A glucosamine 6-phosphate deaminase homologue from S. mutans was expressed, purified and crystallized. Diffraction data have been collected to 2.4 Å resolution.
Keywords: SMU.636, Streptococcus mutans, glucosamine 6-phosphate deaminase
Abstract
The SMU.636 protein from Streptococcus mutans is a putative glucosamine 6-phosphate deaminase with 233 residues. The smu.636 gene was PCR-amplified from S. mutans genomic DNA and cloned into the expression vector pET-28a(+). The resultant His-tagged fusion protein was expressed in Escherichia coli and purified to homogeneity in two steps. Crystals of the fusion protein were obtained by the hanging-drop vapour-diffusion method. The crystals diffracted to 2.4 Å resolution and belong to space group P212121, with unit-cell parameters a = 53.83, b = 82.13, c = 134.70 Å.
1. Introduction
The Gram-positive bacterium Streptococcus mutans is the leading pathogen in human dental caries (tooth decay; Loesche, 1986 ▶) and is associated with non-oral infections such as subacute bacterial endocarditis (Ullman et al., 1988 ▶). The genome of S. mutans UA159, a serotype strain, was completely sequenced in 2002. It is composed of 2 030 936 base pairs containing 1963 ORFs, about 63% of which have been assigned putative functions (Ajdic et al., 2002 ▶).
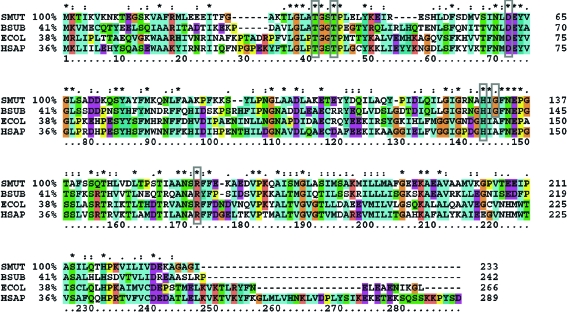
The smu.636 gene of S. mutans encodes a putative protein of 233 residues with a molecular weight of 25.5 kDa. Primary sequence alignment shows that the SMU.636 protein belongs to the glucosamine 6-phosphate deaminase (NagB; EC 3.5.99.6) family (Fig. 1 ▶). NagB performs the deamination and isomerization reactions that convert glucosamine 6-phosphate (GlcN-6-P) to fructose 6-phosphate (F6P) and release ammonia (Warren, 1972 ▶), which is the final step in the specific pathway of GlcNAc utilization. Lying at the central position in peptidoglycan formation and glycolysis, NagB decides the metabolic fate of GlcNAc (Komatsuzawa et al., 2004 ▶). To date, NagB structures from Escherichia coli, Bacillus subtilis and human have been determined (Oliva et al., 1995 ▶; Vincent et al., 2005 ▶; Arreola et al., 2003 ▶). E. coli and human NagBs exist as hexamers and are allosterically regulated, while B. subtilis NagB exists as a monomer. SMU.636 protein shows a higher sequence identity to B. subtilis NagB. Structural analysis of B. subtilis NagB indicates that Thr36, Thr39, Asp67, His138, Gly140 and Arg167 are conserved and compose the catalytic active centre (Vincent et al., 2005 ▶); these residues are also conserved in SMU.636 protein (Fig. 1 ▶).
Figure 1.
Multiple sequence alignment of SMU.636 homologues. The sequences of the glucosamine 6-phosphate deaminases of known structure from B. subtilis (Vincent et al., 2005 ▶), E. coli (Oliva et al., 1995 ▶) and human (Arreola et al., 2003 ▶) are aligned with SMU.636. The conserved residues composing the active centre are indicated by grey boxes and the percentage sequence identities with SMU.636 are indicated after the organism names; asterisks indicate totally conserved residues. The alignment was performed using the program CLUSTALX (Thompson et al., 1997 ▶). BSUB, B. subtilis; ECOL, E. coli; HSAP, Homo sapiens.
The enzymes participating in the biosynthesis of peptidoglycan have long been considered attractive targets for antibacterial agents. The determination of the crystal structure of SMU.636, which is a potential ‘gate’ in this pathway, will help us understand its biological function and provide clues for the design of antimicrobial agents against this dental pathogen.
2. Materials and methods
2.1. Cloning and expression
The SMU.636 gene was amplified by polymerase chain reaction (PCR) from S. mutans genomic DNA using primers 5′-CGCGGATCCCATGAAAACTATTAAAGTAAAAAAT-3′ and 5′-CCGCTCGAGTTAAATTCCTGCTCCTGCTTTT-3′. The amplified fragment was inserted into the BamHI/XhoI-digested expression vector pET-28a(+) (Novagen). An N-terminal His6 tag of 34 amino-acid residues and sequence MGSSHHHHHHSSGLVPRGSHMASMTGGQQMGRGS was added to the gene product. Transformed E. coli strain BL21 (DE3) cells were cultured in lysogeny broth (LB) medium supplemented with 50 µg ml−1 kanamycin at 310 K until the OD600 reached 0.6–0.8. Gene expression was then induced with 1.0 mM isopropyl β-d-thiogalactoside (IPTG) and the culture was incubated for a further 5 h at 303 K. The cells were then harvested by centrifugation at 6700g for 10 min and resuspended in lysis buffer containing 20 mM Tris–HCl, 500 mM NaCl pH 7.5.
