter gene expression as opposed to the wild type untreated DNA, supporting methylation of cytosines in CpG clusters resulting in complete gene silencing (Fig. 1C, bottom panel). We next employed a generic HDAC inhibitor, trichostatin A (0.6 μm). These studies demonstrated that the MyoD-II site when mutated led to a 5-fold activation of gene expression supporting an HDAC-mediated inhibitory role for this site in glut4 gene transcription (Fig. 1D). Mutations of the CpG-II, CpG-III, MEF2, and MyoD-I sites led to a 1.5–2-fold activation suggesting a role in HDAC-mediated inhibition of gene transcription as well. Thus HDAC-mediated repression of either MEF2- and/or MyoD-induced transcription of the glut4 gene is a distinct possibility. Furthermore, in the presence of in vitro glucose and amino acid deprivation, significant suppression of glut4 gene transcription was noted when compared with that observed under control conditions (Fig. 1E).
In Vivo Studies
Skeletal Muscle glut4 mRNA—Fig. 2 demonstrates the 450-day-old female and male skeletal muscle glut4 mRNA in the IUGR versus CON. Although a 30–40% decline in the 450-day-old female SM glut4 mRNA was observed by Northern blot analysis (Fig. 2A), no such change was noted in the age-matched male SM (Fig. 2B). This change in the female was previously also observed at 2 and 60 days of age (4) and was associated with lower pancreatic β-islet cell insulin mRNA concentrations (supplemental Fig. S.1).
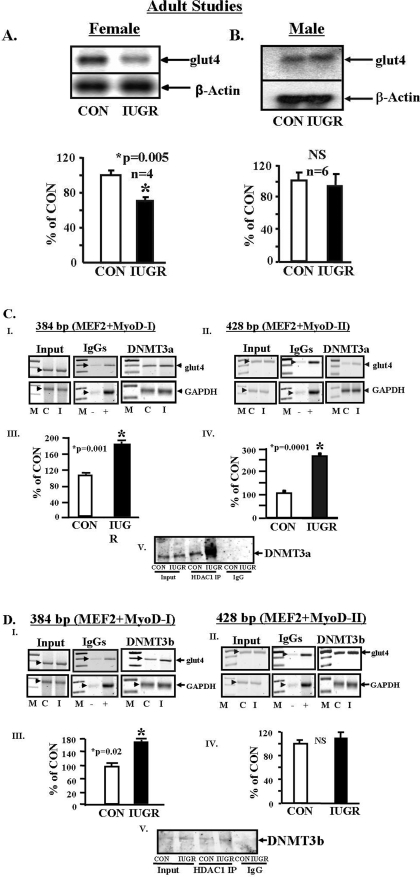
FIGURE 2.
Adult studies. A and B, skeletal muscle glut4 mRNA. Representative Northern blots demonstrating glut4 mRNA (2.8 kb) (top panel) and the internal control β-actin (1.8 kb) mRNA (bottom panel) in skeletal muscle of 450-day-old control (CON) and IUGR female (A) and male (B) rats. The quantification by PhosphorImager analysis of glut4 mRNA in arbitrary units is depicted as a ratio to β-actin and represented as a percent of control (CON). Inter-group differences were established by the Student's t test (*). NS, not significant. C and D, DNA methyltransferases and glut4 5′-upstream region. C, DNMT3a. Panels I and II, representative 2% agarose gels demonstrate the input PCR glut4 and GAPDH control without an antibody (left panels), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panels), and ChIP assay demonstrating the 384-bp PCR glut4 DNA amplification product, which contains the MEF2- and MyoD-I-binding sites and CpG-I and CpG-II regions (panel I) or the 428-bp PCR glut4 DNA amplification product containing MEF2, MyoD-II, and CpG-III regions (panel II), and the 230-bp PCR GAPDH DNA amplification product (served as an internal control) obtained from DNMT3a nuclear immunoprecipitates (IP)(right panels). M = DNA size markers, C = control, and I = IUGR. Arrowheads show the glut4 and GAPDH DNA bands. Panels III and IV, quantification of the amplified 384-bp (panel III) and 428-bp (panel IV) glut4 DNA product as a ratio to that of GAPDH, corrected for the input control and expressed as a percent of CON. Difference between CON and IUGR was assessed by the Student's t test (*). Panel V, representative Western blot demonstrating co-immunoprecipitation (co-IP) experiments where DNMT3a was detected in association with input nuclear protein (Input) obtained from CON and IUGR, anti-HDAC1 IgG chromatin IPs (HDAC1 IP) with increased association of DNMT3a observed in IUGR versus CON, and no DNMT3a detected in nonspecific IgG nuclear chromatin IP (IgG) of CON and IUGR. D, DNMT3b. Panels I and II, representative 2% agarose gels demonstrate the input PCR glut4 and GAPDH control without an antibody (left panels), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panels), and ChIP assay demonstrating the 384-bp PCR glut4 DNA (panel I) or the 428-bp PCR glut4 DNA (panel II) and the 230-bp PCR GAPDH DNA amplification products obtained from DNMT3b nuclear chromatin IPs (right panels). M = DNA size markers, C = control, and I = IUGR. Panels III and IV, quantification of the amplified 384-bp (panel III) and 428-bp (panel IV) glut4 DNA product as a ratio to that of GAPDH corrected for the input control and expressed as a percent of CON. Difference between CON and IUGR was assessed by the Student's t test (*). NS = not significant. Panel V, representative Western blot demonstrating co-IP experiments where DNMT3b was detected in association with input nuclear protein (Input) obtained from CON and IUGR, anti-HDAC1 IgG nuclear chromatin IPs (HDAC1 IP) with slightly increased association of DNMT3b observed in IUGR versus CON, and no DNMT3b detected in IgG nuclear chromatin IPs (IgG) of CON and IUGR.
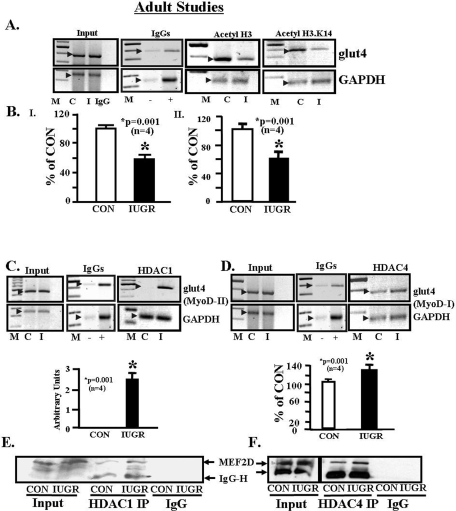
DNA Methylation of the Rat glut4 Gene and DNA Methyltransferases—Bisulfite conversion of genomic DNA obtained from 2-day-old male and female and 450-day-old female skeletal muscle of IUGR and CON demonstrated no differential 5′-cytosine methylation of the three CpG regions or the proximal random CpG sites closer to the transcription start site of the glut4 gene. Instead unmethylated CpGs were observed between the MyoD-I and far beyond the WT-1 recognition sites (CpG 1–20, supplemental Fig. S.2) in both IUGR and CON DNA. In both groups, the cytosines within the CA repeats of the WT-1 region were protected from bisulfite conversion. Similarly, CpG regions in the proximal promoter region (21–27) (supplemental Fig. S.2) were also unmethylated in both IUGR and CON groups. For employing Western blot analysis on nuclear extracts and the ChIP assay (primers and amplification products are shown in supplemental Fig. S.3) to detect protein-DNA interactions, no difference in maintenance methylation mediated by DNMT1 enzyme recruitment to the glut4 promoter region (supplemental Fig. S.4, panels I and II) was observed in the 450-day-old female IUGR and CON. In contrast, an increase in the de novo methylating DNMT3a enzyme recruitment to glut4 promoter whether it contained the MEF2 and MyoD-I or MEF2 -and MyoD-II-binding sites (Fig. 2C, panels I–IV) was noted in the IUGR versus CON. A lesser increase was apparent with the recruitment of DNMT3b to the glut4 promoter containing the MEF2- and MyoD-I-binding sites (Fig. 2D, panels I and III). Furthermore, although an increase in nuclear MeCP2 (methyl CpG-binding protein 2) concentrations was observed (supplemental Fig. S.4, panels III and IV), there was no difference in the recruitment of this protein to the glut4 DNA-protein complex in IUGR versus control (supplemental Fig. S.4, panels V and IV). In contrast, co-immunoprecipitation assays revealed increased protein-protein interaction between DNMT3a and HDAC1 (Fig. 2C, panel V) and DNMT3b and HDAC1 (Fig. 2D, panel V).
