The expression, purification and crystallization of recombinant human mitochondrial phenylalanyl-tRNA synthetase (mitPheRS) are reported. Diffraction data were collected to 2.2 Å resolution and the mitPheRS structure was solved using the molecular-replacement method.
Keywords: phenylalanyl-tRNA synthetase, mitochondria, aminoacylation
Abstract
Human monomeric mitochondrial phenylalanyl-tRNA synthetase (mitPheRS) is an enzyme that catalyzes the charging of tRNA with the cognate amino acid phenylalanine. Human mitPheRS is a chimera of the bacterial α-subunit of PheRS and the B8 domain of its β-subunit. Together, the α-subunit and the ‘RNP-domain’ (B8 domain) at the C-terminus form the minimal structural set to construct an enzyme with phenylalanylation activity. The recombinant human mitPheRS was purified to homogeneity and crystallized in complex with phenylalanine and ATP. The crystals diffracted to 2.2 Å resolution and belonged to space group P212121, with unit-cell parameters a = 55, b = 90, c = 96 Å.
1. Introduction
Covalent attachment of amino acids to their cognate tRNAs is the first step in protein biosynthesis catalyzed by aminoacyl-tRNA synthetases (aaRSs; reviewed by Francklyn et al., 2002 ▶). The aaRSs play a crucial role in the maintenance of faithful translation, promoting close control over the two-step aminoacylation reaction. In some cases, the aaRSs are incapable of discriminating between structurally similar amino acids and misacylation of tRNA with a noncognate amino acid occurs. To enhance the accuracy of aa-tRNA synthesis, an editing activity against mis-activated amino acids and/or mischarged aa-tRNAs has evolved (reviewed by Hendrickson & Schimmel, 2003 ▶; Jakubowsky, 2004 ▶).
In eukaryotes, protein synthesis occurs not only in the cytoplasm but also in different organelles, such as mitochondria and chloroplasts. In the majority of organisms, mitochondrial aaRSs (mit-aaRSs) are encoded in the nucleus and then post-translationally transported into the organelle. In a few cases, mit-aaRSs are identical to their cytosolic counterparts, as in the case of yeast mitValRS (Wang et al., 2003 ▶; Chatton et al., 1988 ▶) and mitHisRS (Natsoulis et al., 1986 ▶), human mitGlyRS (Sissler et al., 2005 ▶) and plant mitThrRS (Souciet et al., 1999 ▶). In other cases, eukaryotic mit-aaRSs exhibit higher homology to bacterial enzymes than to their cytosolic counterparts from the same organism (Spencer et al., 2004 ▶).
Human mitPheRS also exhibits higher homology to the bacterial PheRSs than to the corresponding cytosolic enzyme. Like other PheRS, the human mitochondrial enzyme has the characteristic sequence motifs of a class II aaRS. However, while the bacterial enzyme is an (αβ)2 heterodimer with a total length of 2270 residues (Mosyak et al., 1995 ▶; Goldgur et al., 1997 ▶), the mitPheRS homolog is a 415 amino-acid single-chain enzyme and is in fact a chimera of the α-subunit and the B8 domain from the β-subunit of the bacterial PheRS (Bullard et al., 1999 ▶). It has been suggested (Bullard et al., 1999 ▶) that the mitochondrial enzyme is a simplified version of the synthetase that solely serves the aminoacylation function, while the prokaryotic and eukaryotic cytosolic enzymes have developed additional functions inside the cell (e.g. Dou et al., 2001 ▶; Ivanov et al., 2000 ▶). It might also be speculated that this simplification evolved evolutionarily because of the challenges involved with the import, folding and assembly of a heterodimeric synthetase during the post-translational transport of the protein through the mitochondrial membrane.
