Preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from B. longum is described.
Keywords: galacto-N-biose-/lacto-N-biose I-binding protein, ABC transporter, Bifidobacterium longum
Abstract
A recombinant galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) from Bifidobacterium longum JCM1217 has been prepared and crystallized by the hanging-drop vapour-diffusion method using 10 mg ml−1 purified enzyme, 0.01 M zinc sulfate, 0.1 M MES buffer pH 5.9–6.4 and 20–22%(v/v) PEG MME 550 in the presence of 5 mM disaccharide ligands. Suitable crystals grew after 10 d incubation at 293 K. The crystals belong to space group C2221, with unit-cell parameters a = 106.3, b = 143.6, c = 114.6 Å for the lacto-N-biose I complex and a = 106.4, b = 143.4, c = 115.5 Å for the galacto-N-biose complex, and diffracted to 1.85 and 1.99 Å resolution, respectively.
1. Introduction
Bifidobacteria are considered to be health-promoting bacteria and have therefore attracted a great deal of attention. In addition to direct intake of bifidobacteria as probiotics, several attempts have been made to stimulate the growth of the organisms in the gut ecosystem by supplying bifidogenic compounds as food additives (prebiotics; Collins & Gibson, 1999 ▶; Parkes, 2007 ▶). In most cases, these approaches have employed oligosaccharides and polysaccharides to reflect the diverse abilities of these organisms to utilize carbohydrates, which was substantiated by a genomic analysis of Bifidobacterium longum NCC2705 (Schell et al., 2002 ▶). One of the interesting features arising from this genomic analysis is that the bacterium has evolved glycosidases not only for degradation of plant-derived nondigestible polysaccharides but also for degradation of host-derived polysaccharides. The recent identification of 1,2-α-fucosidase and endo-α-N-acetylgalactosaminidase genes in bifidobacteria further illustrates this point (Katayama et al., 2004 ▶; Fujita et al., 2005 ▶). However, our knowledge regarding the oligosaccharide-degradation pathways in bifidobacterial cells remains fragmentary.
Recently, Kitaoka et al. (2005 ▶) reported the molecular cloning of lacto-N-biose phosphorylase (LNBP) from B. longum JCM1217. This enzyme, which is found in most bifidobacterial strains isolated from infants, catalyzes the phosphorolysis of both lacto-N-biose I (LNB; galactosyl-β1,3-N-acetylglucosamine) and galacto-N-biose (GNB; galactosyl-β1,3-N-acetylgalactosamine) to produce galactose 1-phosphate and the respective N-acetylhexosamines (Derensy-Dron et al., 1999 ▶). Since LNB and GNB are the core disaccharide structures constituting human milk and mucin-type oligosaccharides, respectively, they are supposed to be abundantly present in the gut, especially in the intestines of breast-fed infants (Kunz et al., 2000 ▶; Podolsky, 1985 ▶; Slomiany et al., 1984 ▶). The disaccharides would then be cleaved off from their original sugar chains by the actions of the extracellular lacto-N-biosidase and endo-α-N-acetylgalactosaminidase of bifidobacteria, respectively. LNBP is localized in the cytoplasmic space; therefore, the released disaccharides must be imported into the cells by specific transport systems.
Scrutiny of the B. longum NCC2705 sequence around the lnbp gene led to the assumption that the gene cluster BL1638-1640 encodes the GNB/LNB-uptake system (Kitaoka et al., 2005 ▶). The cluster contains the genes for a typical ABC-type transporter of Gram-positive bacteria and is located just upstream of the lnbp gene. This type of gene organization is also found in Streptococcus mutans, in which the sucrose phosphorylase gene is preceded by the ABC transporter (Russell et al., 1992 ▶). In this study, therefore, as a first step towards elucidating the role of this gene cluster, we cloned a gene encoding a solute-binding protein (SBP), designated BL1638, of the system and characterized its product, since the substrate specificities of ABC transporters are essentially determined by SBPs (Tam & Saier, 1993 ▶).
The SBP of B. longum JCM1217 was found to comprise 438 amino-acid residues with a predicted molecular weight of 46 kDa. Analysis of the primary structure revealed the presence of a lipid-anchor signal at its N-terminus, indicating that it is a membrane-tethered lipoprotein (Sutcliffe & Russell, 1995 ▶). The protein contains the so-called SBP_bac_1 domain (amino-acid residues 9–344; Pfam01547) and exhibits amino-acid sequence similarities to SBPs of the carbohydrate-uptake transporter-1 (CUT1) family in the TCDB database (http://www.tcdb.org/progs/blast.php; Saier et al., 2006 ▶; Schneider, 2001 ▶). The purified protein showed exothermic binding to both LNB and GNB in isothermal titration calorimetry analyses, but was heat-silent toward other carbohydrates tested (manuscript in preparation), indicating the involvement of this transport machinery in GNB/LNB-specific uptake. Here, we describe the expression, purification, crystallization and preliminary X-ray analysis of the GNB/LNB-binding protein (GL-BP) of B. longum JCM1217.