Adult Studies, Transactivating Nuclear Factors
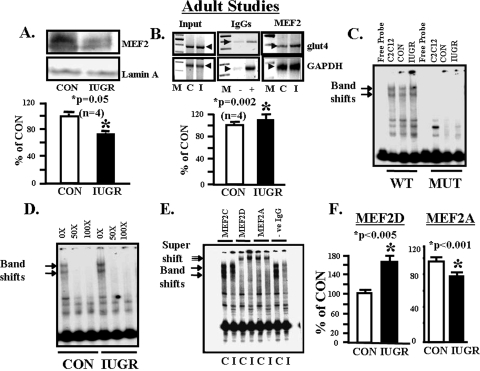
MEF2 Protein Binding to the glut4 Gene—Although nuclear concentrations of total MEF2 protein decreased (Fig. 3A, upper and lower panels), ChIP assays demonstrated increased total MEF2 binding to the glut4 gene (–836 to –452 bp) in the IUGR SM versus control (Fig. 3B, upper and lower panels). Further confirmation by EMSA using SM nuclear extract and a radiolabeled DNA containing the wild type MEF2-binding site revealed a band shift that disappeared with a mutated MEF2-binding probe with C2C12 cells serving as the positive control (Fig. 3C). In addition, this band shift was specifically displaced by excess unlabeled MEF2 oligonucleotide in control and IUGR SM (Fig. 3D). This MEF2 band shift was of higher intensity in the IUGR when compared with CON (Fig. 3, C and D). To tease out the specific MEF2 isoforms engaged in glut4 DNA binding, supershift assays using isoform-specific MEF2 antibodies revealed no supershift in the case of MEF2C as observed previously (43) similar to the absence of IgG, a supershift with MEF2D of increased intensity but diminished intensity of MEF2A in the supershift of IUGR versus control (p < 0.005) (Fig. 3, E and F). This observation supports the existence of an unbalanced contribution by MEF2D or MEF2A homodimers and MEF2A–2D heterodimer in skeletal muscle, with increased MEF2D but decreased MEF2A association with the glut4 gene in the case of IUGR.
FIGURE 3.
Adult studies, MEF2 nuclear protein and glut4 DNA. A, top panel, representative Western blot demonstrating total nuclear MEF2 protein concentrations in CON and IUGR skeletal muscle with the nuclear marker Lamin A protein serving as an internal loading control. Bottom panel, quantification of MEF2 protein concentrations as a ratio to Lamin A protein is depicted as a percent of CON. Inter-group difference was assessed by Student's t test (*). B, top panel, representative 2% agarose gels demonstrate the input PCR glut4 and GAPDH control without an antibody, in the presence of nonspecific (–) and anti-polymerase (+) IgGs and ChIP assay demonstrating the 384-bp PCR glut4 and the 230-bp PCR GAPDH DNA amplification products obtained from total MEF2 nuclear chromatin IPs. M = DNA size markers, C = control, and I = IUGR. Bottom panel, quantification of the ChIP glut4 amplification product represented as a ratio to that of GAPDH, corrected for the input control, and shown as a percent of CON. Inter-group difference is assessed by Student's t test (*). C, representative polyacrylamide gel demonstrating two gel-shifted bands (arrows) in the presence of a 32P-end-labeled DNA probe (free probe) that spans the glut4 gene containing the wild type (WT) MEF2-binding site that are not seen in the presence of MUT MEF2-binding site in C2C12 cell (positive control), CON and IUGR skeletal muscle nuclear extracts. D, representative polyacrylamide gel demonstrating competition for the two gel-shifted bands (arrows) in the presence of a 32P-end-labeled DNA probe that spans the glut4 gene containing the wild type MEF2-binding site and skeletal muscle nuclear extracts from CON and IUGR groups with increasing concentrations (10× to 100×) of unlabeled probe. E, representative polyacrylamide gel demonstrating supershifted bands (double arrow) in the presence of a 32P-end-labeled glut4 DNA probe containing the wild type MEF2-binding site, skeletal muscle nuclear extracts from control (C) and IUGR (I) groups, and anti-MEF2C, anti-MEF2D, anti-MEF2A IgGs and in the absence of IgG (–ve). F, quantification of the MEF2D and MEF2A supershift bands in control (CON) and IUGR groups. Difference between the two groups was assessed by Student's t test (*).
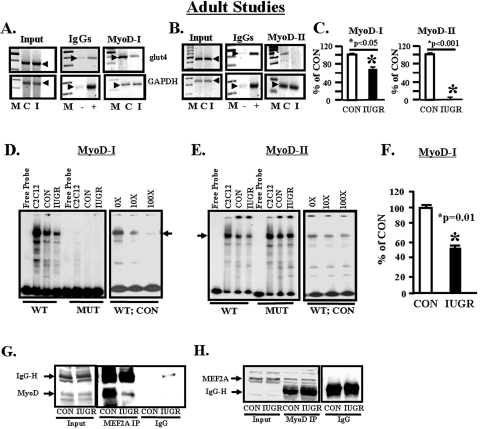
MyoD Protein Binding to the glut4 Gene—In ChIP assays, SM chromatin of CON and IUGR containing the –836 to –452-bp region of the glut4 gene that carried the MyoD-I-binding site bound less of the MyoD nuclear factor in the latter versus the former (Fig. 4, A and C). In contrast MyoD binding to the –473- to –45-bp region of the glut4 gene that contained the MyoD-II-binding site was weaker in CON and absent in the IUGR (Fig. 4, B and C). Using radiolabeled WT and mutated (MUT) MyoD-I and MyoD-II regions of the rat glut4 promoter and control and IUGR SM nuclear extracts, with C2C12 nuclear extracts serving as the positive control, EMSA demonstrated a specific band shift with the WT MyoD-I (Fig. 4D) and WT MyoD-II (Fig. 4E) probes. The band shifts with MyoD-I disappeared with the MUT MyoD-I radiolabeled probe or with competition by excess unlabeled WT MyoD-I oligonucleotide (Fig. 4D). The band shift density in the presence of a MUT MyoD-II radiolabeled probe was higher than the wild type probe and disappeared to a lesser extent with excess unlabeled WT MyoD-II oligonucleotide (Fig. 4E) when compared with the competition with the MyoD-I site (Fig. 4D). The density of the band shift with the MyoD-I radiolabeled probe was decreased in IUGR versus CON (Fig. 4, D–F), but the anti MyoD antibody failed to demonstrate a detectable super-shifted band (data not shown).
FIGURE 4.