The three-dimensional structure of the cytoplasmic Thermus thermophilus PheRS has been determined both in the apo form and with the substrates phenylalanine, PheAMP and tRNAPhe (Fishman et al., 2001 ▶; Goldgur et al., 1997 ▶; Reshetnikova et al., 1999 ▶). It was shown that the amino-acid-binding pocket accommodates not only the cognate amino acid phenylalanine, but also the noncognate tyrosine and the unnatural amino acid p-chlorophenylalanine (Kotik-Kogan et al., 2005 ▶). Both bacterial and eukaryotic cytoplasmic PheRSs have been shown to misactivate l-tyrosine, but do not attach this amino acid stably to tRNAPhe (Roy et al., 2004 ▶; Kotik-Kogan et al., 2005 ▶; Sasaki et al., 2006 ▶). Rather, these enzymes hydrolyze the misacylated Tyr-tRNAPhe in the editing site located at the interface of the B3 and B4 domains. Yeast mitPheRS also misactivates l-tyrosine, although its selectivity is one order of magnitude higher than that of the yeast cytosolic PheRS (Roy et al., 2005 ▶). However, the yeast and human mitPheRSs are unable to deacylate Tyr-tRNAPhe owing to the absence of the editing module associated with the B3 and B4 domains. No trans-editing has been detected in mitochondrial extracts, indicating that the quality control of protein synthesis normally gained by proofreading Tyr-tRNAPhe is missing from mitochondria (Roy et al., 2005 ▶).
The lack of post-transfer editing activity in mitochondrial aaRS may be a general phenomenon, although it may be compensated for to some extent by increased specificity for the cognate amino acid, as has been demonstrated for a human mitLeuRS (Lue & Kelley, 2005 ▶). This suggests that certain stages in the translation of the genetic code in mitochondria might generally be more error-prone than other translational systems. Poor fidelity in translation can lead to the accumulation of misfolded proteins and in turn result in cell death (Lee et al., 2006 ▶). On the other hand, in mitochondria only a small fraction of proteins are synthesized inside the organelle (e.g. 13 proteins in human mitochondria), while the majority of proteins are post-translationally imported following synthesis in the cell cytoplasm. One can speculate that the mitochondria can tolerate the low fidelity in translation, thus gaining a simplified (and less energy-consuming) translational machinery.
Information on the three-dimensional structure of mitPheRS is essential to provide the structural basis of the function of this unusual enzyme. The insights provided by structural studies will shed light on the increased amino-acid specificity and novel mode of tRNA binding to human mitPheRS. The solution of the three-dimensional structure of this enzyme is even more important in view of recent findings showing that at least 150 known human genetic diseases originate from mutations in the mitochondrial translational machinery (Jacobs & Turnbull, 2005 ▶; Wittenhagen & Kelley, 2003 ▶; Schapira, 2006 ▶). Here, we report the purification and crystallization of the human mitPheRS, as well as preliminary crystallographic characterization of the enzyme.
2. Materials and methods
2.1. Protein expression and purification
The residues predicted to be present in the mature form of human mitPheRS were originally cloned in a pET-21c(+) vector as described by Bullard et al. (1999 ▶). This vector produces a C-terminally His-tagged mature mitPheRS protein. However, the construct thus expressed and purified appeared to consist of approximately 20% dimeric and 80% monomeric forms [as judged by the elution of the construct from a 25 ml size-exclusion analytical Superdex 200 FPLC column (GE Healthcare) calibrated with the Sigma Gel Filtration Molecular Weight Marker Kit (No. MWGF70); data not shown]. The His tag was therefore removed from the construct in order to avoid any possible oligomeric heterogeneity that might interfere with crystallization. The PCR product encompassing the gene encoding mitPheRS (415 residues in total) without the His tag was cloned into the pET-21c(+) vector using the XbaI and XhoI restriction sites that had been incorporated into the PCR primers. DNA sequencing was performed on an Applied Biosystems Model 373A DNA sequencer using appropriate primers. The vector was transformed into Escherichia coli Rosetta DE3 strain cells (Novagen). A number of fresh colonies were inoculated into 3 l LB medium supplemented with 50 µg ml−1 ampicillin and cultured at 310 K to an OD600 of 0.6–0.8. The cells were then induced with 0.5 mM IPTG and cultured overnight at 298 K. Cells were harvested by centrifugation at 4000 rev min−1 for 30 min and stored at 193 K.