2. Materials and methods
2.1. Plasmid construction and preparation of the recombinant proteins
A gene encoding a signal peptide-truncated form of GL-BP (amino-acid residues 28–438) was amplified by high-fidelity polymerase chain reaction with KOD-Plus polymerase (Toyobo) using the genomic DNA of B. longum JCM 1217 as a template and the primer pair 5′-GCATATGGACACTGCAGGAGACACGAAGAC-3′ and 5′-GCTCGAGTCACTCGGAAACAGACAGGCCG-3′ (sequences in bold represent the restriction-endonuclease sites). The amplified fragment was digested with NdeI and XhoI and then inserted into the corresponding sites of pET-16b and pET-23b (Novagen), thereby producing a factor Xa-cleavable decahistidine-tagged protein and a tag-free protein, respectively. Each expression plasmid was introduced into Escherichia coli Rosetta(DE3) pLacI cells (Novagen) and the transformants were grown in Luria–Bertani medium containing 100 mg l−1 ampicillin and 20 mg l−1 chloramphenicol at 303 K. When the optical density at 600 nm reached 0.5, isopropyl β-d-thiogalactoside was added to a final concentration of 0.5 mM to induce protein expression. Following additional incubation for 3 h, the cells were harvested and lysed using the Bugbuster protein-extraction reagent (Novagen) and the soluble fraction was obtained by centrifugation at 16 000g for 30 min. In the case of histidine-tagged GL-BP, the protein was first purified by Ni-charged HiTrap chelating column chromatography (GE Healthcare), followed by size-exclusion chromatography using Superdex 200 10/300 GL (GE Healthcare). The protein was subsequently treated with factor Xa and passed through an Ni-charged HiTrap chelating column. Unbound protein was collected and further purified by Superdex 200 10/300 GL column chromatography. After cleavage, the recombinant protein contained residues 28–438 of GL-BP preceded by a histidine residue derived from the expression vector. To obtain tag-free GL-BP, the protein was precipitated by adding ammonium sulfate to 80–90% saturation. The precipitate was dissolved and dialyzed against 10 mM Tris–HCl buffer pH 8.0; the resulting solution was loaded onto a Mono Q 5/50 GL column (GE Healthcare) and the protein was eluted with a linear gradient of 0–1 M NaCl. The purified recombinant proteins were desalted and concentrated (10 mg ml−1) in 10 mM HEPES buffer pH 7.0 using Amicon Ultra and Microcon centrifugal filter devices (Millipore). The homogeneity of both proteins was established by SDS–PAGE analysis (Laemmli, 1970 ▶). The protein concentrations were determined by measuring the absorbance at 280 nm using a theoretical molar absorbance coefficient of 68 870 M −1 cm−1 calculated from the amino-acid composition of GL-BP.
2.2. Crystallization
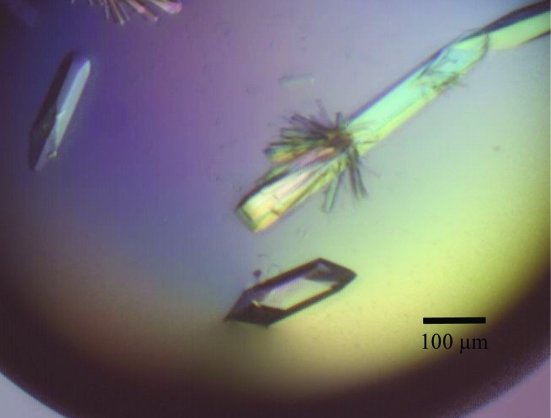
Crystallization was performed by the hanging-drop vapour-diffusion method using 48-well VDX plates (Hampton Research) at 293 K. The initial conditions were screened with the Crystallization Extension Kit (Sigma–Aldrich). Each drop was prepared by mixing 1 µl purified protein solution with the same volume of reservoir solution and was equilibrated against 200 µl reservoir solution. GNB and LNB were added to final concentrations of 5 mM. Crystals were only obtained in the presence of a ligand in drops containing solution No. 27 of the kit [0.01 M zinc sulfate, 0.1 M MES pH 6.5 and 25%(v/v) PEG MME 550]. To improve the reproducibility of crystallization, the pH and PEG MME 550 concentration were changed. Suitable crystals were obtained with a reservoir solution comprising 0.01 M zinc sulfate, 0.1 M MES pH 5.9 and 22%(v/v) PEG MME 550 for tag-removed GL-BP complexed with LNB and with 0.01 M zinc sulfate, 0.1 M MES pH 6.4 and 20%(v/v) PEG MME 550 for tag-free GL-BP complexed with GNB. The optimum conditions resulted in the formation of rod-like crystals after less than 10 d incubation at 293 K (Fig. 1 ▶).