Adult studies, MyoD nuclear protein and glut4 DNA. A, representative 2% agarose gels demonstrate the input PCR glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panel), and ChIP assay demonstrating the 384-bp PCR glut4 DNA and the 230-bp PCR GAPDH DNA amplification products obtained from total MyoD nuclear chromatin IPs (right panel). M = DNA size markers, C = control, and I = IUGR. Arrowheads demonstrate the glut4 and GAPDH DNA bands. B, representative 2% agarose gels demonstrate the input PCR glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) or anti-polymerase II (+) IgGs (middle panel), and ChIP assay demonstrating the 428-bp PCR glut4 DNA and the 230-bp PCR GAPDH DNA amplification products obtained from total MyoD nuclear chromatin IPs (right panel). M = DNA size markers, C = control, and I = IUGR. Arrowheads show the glut4 and GAPDH DNA bands. C, quantification of the ChIP glut4 amplification product represented as a ratio to that of GAPDH, corrected for the input control and shown as a percent of CON. Inter-group difference between MyoD-I and MyoD-II compared with their respective CON was assessed by Student's t test (*). D, representative polyacrylamide gel demonstrating a single gel-shifted band (arrows) in the presence of a 32P-end-labeled DNA probe (free probe) that spans the glut4 gene containing the wild type (WT) MyoD-I-binding site, which is not seen in the presence of MUT MyoD-I-binding site and nuclear extracts obtained from C2C12 cells (positive control), CON, and IUGR skeletal muscle. This gel shift band is competed effectively by excess (10×, 100×) unlabeled DNA probe in the CON skeletal muscle nuclear extract with the WT-labeled DNA as the probe. E, representative polyacrylamide gel demonstrating a single gel-shifted band (arrows) in the presence of a 32P-end-labeled DNA probe (free probe) that spans the glut4 gene containing the wild type (WT) MyoD-II-binding site, which is more intense in the presence of MUT MyoD-II-binding site and nuclear extracts obtained from C2C12 cells (positive control), CON, and IUGR skeletal muscle. This gel shift band is only partially competed by excess (10×, 100×) unlabeled DNA probe in the CON skeletal muscle nuclear extract with the WT-labeled DNA as the probe. F, quantification of the MyoD-I gel shift band in CON and IUGR groups shown as a percent of CON. Difference between the two groups was assessed by Student's t test (*). G and H, MEF2 and MyoD protein-protein interaction. G, representative Western blot demonstrated MyoD protein band (arrow) in CON and IUGR input chromatin in the absence (input) or presence of nuclear MEF2A IP or in the presence of a nonspecific IgG (IgG) alone. H, representative Western blot demonstrated MEF2A protein band (arrow) in CON and IUGR input chromatin in the absence (input) or presence of nuclear MyoD-IP or in the presence of a nonspecific IgG (IgG) alone.
MEF2-MyoD Interactions—Co-immunoprecipitation experiments employing the anti-MyoD antibody in Western blot analysis to detect this protein in MEF2A immunoprecipitates from SM nuclear extracts revealed a decrease in IUGR compared with control (Fig. 4G). In contrast, when the anti-MEF2A antibody was used in Western blot analysis to detect this protein in MyoD immunoprecipitates, no difference was observed between control and IUGR SM (Fig. 4H). These observations suggest that when equal amounts of MEF2A immunoprecipitates were analyzed, a lower amount of MyoD interacted with MEF2A in the IUGR versus control. However, when MyoD immunoprecipitated protein amounts were equalized and analyzed, no differences in MEF2A interaction with MyoD were observed in the two groups. Thus in the IUGR lower amounts of MyoD interacted with MEF2A in the nucleus.
Adult Studies, Histone Modifications
Histone Acetylation of the glut4 Gene—Employing intact chromatin from CON and IUGR SM, the ChIP assay revealed interaction between –836- to –452-bp region of the glut4 gene, which contained the MEF2- and MyoD-I-binding sites and the two CpG regions (CpG-I and CpG-II) and acetylated H3 (Fig. 5, A and B) and H4 proteins (data not shown). However, a decrease in acetylated H3 (p < 0.001) (Fig. 5, A and B, panel I) and not H4 (data not shown) that associated with the glut4 gene was observed in IUGR versus CON. Further teasing of histone 3 N-tail amino acid residues revealed decreased acetylation of lysine 14 (Fig. 5, A and B, panel II) in IUGR versus CON (p < 0.001), with no difference in that of lysine 9, 18, or 27 (data not shown).
FIGURE 5.
Adult studies, acetylated histones associated with the glut4 gene. A, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (2nd panel), and ChIP assay demonstrating the PCR amplification products of the 384-bp glut4 DNA or 230-bp GAPDH DNA (internal control) from CON (C) and IUGR (I) skeletal muscle chromatin in the presence of either anti-acetyl-histone 3 IgG (3rd panel) or the anti-acetyl-histone 3.lysine 14 amino acid IgG (right panel). B, quantification of the PCR glut4 amplification product in the acetyl H3 chromatin IP (panel I) and the acetyl-H3.K14 chromatin IP (panel II) as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*). Histone deacetylase enzyme interaction with the glut4 gene is shown. C, top panel, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panel), and ChIP assay demonstrating the PCR amplification products of the 428-bp glut4 DNA or 230-bp GAPDH DNA from CON (C) and IUGR (I) skeletal muscle chromatin in the presence of anti-HDAC1 IgG. Bottom panel, quantification of the PCR glut4 amplification product in the HDAC1 chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed in arbitrary units because the product was undetectable in CON. Inter-group difference was established by the Student's t test (*) after arbitrarily assigning a value of 0.1 to CON. D, top panel, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panel) and ChIP assay demonstrating the PCR amplification products of the 384-bp glut4 DNA or 230-bp GAPDH DNA from CON (C) and IUGR (I) skeletal muscle chromatin in the presence of anti-HDAC4 IgG. Bottom panel, quantification of the PCR glut4 amplification product in the HDAC4 chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*). MEF2D and HDAC interaction is shown. E, representative Western blot demonstrates MEF2D protein band (arrow) in CON and IUGR input nuclear protein in the absence (input) or presence of HDAC1 IP (HDAC1 IP) or in the presence of a nonspecific IgG (IgG) alone. F, representative Western blot demonstrates MEF2D protein band (arrow) in CON and IUGR input nuclear protein in the absence (input) or presence of nuclear HDAC4 IP (HDAC4 IP), or in the presence of a nonspecific IgG (IgG) alone.
Histone De-acetylation of the glut4 Gene—The next set of experiments was performed to determine whether HDACs were involved in SM glut4 gene expression. The results of our in vitro experiments using a generic HDAC inhibitor (Fig. 1D) motivated the in vivo ChIP experiments using IUGR and CON SM chromatin containing either the –473- to –45-bp region that includes the MEF2- and MyoD-II-binding sites (428 bp) or the –836- to –452-bp region that includes the MEF2- and MyoD-I (384 bp)-binding sites of the glut4 gene. The former region recruited HDAC1 in the IUGR but not in CON (p < 0.001) (Fig. 5C), whereas the latter region failed to interact with HDAC1 (data not shown) but demonstrated enhanced association with HDAC4 in IUGR versus CON (p < 0.001) (Fig. 5D). No differences in glut4 gene interaction with any of the other HDACs was observed between IUGR and CON (data not shown). These observations support the need for two classes of HDACs (HDAC4 and HDAC1) in the assembly of a transcription repressor complex (44) with HDAC1 being the critical instigator in IUGR.