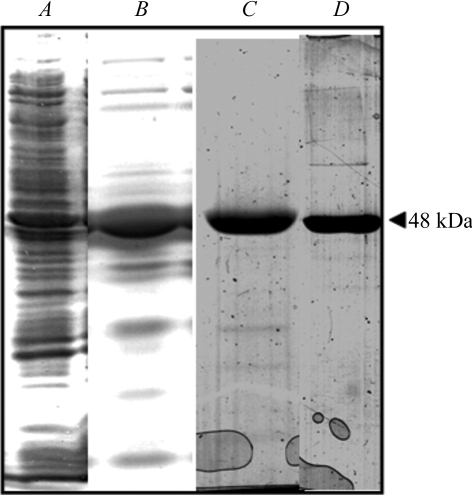
For protein purification, the cells were resuspended in lysis buffer (25 mM Tris–HCl pH 8, 400 mM NaCl, 7 mM MgCl2, 10% glycerol, 10 mM 2-mercaptoethanol). After disruption by pulsed sonication, cell debris was removed by ultracentrifugation at 30 000 rev min−1 for 0.5 h. DNA was then precipitated by a 10 min incubation with 1.3% streptomycin sulfate followed by ultracentrifugation at 30 000 rev min−1 for 0.5 h. Because of the pI of the mitPheRS (the theoretical value is 6.4), the protein was poorly retained on the DEAE-Sepharose column (Bullard et al., 1999 ▶). Satisfactory binding of mitPheRS to the DEAE resin was achieved by increasing the pH value of the buffer to 8.6. The clear cell extract (892 mg protein) was dialyzed against DEAE-loading buffer (20 mM Tris–HCl pH 8.6, 20 mM NaCl, 7 mM MgCl2, 10% glycerol, 10 mM 2-mercaptoethanol), applied onto a 50 ml DEAE-Sepharose CL-6B 250 × 25 mm ion-exchange column (GE Healthcare) and washed with 250 ml DEAE-loading buffer at 277 K. The retained mitPheRS was eluted from the DEAE-Sepharose column using a linear 500 ml gradient (20–200 mM) of NaCl in DEAE-loading buffer. Fractions (9 ml) were collected at a flow rate of 1.5 ml min−1 at 277 K. The mitPheRS eluted at about 100 mM NaCl. Fractions containing mitPheRS were identified by SDS–PAGE (Fig. 1 ▶), pooled, dialyzed against heparin-loading buffer (20 mM Tris–HCl pH 8, 20 mM NaCl, 7 mM MgCl2, 10 mM 2-mercaptoethanol) and loaded onto a 5 ml HiTrap heparin-affinity column (25 × 16 mm, GE Healthcare). MitPheRS was well retained by the column and was eluted as a single peak using a linear 400 ml gradient (20–400 mM) of NaCl in heparin-loading buffer. The flow rate was 1 ml min−1 and 7 ml fractions were collected. Fractions containing mitPheRS were identified by SDS–PAGE and pooled.
Figure 1.
SDS–PAGE analysis of the overproduced human mitPheRS following the purification procedure. Samples were analyzed on 12%(w/v) polyacrylamide gels and the proteins were visualized by staining with Coomassie Blue. All lanes were loaded with 20 mg protein. Lane A, crude lysate; lane B, material following purification on a DEAE-Sepharose column; lane C, sample of the mitPheRS preparation following purification on the heparin column; lane D, purified mitPheRS obtained by purification on Superdex-200.
MitPheRS was then concentrated by adding 35% ammonium sulfate to the pooled fractions and trapping the protein on a 1 ml TSK hydrophobic interaction column (Tosoh). MitPheRS was eluted from this column with heparin-loading buffer in a final volume of about 2 ml and then loaded onto a 600 × 16 mm size-exclusion HiLoad Superdex 200 column (GE Healthcare). Elution was performed with size-exclusion buffer (50 mM Tris–HCl pH 8, 1 M NaCl, 5 mM MgCl2, 5% glycerol, 5 mM 2-mercaptoethanol). The purified protein eluted from this column in a single symmetrical peak at a retention volume of 79 ml. The Sigma Gel Filtration Molecular Weight Markers Kit (No. MWGF70) was used to determine the molecular weight of the mitPheRS. The retention volume of mitPheRS (79 ml) corresponded to a protein with an approximate molecular weight of 48 kDa, which in turn indicated that the purified mitPheRS was monomeric. It was then concentrated on a 1 ml TSK hydrophobic interaction column, as described above, to a final concentration of 6 mg ml−1 and dialyzed against crystallization buffer (20 mM Tris–HCl pH 8, 100 mM NaCl, 7 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM EDTA and 0.0065% NaN3). The yield of mitPheRS was about 4 mg from 1 l LB medium. The protein was stored in small aliquots and flash-frozen at 193 K; it was stable when thawed.