Figure 1.
Crystals of GL-BP. The crystals were grown in a buffer system consisting of 0.01 M zinc sulfate, 0.1 M MES pH 5.9 and 22%(v/v) PEG MME 550 in the presence of lacto-N-biose I. Crystals of the GL-BP–GNB complex had similar morphology and size.
2.3. Data collection
Diffraction data were collected using a charge-coupled device (CCD) camera on the BL-17A station at the Photon Factory and the NW12A station at the Photon Factory AR, High Energy Accelerator Research Organization (KEK), Tsukuba, Japan (λ = 1.000 Å). The crystals were flash-cooled in a stream of nitrogen maintained at 100 K. Cryoprotection was not necessary because of the high PEG MME 550 concentration. For the crystal complexed with LNB a total of 360 frames were collected with 0.5° oscillations and 1 s exposures at the NW12A station and for the crystal complexed with GNB a total of 120 frames were collected with 1.0° oscillations and 5 s exposures at the BL-17A station. Diffraction images were indexed, integrated and scaled using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶). The crystals of GL-BP were found to belong to the C-centred orthorhombic space group C2221 and diffract to resolutions of 1.85 and 1.99 Å for the GNB and LNB complexes, respectively. The data-collection statistics are presented in Table 1 ▶.
Table 1. Crystallographic data-collection statistics.
Values in parentheses are for the highest resolution shell.
| LNB complex | GNB complex | |
|---|---|---|
| X-ray source | PF-AR NW12A | PF BL-17A |
| Wavelength (Å) | 1.000 | 1.000 |
| Space group | C2221 | C2221 |
| Unit-cell parameters | ||
| a (Å) | 106.3 | 106.4 |
| b (Å) | 143.6 | 143.4 |
| c (Å) | 114.6 | 115.5 |
| Resolution (Å) | 50.0–1.85 (1.92–1.85) | 50.0–1.99 (2.06–1.99) |
| Measured reflections (I > 3σ) | 532644 | 278763 |
| Unique reflections | 74117 (7268) | 60560 (5985) |
| Completeness (%) | 99.0 (98.3) | 99.8 (99.9) |
| Redundancy | 7.2 (6.3) | 4.6 (4.4) |
| Mean I/σ(I) | 42.2 (5.2) | 21.7 (3.5) |
| Rmerge† (%) | 4.7 (26.4) | 6.3 (34.5) |
R
merge = 
 , where I(h, i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the average value over multiple measurements.
, where I(h, i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the average value over multiple measurements.
3. Conclusions
Assuming that B. longum cells have GNB and LNB transporters, we scrutinized the genomic sequence of B. longum NCC2705 and identified open reading frames BL1638–1640 as a good candidate region for the system. The homologous gene to BL1638 was isolated from B. longum JCM1217 and its product (GL-BP) was analyzed. The protein only formed crystals in the presence of its ligands GNB and LNB, strongly suggesting that the protein is bound to the disaccharides in the crystals.
GL-BP eluted as a monomeric form in size-exclusion chromatography and thus should act as a monomer in the machinery, like other SBPs (Schneider, 2001 ▶). Assuming the presence of two molecules of GL-BP per asymmetric unit, the calculated V M value (Kantardjieff & Rupp, 2003 ▶) and solvent content were 2.49 Å3 Da−1 and 50.62%, respectively. The native Patterson map did not show a strong peak, indicating that the molecules in the asymmetric unit are not related by pseudo-translation. A plot of self-rotation functions calculated in polar coordinates with the rotation angle set to 180° displayed strong peaks perpendicular to the crystallographic c axis. These results indicate that there are two molecules in the asymmetric unit related by a noncrystallographic twofold axis.