HDAC and MEF2D Protein-Protein Interactions—Because MyoD failed to demonstrate specific binding of DNA containing the MyoD-II-binding site (Fig. 4E), and HDAC1 failed to associate with DNA containing the MyoD-I-binding site of the glut4 gene (data not shown), we questioned the association of HDAC1 with MEF2 proteins rather than the MyoD protein by co-immunoprecipitation experiments. HDAC1 formed a complex with the MEF2D isoform in IUGR and CON SM nuclear extracts. The anti-MEF2D antibody detected increased amounts of the protein in HDAC1 immunoprecipitates obtained from IUGR versus CON SM (Fig. 5E). Because HDAC4 associated with DNA containing the MEF2- and MyoD-I- and not MyoD-II-binding sites, co-immunoprecipitation confirmed higher association of MEF2D with HDAC4 in the IUGR versus CON (Fig. 5F). Thus, MEF2D is the common nuclear factor that binds HDAC1 and HDAC4. The increased interaction of MEF2D, a transcriptional inhibitor, with HDAC1 and HDAC4 (repressors) (45) along with decreased interaction of MEF2A, a transcriptional activator with MyoD (potential co-activator) (Fig. 4G) (19–21, 43, 46) in IUGR, sets the stage for repression of the glut4 gene transcription.
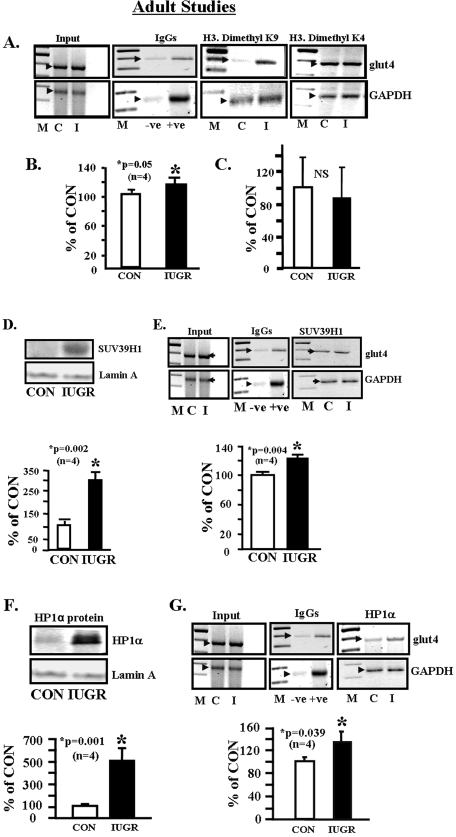
Histone Methylation of the glut4 Gene—Increased dimethylation of the histone 3 N-tail lysine 9 (inhibition) (p < 0.05) and not lysine 4 (activation) amino acid residues was associated with the IUGR SM chromatin, containing the –836 to –452-bp region of the glut4 gene, versus CON (Fig. 6, A–C). This was associated with increased nuclear SUV39H1 histone 3.K9 methylase enzyme concentrations (p < 0.002) (Fig. 6D), resulting in increased recruitment to the glut4 gene (p = 0.004) (Fig. 6E), in IUGR SM versus CON. Nuclear concentrations of heterochromatin protein HP1α (repressor) were higher (p < 0.001) (Fig. 6F) with increased recruitment to the glut4 gene (p < 0.039) in IUGR versus CON (Fig. 6G).
FIGURE 6.
Adult studies, dimethylated histone 3 lysine residues associated with the glut4 gene. A, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (2nd panel), and ChIP assay demonstrating the PCR amplification product of the 384-bp glut4 DNA or 230-bp GAPDH DNA from CON (C) and IUGR (I) skeletal muscle chromatin in the presence of either anti-dimethyl H3.K9 IgG (3rd panel) or anti-dimethyl H3.K4 IgG (right panel). B, quantification of the PCR glut4 amplification product in the dimethyl H3.K9 chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*). C, quantification of the PCR glut4 amplification product in the dimethyl H3.K4 chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test, NS = not significant. Histone 3, lysine 9 methylase associated with the glut4 gene. D, top panel, representative Western blot demonstrating total nuclear Suv39H1 (H3.K9 dimethylase) protein concentrations in CON and IUGR skeletal muscle with the nuclear marker Lamin A protein serving as an internal loading control. Bottom panel, quantification of Suv39H1 protein concentrations as a ratio to Lamin A protein is depicted as a percent of CON. Inter-group difference was assessed by Student's t test (*). E, top panel, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panel), and ChIP assay demonstrating the PCR amplification product of the 384-bp glut4 DNA or 230-bp GAPDH DNA (internal control) from CON (C) and IUGR (I) skeletal muscle chromatin in the presence of anti-Suv39H1 methylase IgG. Bottom panel, quantification of the PCR glut4 amplification product in the Suv39H1 methylase chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*). Heterochromatin protein 1α (HP1α) association with the glut4 gene. F, top panel, representative Western blot demonstrating total nuclear HP1α concentrations in CON and IUGR skeletal muscle with the nuclear marker Lamin A protein serving as an internal loading control. Bottom panel, quantification of HP1α concentrations as a ratio to Lamin A protein is depicted as a percent of CON. Inter-group difference was assessed by Student's t test (*). G, top panel, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 and GAPDH control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panel), and ChIP assay demonstrating the PCR amplification product of the 384-bp glut4 DNA or 230-bp GAPDH DNA (internal control) from CON (C) and IUGR (I) skeletal muscle chromatin in the presence of anti-HP1α IgG. Bottom panel, quantification of the PCR glut4 amplification product in the HP1α chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*).
Postnatal Studies, Transactivating Nuclear Factors and Histone Modifications
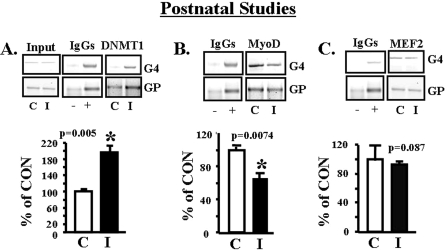
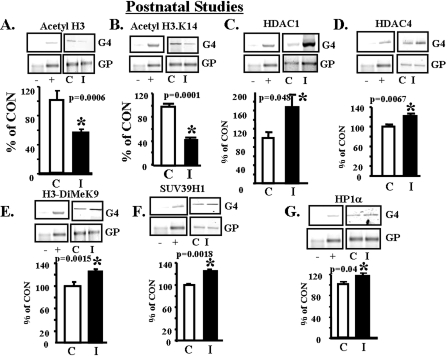
To determine whether the histone modifications of the 450-day-old IUGR SM glut4 reflect epigenetic modifications, we examined the immediate postnatal period (2 days). ChIP assays revealed increased recruitment of DNMT1 (p = 0.005) (Fig. 7A) but not DNMT3a and DNMT3b enzymes to the –836- to –452-bp region (MEF2 and MyoD-I) of the glut4 gene in IUGR versus CON (supplemental Fig. S.5, A and B). This is despite no differential methylation of CpG islands in the glut4 promoter of IUGR versus CON (supplemental Fig. S.2). A decline in MyoD (p = 0.0074) (Fig. 7B) with no change in total MEF2 (Fig. 7C) binding to the glut4 gene was also observed at this early age, when SM glut4 expression is significantly lower than in the adult (4). Similar to the adult, the postnatal female IUGR offspring demonstrated de-acetylation of H3 (p = 0.0006) (Fig. 8A), particularly the H3.K14 residue (p = 0.0001) (Fig. 8B). This was mediated by increased recruitment of HDAC1 (p = 0.048) (Fig. 8C) and HDAC4 (p = 0.0067) (Fig. 8D). Furthermore, increased dimethylation of the histone 3 N-tail lysine 9 amino acid residue (p = 0.0015) (Fig. 8E) with increased recruitment of SUV39H1, the H3.K9 methylase enzyme (p = 0.0018) (Fig. 8F), and the HP1α repressor protein (p = 0.04) (Fig. 8G) to the glut4 gene in IUGR was observed.