The protein concentration was determined by the Bradford assay using bovine serum albumin as a standard. SDS–PAGE was performed on 12% slab gels with 5% stacking gels (Bio-Rad MiniProtein II system) and stained with Coomassie Brilliant Blue.
2.2. Aminoacylation assay
MitPheRS activity was determined in the tRNA-aminoacylation reaction using a trichloroacetic acid (TCA) precipitation assay. Reaction mixtures contained 25 mM Tris–HCl pH 8.5, 10 mM MgCl2, 5 mM ATP, 2 µM l-[3H]-phenylalanine (54 Ci mmol−1) and 5 mg ml−1 baker’s yeast tRNAbulk (Sigma). The reaction was started by the addition of an amount of protein that causes a linear increase in phenylalanylation within the first 2 min. The reaction was carried out at 310 K. The velocity of the esterification was measured by the rate of l-[3H]-phenylalanine incorporation into tRNA. At the appropriate times, aliquots were spotted onto paper filters impregnated with TCA. The paper filters were then washed and dried, after which the TCA-insoluble radioactivity was measured by liquid-scintillation counting.
2.3. Crystallization
Once purified, mitPheRS was screened against Crystal Screens I and II, Index and SaltRx sparse-matrix crystallization screens (Hampton Research, USA). Crystallization assays were carried out with the apo form of the enzyme and three holo forms: mitPheRS–phenylalanine, mitPheRS–phenylalanine–ATP and mitPheRS–ATP. Screening was performed at 292 and 277 K in the standard 2 µl hanging-drop setup (1 µl protein mixed with 1 µl reservoir solution; 700 µl reservoir solution), as well as in 0.3 µl (0.15 µl protein mixed with 0.15 µl reservoir solution; 70 µl reservoir solution) sitting drops using the Mosquito Crystallization Robot (TTP LabTech; Albeck et al., 2006 ▶). Crystals of the mitPheRS complex with phenylalanine and ATP appeared in 0.3 µl sitting drops containing 1.8 M sodium acetate buffer pH 7, 100 mM bis-Tris propane pH 7 at 292 K. The crystals appeared over two months and had a needle shape; their size was less then 0.03 mm in the longest direction.
Variation of the amount of the ligands and the use of additives (Additive Screens I and II, Hampton Research) and seeding were used to optimize the crystallization conditions. Final crystals were obtained in 3.5 µl hanging drops containing 3.1 mg ml−1 (65 µM) mitPheRS, 360 µM ATP, 360 µM phenylalanine, 1 mM DTT, 60 mM MgCl2, 60 mM bis-Tris propane pH 7 and 1 M sodium acetate pH 7 incubated over a 0.5 ml reservoir solution containing 2 mM DTT, 100 mM bis-Tris propane pH 7 and 1.8 M sodium acetate pH 7 at 292 K and streak-seeded from the previous crystallization experiments. MitPheRS was mixed with the ligands immediately prior to setting up the crystallization drops.
2.4. Data collection and processing
For data collection at cryogenic temperature (100 K), the crystals were transferred to a mother-liquor solution containing 25% glycerol, mounted on a cryogenic loop (Teng, 1990 ▶) and flash-cooled using an Oxford Cryostream (Cosier & Glaser, 1986 ▶) low-temperature device. A complete data set to 2.2 Å was collected from a single crystal using a Rigaku R-AXIS IV++ imaging-plate area detector mounted on a Rigaku RU-H3R rotating-anode generator equipped with a multilayer focusing mirror from Osmic and Cu X-rays (λ = 1.5418 Å). The diffraction data were integrated, scaled and reduced using the HKL-2000 (Otwinowski & Minor, 1997 ▶) program package (Table 1 ▶).
Table 2. Purification of the overexpressed human mitPheRS from 18 g E. coli Rosetta + pET-21_mitPheRS.
| Protein (mg) | Total activity† (U min−1) | Specific activity† (U min−1 mg−1) | Purification factor | Yield (%) | |
|---|---|---|---|---|---|
| Crude extract | 892 | 6000 | 6.7 | 100 | |
| DEAE Sepharose | 68 | 4500 | 66 | 9.8 | 75 |
| HiTrap heparin | 12 | 2180 | 180 | 27 | 36 |
| Superdex 200 | 10 | 2000 | 200 | 30 | 33 |
One unit of mitPheRS activity is defined as the amount of enzyme necessary to catalyze the attachment of 1 nmol L-[3H]-phenylalanine to the tRNA in 1 min at 310 K.