The protein shows sequence similarities to SBPs of the CUT1 family; within this family the crystal structures of bacterial (E. coli) and archeal (Pyrococcus furiosus) maltose/maltodextrin-binding proteins (E. coli, 396 amino acids, 21% identity, PDB code 1anf, Quiocho et al., 1997 ▶; P. furiosus, 381 amino acids, 33% identity, PDB code 1elj, Evdokimov et al., 2001 ▶), alginate-binding protein from Sphingomonas sp. (516 amino acids, 25% identity, PDB code 1j1n; Mishima et al., 2003 ▶), (un)saturated hexuronate-binding protein from Yersinia enterocolitica (430 amino acids, 30% identity, PDB code 2uvh; Abbott & Boraston, 2007 ▶) and trehalose/maltose-binding protein from Thermococcus litoralis (450 amino acids, 16% identity, PDB code 1eu8; Diez et al., 2001 ▶) have been solved in apo and/or liganded forms. Comparisons of the binding coordinates as well as the ligand-mediated thermodynamics of these proteins would be of great importance towards better understanding of the structure–function relationships of the SBPs of ABC transporters. All attempts at molecular replacement using known SBP structures as search models failed and multiwavelength anomalous diffraction phasing using selenomethionine-substituted GL-BP is currently in progress.
Acknowledgments
We wish to thank the staff of the Photon Factory, KEK for data collection. This work was supported in part by a Grant-in-Aid from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN). JW was supported by a grant from the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science and Technology to the Graduate School of Biostudies and Institute for Virus Research, Kyoto University.
References
- Abbott, D. W. & Boraston, A. B. (2007). J. Mol. Biol.369, 759–770. [DOI] [PubMed]
- Collins, M. D. & Gibson, G. R. (1999). Am. J. Clin. Nutr.69, 1052S–1057S. [DOI] [PubMed]
- Derensy-Dron, D., Krzewinski, F., Brassart, C. & Bouquelet, S. (1999). Biotechnol. Appl. Biochem.29, 3–10. [PubMed]
- Diez, J., Diederichs, K., Greller, G., Horlacher, R., Boos, W. & Welte, W. (2001). J. Mol. Biol.305, 905–915. [DOI] [PubMed]
- Evdokimov, A. G., Anderson, D. E., Routzahn, K. M. & Waugh, D. S. (2001). J. Mol. Biol.305, 891–904. [DOI] [PubMed]
- Fujita, K., Oura, F., Nagamine, N., Katayama, T., Hiratake, J., Sakata, K., Kumagai, H. & Yamamoto, K. (2005). J. Biol. Chem.280, 37415–37422. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Katayama, T., Sakuma, A., Kimura, T., Makimura, Y., Hiratake, J., Sakata, K., Yamanoi, T., Kumagai, H. & Yamamoto, K. (2004). J. Bacteriol.186, 4885–4893. [DOI] [PMC free article] [PubMed]
- Kitaoka, M., Tian, J. & Nishimoto, M. (2005). Appl. Environ. Microbiol.71, 3158–3162. [DOI] [PMC free article] [PubMed]
- Kunz, C., Rudloff, S., Baier, W., Klein, N. & Strobel, S. (2000). Annu. Rev. Nutr.20, 699–722. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Mishima, Y., Momma, K., Hashimoto, W., Mikami, B. & Murata, K. (2003). J. Biol. Chem.278, 6552–6559. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Parkes, G. C. (2007). Nurs. Stand.21, 43–47. [DOI] [PubMed]
- Podolsky, D. K. (1985). J. Biol. Chem.260, 8262–8271. [PubMed]
- Quiocho, F. A., Spurlino, J. C. & Rodseth, L. E. (1997). Structure, 5, 997–1015. [DOI] [PubMed]
- Russell, R. R. B., Aduse-Opoku, J., Sutcliffe, I. C., Tao, L. & Ferretti, J. (1992). J. Biol. Chem.267, 4631–4637. [PubMed]
- Saier, M. H. Jr, Tran, C. V. & Barabote, R. D. (2006). Nucleic Acids Res.34, D181–D186. [DOI] [PMC free article] [PubMed]
- Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M. C., Desiere, F., Bork, P., Delley, M., Pridmore, R. D. & Arigoni, F. (2002). Proc. Natl Acad. Sci. USA, 99, 14422–14427. [DOI] [PMC free article] [PubMed]
- Schneider, E. (2001). Res. Microbiol.152, 303–310. [DOI] [PubMed]
- Slomiany, B. L., Zdebska, E. & Slomiany, A. (1984). J. Biol. Chem.259, 2863–2869. [PubMed]
- Sutcliffe, I. C. & Russell, R. R. B. (1995). J. Bacteriol.177, 1123–1128. [DOI] [PMC free article] [PubMed]
- Tam, R. & Saier, M. H. Jr (1993). Microbiol. Rev.57, 320–346. [DOI] [PMC free article] [PubMed]