FIGURE 7.
Postnatal studies, DNMT1, MyoD, and MEF2 binding to the glut4 gene. A–C, top panel, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 (G4) and GAPDH (GP) control without an antibody (left panel), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (middle panel), and ChIP assay demonstrating the PCR amplification product of the 384-bp glut4 DNA or 230-bp GAPDH DNA from 2-day CON (C) and IUGR (I) skeletal muscle chromatin in the presence of either anti-DNMT1 (A), anti-MyoD (B), or anti-total MEF2 (C) IgG. Bottom panels, quantification of the PCR glut4 amplification product in either DNMT1 (A), MyoD (B), or total MEF2 (C) chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*).
FIGURE 8.
Postnatal studies, histone modifications, de-acetylation and di-methylation. A–G, top panels, representative 2% agarose gels demonstrate the input chromatin PCR-amplified glut4 (G4) and GAPDH (GP) control without an antibody (not shown), in the presence of nonspecific (–) and anti-polymerase II (+) IgGs (left panels) and ChIP assay (right panels) demonstrating the PCR amplification product of the 384-bp glut4 DNA or 230-bp GAPDH DNA (internal control) from 2-day CON (C) and IUGR (I) skeletal muscle chromatin in the presence of either anti-acetyl H3 (A), anti-acetyl H3.K14 (B), anti-HDAC1 (C), anti-HDAC4 (D), anti-H3-dimethyl (DiMe) K9 (E), anti-SUV39H1 methylase (F), or anti-HP1α IgG (G). Bottom panels, quantification of the PCR glut4 amplification product in the acetyl H3 (A), acetyl H3.K14 (B), HDAC1 (C) HDAC4 (D), H3-dimethyl K9 (E), SUV39H1 methylase (F), or HP1α (G) chromatin IP as a ratio to the GAPDH DNA product corrected for the input control and expressed as a percent of CON. Inter-group difference was established by the Student's t test (*).
DISCUSSION
Previous investigations have demonstrated variable results regarding IUGR-induced changes in skeletal muscle GLUT4 mRNA and protein concentrations. Different rat models of the IUGR adult offspring, such as the utero-placental insufficiency or selective protein restriction, have shown no change in the former (47) and a minimal decrease in the latter (6) of skeletal muscle total GLUT4 protein concentrations. In contrast, our prior investigations and those of others employing total calorie restriction-induced IUGR have demonstrated significantly decreased skeletal muscle GLUT4 mRNA and protein concentrations as early as 2 days of age persisting into adult stages (4, 14). This latter observation was replicated in the young adult IUGR human skeletal muscle, lending credence to this finding (6). To sort out this discrepancy between studies, we systematically assessed sex-specific differences in this study. Investigations using the utero-placental insufficiency did not make a sex-dependent distinction; the protein restriction rat models used males (6, 48), and the total calorie restriction studies employed females (4, 14). In keeping with prior results at earlier stages of development (2, 60, and 70 days) (4, 14), at 450 days the females demonstrated a significant decrease in SM total glut4 mRNA concentrations. In contrast the 450-day-old male IUGR offspring demonstrated no change in SM total glut4 mRNA concentrations, supporting sex-specific differences. This sex-specific difference may stem from early developmental perturbations in pancreatic β-islet cell insulin synthesis/secretion and circulating insulin concentrations that are sex-dependent (48). The young females demonstrate persistent postnatal hypoinsulinemia that regulates skeletal muscle glut4 transcription, although the males normalize their insulin concentrations much earlier. Based on this observation and our previous trans-generational persistence of the insulin-resistant phenotype in females (25), we pursued transcriptional studies focused on the adult and immediate postnatal female IUGR offspring. In addition, we employed in vitro experimentation to demonstrate mechanistic links underlying the associative in vivo observations.
We next investigated epigenetic mechanisms responsible for heritable changes in gene expression underlying cellular memory retention. The two processes that underlie epigenesis are DNA methylation, which occurs at the 5′-position of cytosine residues within the nonrandomly distributed CpG dinucleotides throughout the mammalian genome and histone N-tail post-translational modifications. CpG methylation can regulate gene expression by modulating the binding of methyl-sensitive DNA-binding proteins, thereby altering regional chromatin conformation and locus accessibility (26). Histone modifications can alter the positioning of histone-DNA interactions thereby also affecting chromatin conformation and potentiating co-repressor complex formation (29, 30).
Generally DNA methylation in the promoter results in inhibition of gene expression. Methylation of specific CpG islands, particularly at critical epigenetically labile genomic regions, with methylation-sensitive transcription factors contribute toward gene silencing. Three potential epigenetic susceptibility targets for environmentally induced effects are transposable elements, the promoter region of housekeeping genes, and cisacting regulatory elements of imprinted genes (31). These genomic targets contain CpG islands that are normally methylated and if not lend toward metastability, unmethylation, or differential methylation. Differential DNA methylation has been demonstrated in promoters of genes involved in metabolism, including glut4 (49) and uncoupling protein 2 gene (50). A general defect in DNA methylation has been suggested in diabetes, and dietary factors can perturb DNA methylation (51–53). The influence of perinatal diet on DNA methylation status and gene expression has been elegantly demonstrated using the Avy-dependent coat color and obesity phenotypic readout. This involved CpG methylation in the intracisternal A particle retrotransposan present upstream of the transcription start site in the Agouti gene (31, 54). Maternal methyl dietary supplementation with extra folic acid, vitamin B12, choline, and betaine or genistein alone shifted the agouti yellow coat color distribution of the offspring toward pseudoagouti with protection from the obesity phenotype (31). This shift is caused by hypermethylation during early embryonic development at each of the seven Avy pseudoexon 1A CpG sites that was evident in all tissues arising from various cellular lineages and was maintained over time through development (31).
Others have reported global genomic hypermethylation or hypomethylation in livers of the IUGR offspring (53, 55). In both cases, whether IUGR was because of selective nutrient restriction or an ischemic event, false positives clouded the results in lieu of methylated CpG sequence confirmation (53, 55). Recently livers of protein-restricted 28-day IUGR male and female offspring with that of controls were examined by methylation-sensitive PCR. Relative hypomethylation of the glucocorticoid receptor 110 and the peroxisome proliferator-activated receptor α promoter, with enhanced expression of the corresponding gene in the IUGR, was observed (56). Of importance was the observation that these changes were transmitted to the F2 generation and could be reversed by excess folic acid supplementation in the protein-restricted diet of the F0 generation (56, 57). Another study demonstrated that a methyl-deficient diet of 3 weeks led to bisulfite-converted and sequencing-confirmed hypomethylation of CpGs in the hepatic glutathione S-transferase π promoter region that increased gene expression (58). Because glucocorticoid receptor 110, peroxisome proliferator-activated receptor α, or glutathione S-transferase π genes are not housekeeping genes, whether they contain retrotransposans and demonstrate hypomethylation of CpGs within these regions, thereby resulting in diet-induced metastable epialleles or constitute differentially methylated imprinted genes, needs clarification (56–58).