3. Results and discussion
Mature untagged monomeric human mitPheRS protein composed of 415 amino acids and with a molecular weight of 48 kDa and a theoretical pI of 6.4 has been overexpressed in E. coli cells and purified using three successive chromatographic steps: (i) ion exchange on Fast Flow DEAE-Sepharose resin, (ii) affinity chromatography on heparin resin and (iii) size exclusion on Superdex-200 (Fig. 1 ▶). Using this purification procedure, we achieved a 30-fold purification of mitPheRS (Table 2 ▶). The total yield of mitPheRS provided about 4 mg of purified protein from 1 l LB medium. The purity of human mitPheRS was assessed by SDS–PAGE, where it migrated as a single ∼50 kDa band.
Table 1. Data-collection statistics.
Values in parentheses are for the outermost shell.
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 54.9, b = 90.1, c = 95.8, α = 90, β = 90, γ = 90 |
| Resolution (Å) | 50–2.2 (2.24–2.2) |
| Completeness (%) | 100 (100) |
| 〈I〉/〈σ(I)〉 | 20.7 (3.1) |
| Rmerge (based on intensities) | 0.07 (0.44) |
| Redundancy | 4.4 (4.4) |
| No. of unique reflections | 24973 (1198) |
No crystals of the human mitPheRS apo form were detected in any of the sparse-matrix crystallization screens tested. Crystals of mitPheRS complexed with phenylalanine and ATP appeared in a single screening condition and could be grown over the course of a few days when streak-seeded from the initial successful crystallization screening experiment. The crystals had a bipyramidal morphology, diffracted to 2.7 Å resolution and exhibited orthorhombic symmetry. Further refinement of the crystallization conditions by the inclusion of 60 mM MgCl2 in the mother liquor yielded crystals with prismatic morphology and orthorhombic symmetry and with unit-cell parameters that were virtually the same as those of the bipyramidal crystals (Fig. 2 ▶). These crystals appeared within a week and grew to their final dimensions (0.16 × 0.1 × 0.05 mm) in the following three weeks. The crystals show a diffraction limit of beyond 2.2 Å resolution. A complete data set was collected to 2.2 Å with an R merge of 0.071% from a single crystal (see Table 1 ▶ for data-collection statistics). Examination of the systematic absences indicated that the crystals belong to the orthorhombic space group P212121, with unit-cell parameters a = 55, b = 90, c = 96 Å. This yields a calculated V M value of 2.45 Å3 Da−1 and a solvent content of 49.9%, assuming the presence of one molecule in the asymmetric unit (Matthews, 1968 ▶).
Figure 2.
Diffraction-quality crystals of human mitPheRS grown using the hanging-drop method with the use of streak-seeding. Crystals reached their maximum dimensions of 0.16 × 0.1 × 0.08 mm within 2–3 weeks.
The mitPheRS structure has been solved using the molecular-replacement method implemented in Phaser (McCoy et al., 2005 ▶). The crystal structure of bacterial phenylalanyl-tRNA synthetase from T. thermophilus (PDB code 1pys) was used as the search model. Crystallographic refinement of human mitPheRS is now in progress.
Acknowledgments
This work was supported by BSF grant No. 2005209 to MS. This work was partially supported by the Kimmelman Center for Biomolecular Structure and Assemblies.
References
- Albeck, S. et al. (2006). Acta Cryst. D62, 1184–1195. [DOI] [PMC free article] [PubMed]
- Bullard, J. M., Cai, Y.-C., Demeler, B. & Spremulli, L. L. (1999). J. Mol. Biol.288, 567–577. [DOI] [PubMed]
- Chatton, B., Walter, P., Ebel, J.-P., Lacroute, F. & Fasiolo, F. (1988). J. Biol. Chem.263, 52–57. [PubMed]
- Cosier, J. & Glaser, A. M. (1986). J. Appl. Cryst.19, 105–107.