2.2. Protein purification and crystallization
The cells were disrupted by sonication on ice. The lysate was centrifuged at 34 700g at 277 K for 30 min twice to exclude the debris. The supernatant was loaded onto a 5 ml HiTrap Ni column (GE Healthcare) equilibrated with buffer A (20 mM Tris–HCl, 0.5 M NaCl pH 7.5). Unbound proteins were eluted with buffer A and loosely bound proteins were eluted with 5% buffer B (20 mM Tris–HCl, 0.5 M NaCl, 0.5 M imidazole pH 7.5) in buffer A. The tightly bound proteins were then eluted with buffer B. The protein was further purified to homogeneity using a HiLoad Superdex 75 column (GE Healthcare) with buffer C (20 mM Tris–HCl, 150 mM NaCl pH 7.5). The purity of the protein was examined by SDS–PAGE at each step.
The purified protein from gel filtration was concentrated to 10 mg ml−1 in the final elution buffer by ultrafiltration (Ultra-15, 10 kDa cutoff, Millipore Amicon) and used in crystallization experiments. Initial crystallization experiments were performed at 289 K by the hanging-drop vapour-diffusion method using Crystal Screen I, Crystal Screen II and Index Screen (Hampton Research, CA, USA). 1 µl protein solution was mixed with 1 µl reservoir solution and the droplets were equilibrated against 500 µl reservoir solution.
2.3. X-ray data collection and processing
The crystal was flash-cooled and maintained at 100 K in a cold nitrogen-gas stream during data collection; no cryoprotectant was needed. X-ray diffraction data were collected on a Bruker SMART 6000 CCD detector using Cu Kα radiation from a Bruker–Nonius FR591 rotating-anode generator. A total of 800 frames were collected with 0.3° ϕ oscillation per frame; the exposure time per frame was 100 s. Data were processed using the Bruker PROTEUM online software suite.
3. Results
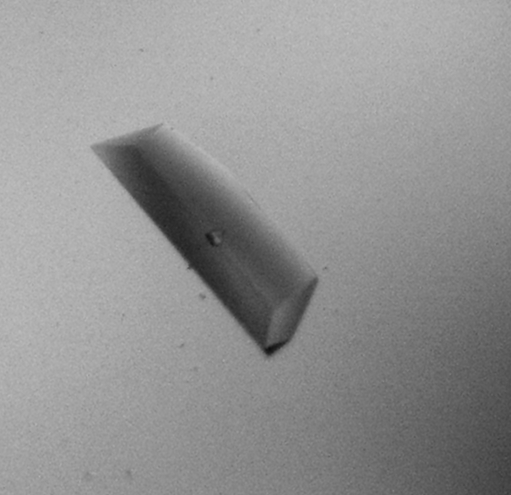
The target protein was purified to homogeneity after gel filtration; only one band was visible on the SDS–PAGE gel. Microcrystals were observed under several conditions and further optimization was applied based on condition No. 66 of Index Screen, which consisted of 0.2 M ammonium sulfate, 0.1 M bis-Tris pH 5.5, 25%(w/v) PEG 3350. After reducing the protein concentration to 5 mg ml−1, diffraction-quality crystals appeared within 2 d under the original conditions (Fig. 2 ▶).
Figure 2.
Crystal of S. mutans SMU.636. Approximate dimensions are 0.1 × 0.2 × 0.8 mm.
The crystal diffracted to 2.4 Å and belonged to space group P212121, with unit-cell parameters a = 53.83, b = 82.13, c = 134.70 Å. There could be two or three molecules in each asymmetric unit, which would give V M values of 3.1 or 2.0 Å3 Da−1, corresponding to solvent contents of 59% and 39%, respectively (Matthews, 1968 ▶). Statistics of data collection are summarized in Table 1 ▶.
Table 1. Data-collection statistics of SMU636.
Values in parentheses are for the highest resolution shell.
| Resolution (Å) | 50–2.4 (2.54–2.4) |
| Completeness (%) | 95.6 (89.5) |
| Rsym† (%) | 9.1 (32.5) |
| Mean I/σ(I) | 5.5 (2.0) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 53.83, b = 82.13, c = 134.70 |
| No. of observed reflections | 196175 |
| No. of unique reflections | 23007 |
R
sym = 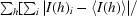
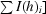 , where I(h)i is the ith observation of reflection h and 〈I(h)〉 is the mean intensity of all observations of h.
, where I(h)i is the ith observation of reflection h and 〈I(h)〉 is the mean intensity of all observations of h.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (30530190). Peking University’s 985 and 211 grants are also greatly acknowledged. YHL is the recipient of a Fok Ying Tong Education Foundation grant (94017).
References
- Ajdic, D. et al. (2002). Proc. Natl Acad. Sci. USA, 99, 14434–14439.
- Arreola, R., Valderrama, B., Morante, M. L. & Horjales, E. (2003). FEBS Lett.551, 63–70. [DOI] [PubMed]
- Komatsuzawa, H., Fujiwara, T., Nishi, H., Yamada, S., Ohara, M., McCallum, N., Berger-Bachi, B. & Sugai, M. (2004). Mol. Microbiol.53, 1221–1231. [DOI] [PubMed]
- Loesche, W. J. (1986). Microbiol. Rev.50, 353–380. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Oliva, G., Fontes, M. R., Garratt, R. C., Altamirano, M. M., Calcagno, M. L. & Horjales, E. (1995). Structure, 3, 1323–1332. [DOI] [PubMed]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997). Nucleic Acids Res.25, 4876–4882. [DOI] [PMC free article] [PubMed]
- Ullman, R. F., Miller, S. J., Strampfer, M. J. & Cunha, B. A. (1988). Heart Lung, 17, 209–212. [PubMed]
- Vincent, F., Davies, G. J. & Brannigan, J. A. (2005). J. Biol. Chem.280, 19649–19655. [DOI] [PubMed]
- Warren, L. (1972). In Glycoproteins, edited by A. Gottschalk. Amsterdam: Elsevier.