Based on such investigations, in the case of IUGR skeletal muscle glut4, we anticipated hypermethylation of key CpG islands within the glut4 promoter region, similar to the results of our in vitro studies with C2C12 cells. The bisulfite conversion assay at birth and later in life at 450 days revealed no differential CpG methylation in the IUGR but rather hypomethylation of the glut4 promoter region in IUGR and CON. This finding at face value suggests no role for DNA methylation in the glut4 promoter causing diminished SM glut4 transcription in the IUGR offspring. Prior in vitro studies demonstrated methylation of CpGs at –11 and –30 bp of the glut4 promoter region in murine 3T3-preadipocytic cells but demethylation in mature adipocytes. This differential methylation explained silenced glut4 gene expression in the undifferentiated pre-adipocytic stage with induced glut4 expression in differentiated adipocytes (49). Furthermore, three CpG sites at –58, –63, and –75 bp were highly demethylated in both cell stages. However, this observation has not been confirmed in vivo nor was the methylated protein that binds the –40- to –1-bp region identified (49). In this study, we explored differential methylation not only in the CpG islands that were in close proximity to the MEF2/MyoD recognition sites but also the CpG sites scattered throughout the promoter (numbered 18–28), including the ones examined in this previous in vitro investigation (49). None of these sites demonstrated differential methylation in the IUGR versus CON. Thus, although it is attractive to demonstrate hypermethylation of CpG regions within gene promoters and assign some responsibility to this change for the ultimate phenotype, the reality is that most CpG islands within inducible promoters are hypomethylated. Diet-induced developmental hypermethylation in promoters is encountered in the presence of transposans as in the Avy allele affecting all tissues or may affect imprinted genes (31). When tissue-specific differential methylation of CpGs in gene promoters is encountered, the tissue fails to express a gene throughout development based on complete silencing (31). In the IUGR we did not encounter complete GLUT4 silencing in skeletal muscle but rather a perinatal diet-induced suppression of gene expression. This may explain why we did not encounter differential DNA methylation of the glut4 promoter in the IUGR.
In the case of other imprinted genes such as the insulin-like growth factor 2, differentially methylated regions have been detected far upstream of the entire gene in question, and long range chromatin interactions were discovered to modify downstream gene transcription (59). Whether a similar situation exists in the case of IUGR glut4 expression cannot be ruled out by our studies that focused primarily on the gene promoter region. Furthermore, whether DNA methylation may occur in promoters of key transcription factors that regulate glut4 expression such as MyoD/MEF2 needs future investigation. The MeCP2 nuclear protein that binds methylated DNA (60) and increased in the IUGR SM nucleus failed to show significantly increased association with the glut4 promoter in the IUGR. In the absence of differential DNA methylation in the glut4 promoter, increased recruitment of DNMT1/DNMT3a and DNMT3b to the glut4 promoter suggests a role for these enzyme isoforms beyond DNA methyltransferase activity that promotes DNA methylation. Previous investigations have demonstrated that DNMT1/DNMT3a can interact with other repressor proteins, including HDAC1, SUV39H1, and HP1 isoforms, thereby participating in a co-repressor complex, targeted at histone deacetylation and methylation, suppressing gene transcription (28). Similarly in the IUGR, DNMT3a with DNMT3b in the adult, and perhaps DNMT1 at an earlier age, participates in a co-repressor complex suppressing SM glut4 transcription.
This is the first study delineating molecular changes responsible for skeletal muscle glut4 gene transcription in the postnatal and adult female IUGR offspring. Tissue-specific gene transcription is reliant on specific nuclear trans-activator directed gene activation. In the case of skeletal muscle, both MyoD and MEF2A are trans-activators of the glut4 gene conferring tissue specificity and hormonal/metabolic regulation (19–21, 43). The MEF2 family of transcription factors consists of four isoforms, A to D. Except for MEF2B, the remaining isoforms are highly expressed in mature skeletal muscle (61). Previous investigations have established that MEF2A activation is regulated by heterodimer formation with other MEF2 isoforms influenced by p38-dependent phosphorylation and sumoylation (62, 63). MEF2D in particular forms a heterodimer with MEF2A, thereby inhibiting MEF2A-directed transcription of downstream genes, including glut4 (46). MEF2C can also form heterodimers with MEF2A, although this isoform may activate or inhibit gene transcription under differing circumstances (16, 43, 64). This study demonstrates a key role for enhanced skeletal muscle MEF2D DNA binding and diminished MEF2A DNA binding in inhibiting glut4 transcription in the IUGR offspring.
Association of MEF2D with class II HDACs (HDAC4) can promote sumoylation and perhaps phosphorylation of MEF2D (63) thereby potentiating transcriptional repression. The class II HDACs possess two distinct domains, the C-terminal domain with catalytic activity in vivo and the N-terminal domain that exerts co-repressor activity via de-acetylase enzyme activity. Furthermore, MEF2 proteins are known to interact with class II (HDAC4, -5, -7, and -9) HDACs that are expressed in a tissue-specific manner and are implicated in skeletal muscle differentiation (45, 65). Similar interaction with ubiquitously expressed relatively compact proteins of class I (HDAC1, -2, and -3) HDACs can occur simultaneously, thereby modifying downstream gene transcription (44). Thus, association of these two HDACs, i.e. 4 (class II) and 1 (class I) with MEF2D, can promote its repressor effect on glut4 gene transcription. Despite interacting with the MEF2 DNA binding domains, HDAC repressors do not inhibit MEF2A DNA binding nor directly bind DNA themselves. Rather HDACs inhibit transcription by de-acetylating lysine side chains on chromatin-forming histones (66). In the de-acetylated state, positively charged histone tails interact with negatively charged DNA phosphate backbone. This close electrostatic interaction physically restricts the access of transcriptional activators to DNA, thereby inhibiting transcription (67). MEF2 isoforms and HDAC when physically associated inhibit MEF2-mediated downstream gene transcription (45). Thus in the case of IUGR, whereas total MEF2 binding increased, MEF2A DNA binding diminished. The decreased interaction between MEF2A and MyoD with diminished binding of MyoD to glut4 DNA sets the stage for repressed gene transcription in the IUGR.
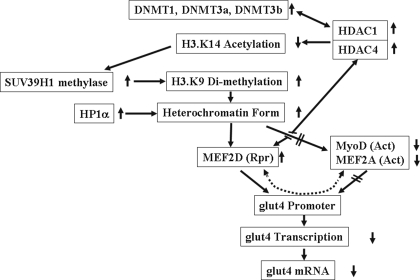
HDAC1/4-MEF2D-directed repression explains findings of adult tissue-specific diminished glut4 gene transcription. The persistence of changes in gene transcription because of cellular memory in response to perinatal nutritional perturbations (4, 25) remains unexplained. In the case of skeletal muscle glut4 as with many other genes, what is encountered with perinatal nutrient restriction is not complete silencing but rather a diminution in gene expression (metabolic knockdown). This imprint of reduced glut4 gene expression is mediated by the recruitment of DNMT1/DNMT3a and DNMT3b enzymes into a co-repressor complex, which attracts histone de-acetylases (HDAC1 and HDAC4) and histone methylases (HMT; e.g. SUV39H1) resulting in histone modifications. Histone modifications consisting of de-acetylation of H3.K14 with a hierarchical progression into di-methylation of H3.K9 contributes to heterochromatin formation. This further recruits repressor proteins such as the chromodomain containing HP1α (68) and MEF2D into the co-repressor complex that associates with the glut4 promoter. In addition, heterochromatin precludes the glut4 promoter DNA binding of activators (MyoD and MEF2A). This collectively sets the stage for glut4 gene repression in the IUGR (Fig. 9). Despite low SM GLUT4 expression in the immediate postnatal period (4), key changes in nuclear protein-glut4 DNA interaction and critical histone modifications were already existent at birth and merely persisted in the adult female IUGR offspring.
FIGURE 9.
Schematic representation of the projected combinatorial progression of epigenetic changes leading to a decrease in SM glut4 transcription and expression in the female adult IUGR offspring. Rpr = repressor, Act = activator.