- Dou, X., Limmer, S. & Kreutzer, R. (2001). J. Mol. Biol.305, 451–458. [DOI] [PubMed]
- Fishman, R., Ankilova, V., Moor, N. & Safro, M. (2001). Acta Cryst. D57, 1534–1544. [DOI] [PubMed]
- Francklyn, C., Perona, J. J., Pütz, J. & Hou, Y. M. (2002). RNA, 8, 1363–1372. [DOI] [PMC free article] [PubMed]
- Goldgur, Y., Mosyak, L., Reshetnikova, L., Ankilova, V., Lavrik, O., Khodyreva, S. & Safro, M. (1997). Structure, 5, 59–68. [DOI] [PubMed]
- Hendrickson, T. L. & Schimmel, P. (2003). Translational Mechanisms, edited by J. Lapointe & L. Brakier-Gingras, pp. 34–64. Georgetown, TX, USA: Landes Bioscience.
- Ivanov, K. A., Moor, N. A., Ankilova, V. N. & Lavrik, O. I. (2000). Biochemistry (Mosc.), 65, 436–441. [PubMed]
- Jacobs, H. T. & Turnbull, D. M. (2005). Trends Genet.21, 312–314. [DOI] [PubMed]
- Jakubowsky, H. (2004). The Aminoacyl-tRNA Synthetases, edited by M. Ibba, C. Francklyn & S. Cusack, pp. 384–396. Georgetown, TX, USA: Landes Bioscience.
- Kotik-Kogan, O., Moor, N., Tworowski, D. & Safro, M. (2005). Structure, 13, 1799–1807. [DOI] [PubMed]
- Lee, J. W., Beebe, K., Nangle, L. A., Jang, J., Longo-Guess, C. M., Cook, S. A., Davisson, M. T., Sundberg, J. P., Schimmel, P. & Ackerman, S. L. (2006). Nature (London), 443, 50–55. [DOI] [PubMed]
- Lue, S. W. & Kelley, S. O. (2005). Biochemistry, 44, 3010–3016. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Matthews, B. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mosyak, L., Reshetnikova, L., Goldgur, Y., Delarue, M. & Safro, M. G. (1995). Nature Struct. Biol.2, 537–547. [DOI] [PubMed]
- Natsoulis, G., Hilger, F. & Fink, G. R. (1986). Cell, 46, 235–243. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Reshetnikova, L., Moor, N., Lavrik, O. & Vassylyev, D. (1999). J. Mol. Biol.287, 555–568. [DOI] [PubMed]
- Roy, H., Ling, J., Alfonzo, J. & Ibba, M. (2005). J. Biol. Chem.280, 38186–38192. [DOI] [PubMed]
- Roy, H., Ling, J., Irnov, M. & Ibba, M. (2004). EMBO J.23, 4639–4648. [DOI] [PMC free article] [PubMed]
- Sasaki, H. M., Sekine, S., Sengoku, T., Fukunaga, R., Hattori, M., Utsunomiya, Y., Kuroishi, C., Kuramitsu, S., Shirouzu, M. & Yokoyama, S. (2006). Proc. Natl Acad. Sci. USA, 103, 14744–14749. [DOI] [PMC free article] [PubMed]
- Schapira, A. (2006). Lancet, 368, 70–82. [DOI] [PubMed]
- Sissler, M., Pütz, J., Faziolo, F. & Florentz, C. (2005). The Aminoacyl-tRNA Synthetases, edited by M. Ibba, C. Francklyn & S. Cusack, pp. 271–277. Georgetown, TX, USA: Landes Bioscience.
- Souciet, G., Menand, B., Ovesna, J., Cosset, A., Dietrich, A. & Wintz, H. (1999). Eur. J. Biochem.266, 848–854. [DOI] [PubMed]
- Spencer, A. C., Heck, A., Takeuchi, N., Watanabe, K. & Spremulli, L. L. (2004). Biochemistry, 43, 9743–9754. [DOI] [PubMed]
- Teng, T.-Y. (1990). J. Appl. Cryst.23, 387–391.
- Wang, C. C., Chang, K. J., Tang, H. L., Hsieh, C. J. & Schimmel, P. (2003). Biochemistry, 42, 1646–1651. [DOI] [PubMed]
- Wittenhagen, L. M. & Kelley, S. O. (2003). Trends Biochem. Sci.28, 605–611. [DOI] [PubMed]