Thus, similar to DNA methylation, histone methylation lends a certain amount of stability to the chromatin structure that can be inherited as an imprint mediating cellular memory. In the IUGR female offspring, differential di-methylation of H3.K9 may contribute toward the epigenetic transgenerational transmission of the insulin-resistant phenotype (25). Although we focused on SUV39H1 methylase, other histone methylases (SUV39H2, ESET/SETDB1, EuHMTase/GLP, CLL8, and RIZ1) can also methylate H3.K9 in the mammalian system (26, 30). More recently demethylases (JHDM2a, JHDM2b, JMJD2B, and JMJD2D) have also been shown to further modify the histone methylation imprint at H3.K9 (30). Histone modifications can either disrupt chromatin contacts or enhance the recruitment of non-histone proteins to chromatin. The presence of these proteins on histones can dictate higher order chromatin structure in which DNA is packaged within the nucleus, and it can orchestrate the ordered recruitment of enzyme complexes to manipulate DNA. Thus histone modifications have the potential to influence many fundamental biological processes, some of which can be epigenetically inherited.
In summary, we have demonstrated a role for de-acetylation and di-methylation of specific amino acid residues in the N-tail of histone 3. These histone modifications lend toward co-repressor complex formation, while interfering with formation of a co-activator complex. These epigenetic changes collectively decrease glut4 transcription at birth, persisting in the adult. This diminution in glut4 gene transcription is associated with the sex-specific decrease in skeletal muscle GLUT4 mRNA and protein concentrations (4, 14). Perturbations in GLUT4 expression along with previously described alterations in post-translational translocation of the GLUT4 protein (15) may be adaptive in response to diminished pancreatic β-islet insulin production. These molecular mechanisms contribute toward the development of gestational diabetes and type 2 diabetes mellitus in the IUGR adult female offspring.
Supplementary Material
Acknowledgments
We thank Melissa Spencer (David Geffen School of Medicine, UCLA) for providing the C2C12 cell line, and we acknowledge Myung-Jun Kim for the pancreatic insulin I Northern blots.
This work was supported, in whole or in part, by National Institutes of Health Grants HD 41230, HD 25024, HD 46979, and HD 33997. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4 and Figs. S.1–S.5.
Footnotes
The abbreviations used are: IUGR, intrauterine and postnatal growth restriction; SM, skeletal muscle; CON, control; DNMT1, DNA methyltransferase 1; HDAC, histone deacetylase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WT, wild type; MUT, mutated; ChIP, chromatin immunoprecipitation; IP, immunoprecipitation; EMSA, electromobility shift assay; Pipes, 1,4-piperazinediethanesulfonic acid.
References
- 1.Holemans, K., Verhaeghe, J., Dequecker, J., and Van Assche, F. A. (1995) J. Soc. Gynecol. Investig. 3 71–77 [DOI] [PubMed] [Google Scholar]
- 2.Martin-Gronert, M. S., and Ozanne, S. E. (2007) J. Intern. Med. 261 437–452 [DOI] [PubMed] [Google Scholar]
- 3.Garg, M., Thamotharan, M., Rogers, L., Bassilian, S., Lee, W. N. P., and Devaskar, S. U. (2006) Am. J. Physiol. 290 1218–1226 [DOI] [PubMed] [Google Scholar]
- 4.Thamotharan, M., Shin, B. C., Suddirikku, D. T., Thamotharan, S., Garg, M., and Devaskar, S. U. (2005) Am. J. Physiol. 288 E935–E947 [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Twinn, D. S., Wayman, A., Ekizoglou, S., Martin, M. S., Hales, C. N., and Ozanne, S. E. (2005) Am. J. Physiol. 288 R368–R373 [DOI] [PubMed] [Google Scholar]
- 6.Ozanne, S. E., Jensen, C. B., Tingey, K. J., Storgaard, H., Madsbad, S., and Vaag, A. A. (2005) Diabetologia 48 547–552 [DOI] [PubMed] [Google Scholar]
- 7.Fueger, P. T., Shearer, J., Bracy, D. P., Posey, K. A., Pencek, R. R., McGuiness, O. P., and Wasserman, D. H. (2005) J. Physiol. (Lond.) 562 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baly, D. L., and Horuk, R. (1988) Biochim. Biophys. Acta 947 571–590 [DOI] [PubMed] [Google Scholar]
- 9.Kahn, B. B. (1992) J. Clin. Investig. 89 1367–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallberg-Henriksson, H., and Zierath, J. R. (2001) 18 205–211 [DOI] [PubMed]
- 11.Richardson, J. M., and Pessin, J. E. (1993) J. Biol. Chem. 268 21021–21027 [PubMed] [Google Scholar]
- 12.Mitsumoto, Y., Burdett, E., Grant, A., and Klip, A. (1991) Biochem. Biophys. Res. Commun. 175 652–659 [DOI] [PubMed] [Google Scholar]
- 13.Zisman, A., Peroni, O. D., Abel, E. D., Michael, M. D., Mauvais-Jarvis, F., Lowell, B. B., Wojtaszewski, J. F., Hirshman, M. F., Virkamaki, A., Goodyear, L. J., Kahn, C. R., and Kahn, B. B. (2000) Nat. Med. 6 924–928 [DOI] [PubMed] [Google Scholar]
- 14.Agote, M., Goya, L., Ramos, S., Alvarez, C., Gavette, M. L., Pascual-Leone, A. M., and Escriva, F. (2001) Am. J. Physiol. 281 E1101–E1109 [DOI] [PubMed] [Google Scholar]
- 15.Oak, S. A., Tran, C., Pan, G., Thamotharan, M., and Devaskar, S. U. (2006) Am. J. Physiol. 290 E1321–E1330 [DOI] [PubMed] [Google Scholar]
- 16.Thai, M. V., Guruswamy, S., Cao, K. T., Pessin, J. E., and Olson, A. L. (1998) J. Biol. Chem. 273 14285–14292 [DOI] [PubMed] [Google Scholar]
- 17.Liu, M. L., Olson, A. L., Edgington, N. P., Moye-Rowley, W. S., and Pessin, J. E. (1994) J. Biol. Chem. 285 28514–28521 [PubMed] [Google Scholar]
- 18.Moreno, H., Serrano, A. L., Santalucia, T., Guma, A., Canto, C., Brand, N. J., Palacin, M., Schiaffino, S., and Zorzano, A. (2003) J. Biol. Chem. 278 40557–40564 [DOI] [PubMed] [Google Scholar]
- 19.Santalucia, T., Moreno, H., Palacin, M., Yacoub, M. H., Brand, N. J., and Zorzano, A. (2001) J. Mol. Biol. 314 195–204 [DOI] [PubMed] [Google Scholar]
- 20.Kaushal, S., Schneider, J. W., Nadal-Ginard, B., and Mahdavi, V. (1994) Science 266 1236–1240 [DOI] [PubMed] [Google Scholar]
- 21.Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1995) Cell 83 1125–1136 [DOI] [PubMed] [Google Scholar]
- 22.Santalucia, T., Moreno, H., Palacin, M., Yacoub, M. H., Brand, N. J., and Zorzano, A. (2005) Acta Physiol. Scand. 183 43–58 [DOI] [PubMed] [Google Scholar]
- 23.Zambrano, E., Martinez-Samayoa, P. M., Bautista, C. J., Deas, M., Guileen, L., Rodriquez-Gonzalez, G. L., Guzman, C., Larrea, F., and Nathanielsz, P. W. (2005) J. Physiol. (Lond.) 566 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boloker, J., Gertz, S. J., and Simmons, R. A. (2002) Diabetes 51 1499–1506 [DOI] [PubMed] [Google Scholar]
- 25.Thamotharan, M., Garg, M., Oak, S., Rogers, L. M., Pan, G., Sangiorgi, F., Lee, P. W. N., and Devaskar, S. U. (2007) Am. J. Physiol. 292 E1270–E1279 [DOI] [PubMed] [Google Scholar]
- 26.Devaskar, S. U., and Raychaudhuri, S. (2007) Pediatr. Res. 61 R1–R4 [DOI] [PubMed] [Google Scholar]
- 27.Bernstein, B. E., Meissner, A., and Lander, E. S. (2007) Cell 128 669–681 [DOI] [PubMed] [Google Scholar]
- 28.Fuks, F., Hurd, P. J., Deplus, R., and Kouzarides, T. (2003) Nucleic Acids Res. 31 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, B., Carey, M., and Workman, J. L. (2007) Cell 128 707–719 [DOI] [PubMed] [Google Scholar]
- 30.Kourzarides, T. (2007) Cell 128 693–705 [DOI] [PubMed] [Google Scholar]
- 31.Dolinoy, D. C., Das, R., Weidman, J. R., and Jirtle, R. L. (2007) Pediatr. Res. 61 R30–R37 [DOI] [PubMed] [Google Scholar]
- 32.Lu, J., McKinsey, T. A., Nicol, R. L., and Olson, E. N. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu, Y., and Weng, Z. (2005) Genome Inform. 16 68–72 [PubMed] [Google Scholar]
- 35.Takai, D., and Jones, P. A. (2003) In Silico Biol. 2 35–40 [PubMed] [Google Scholar]
- 36.Rajakumar, R. A., Thamotharan, S., Menon, R. K., and Devaskar, S. U. (1998) J. Biol. Chem. 273 27474–27483 [DOI] [PubMed] [Google Scholar]
- 37.Nudel, U., Zaket, R., Shani, M., Neuman, S., Levy, Z., and Yaffe, D. (1983) Nucleic Acid Res. 11 1759–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devaskar, S. U., Giddings, S. J., Rajakumar, P. A., Carnaghi, L. R., Menon, R. K., and Zahm, D. S. (1994) J. Biol. Chem. 269 8445–8454 [PubMed] [Google Scholar]
- 39.Rajakumar, R. A., Thamotharan, S., Raychaudhuri, N., Menon. R. K., and Devaskar, S. U. (2004) J. Biol. Chem. 279 26768–26779 [DOI] [PubMed] [Google Scholar]
- 40.Pattyn, F., Hoebeeck, J., Robbrecht, P., Michels, E., De Paepe, A., Bottu, G., Coornaert, D., Herzog, R., Speleman, F., and Vandesompele, J. (2006) BMC Bioinformatics 7 496, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, G., Balamotis, M. A., Stevens, J. L., Yamaguchi, Y., Handa, H., and Berk, A. J. (2005) Mol. Cell 17 683–694 [DOI] [PubMed] [Google Scholar]
- 42.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 43.Mora, S., and Pessin, J. E. (2000) J. Biol. Chem. 275 16323–16328 [DOI] [PubMed] [Google Scholar]
- 44.Huang, E. Y., Zhang, J., Miska, E. A., Guenther, M. G., Kouzarides, T., and Lazar, M. A. (2000) Genes Dev. 14 45–54 [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, J., McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2000) Mol. Cell 6 233–244 [DOI] [PubMed] [Google Scholar]
- 46.Knight, J. B., Eyster, C. A., Griesel, B. A., and Olson, A. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadiq, H. F., Das, U. G., Tracy, T. F., and Devaskar, S. U. (1999) Brain Res. 823 96–103 [DOI] [PubMed] [Google Scholar]
- 48.Chamson-Reig, A., Thyssen, S. M., Arany, E., and Hill, D. J. (2006) J. Endocrinol. 191 83–92 [DOI] [PubMed] [Google Scholar]
- 49.Yokomori, N., Tawata, M., and Onaya, T. (1999) Diabetes 48 685–690 [DOI] [PubMed] [Google Scholar]
- 50.Carretero, M. V., Torres, L., Latasa, U., Garcia-Trevijano, E. R., Prieto, J., Mato, J. M., and Avila, M. A. (1998) FEBS Lett. 439 55–58 [DOI] [PubMed] [Google Scholar]
- 51.Poirier, L. A., Brown, A. T., Fink, L. M., Wise, C. K., Randolph, C. J., de Longchamp, R. R., and Fonseca, V. A. (2001) Metabolism 50 1014–1018 [DOI] [PubMed] [Google Scholar]
- 52.Waterland, R. A., and Jirtle, R. A. (2004) Nutrition 20 63–68 [DOI] [PubMed] [Google Scholar]
- 53.Rees, W. D., Hay, S. M., Brown, D. S., Antipatis, C., and Palmer, R. M. (2000) J. Nutr. 130 1821–1826 [DOI] [PubMed] [Google Scholar]
- 54.Morgan, H. E., Sutherland, H. G., Martin, D. I., and Whitelaw, E. (1999) Nat. Genet. 23 314–318 [DOI] [PubMed] [Google Scholar]
- 55.MacLennan, N. K., James, S. J., MeInyk, S., Piroozi, A., Jernigan, S., Hsu, J. L., Janke, S. M., Pham, T. D., and Lane, R. H. (2004) Physiol. Genomics 18 43–50 [DOI] [PubMed] [Google Scholar]
- 56.Lillycrop, K. A., Slater-Jefferies, J. L., Hanson, M. A., Godfrey, K. M., Jackson, A. A., and Burdge, G. C. (2007) Br. J. Nutr. 97 1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burgde, G. C., Slater-Jefferies, J. L., Torrens, C., Phillips, E. S., Hanson, M. A., and Lillycrop, K. A. (2007) Br. J. Nutr. 97 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinmetz, K. L., Pogribny, I. P., James, S. J., and Pitot, H. C. (1998) Carcinogenesis 19 1487–1494 [DOI] [PubMed] [Google Scholar]
- 59.Ling, J. Q., and Hoffman, A. (2007) Pediatr. Res. 61 R11–R16 [DOI] [PubMed] [Google Scholar]
- 60.Lewis, J. D., Meehan, R. R., Henzel, W. J., Maurer-Fogy, I., Jeppesen, P., Klein, F., and Bird, A. (1992) Cell 69 905–914 [DOI] [PubMed] [Google Scholar]
- 61.Black, B. L., and Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14 167–196 [DOI] [PubMed] [Google Scholar]
- 62.Cox, D. M., Du, M., Marback, M., Yang, E. C., Chan, J., Siu, K. W., and McDermott, J. C. (2003) J. Biol. Chem. 278 15297–15303 [DOI] [PubMed] [Google Scholar]
- 63.Gregoire, S., Tremblay, A. M., Xiao, L., Yang, Q., Ma, K., Nie, J., Mao, Z., Wu, Z., Giguere, V., and Yang, X. J. (2006) J. Biol. Chem. 281 4423–4433 [DOI] [PubMed] [Google Scholar]
- 64.Michael, L. F., Wu, Z., Cheatham, R. B., Puigserver, P., Adelmant, G., Lehmann, J. J., Kelly, D. P., and Spiegelman, B. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miska, E. A., Karlsson, C., Langley, E., Neilsen, S. J., Pines, J., and Kouzarides, T. (1999) EMBO J. 18 5099–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002) Trends Biochem. Sci. 27 40–47 [DOI] [PubMed] [Google Scholar]
- 67.McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2001) Curr. Opin. Genet. Dev. 11 497–504 [DOI] [PubMed] [Google Scholar]
- 68.Zhang, C. L., McKinsey, T. A., and Olson, E. N. (2002) Mol. Cell. Biol. 22 7302–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.