Abstract
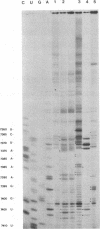
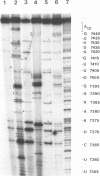
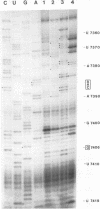
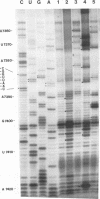
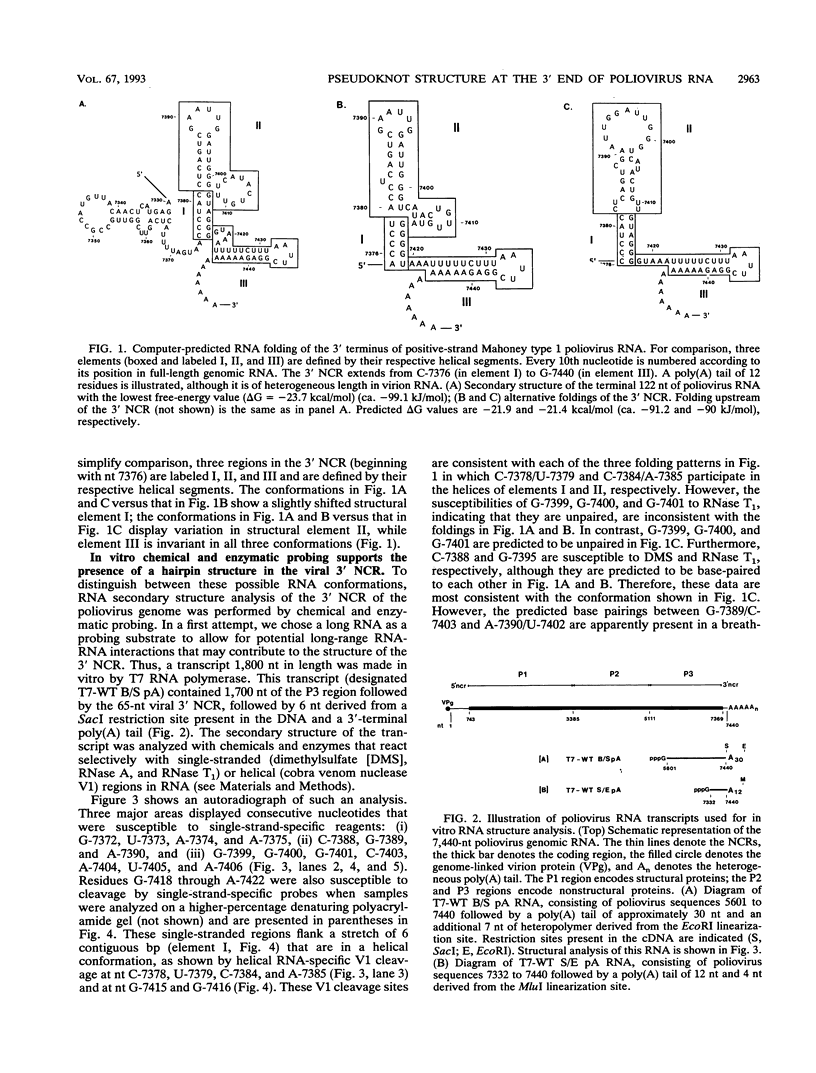
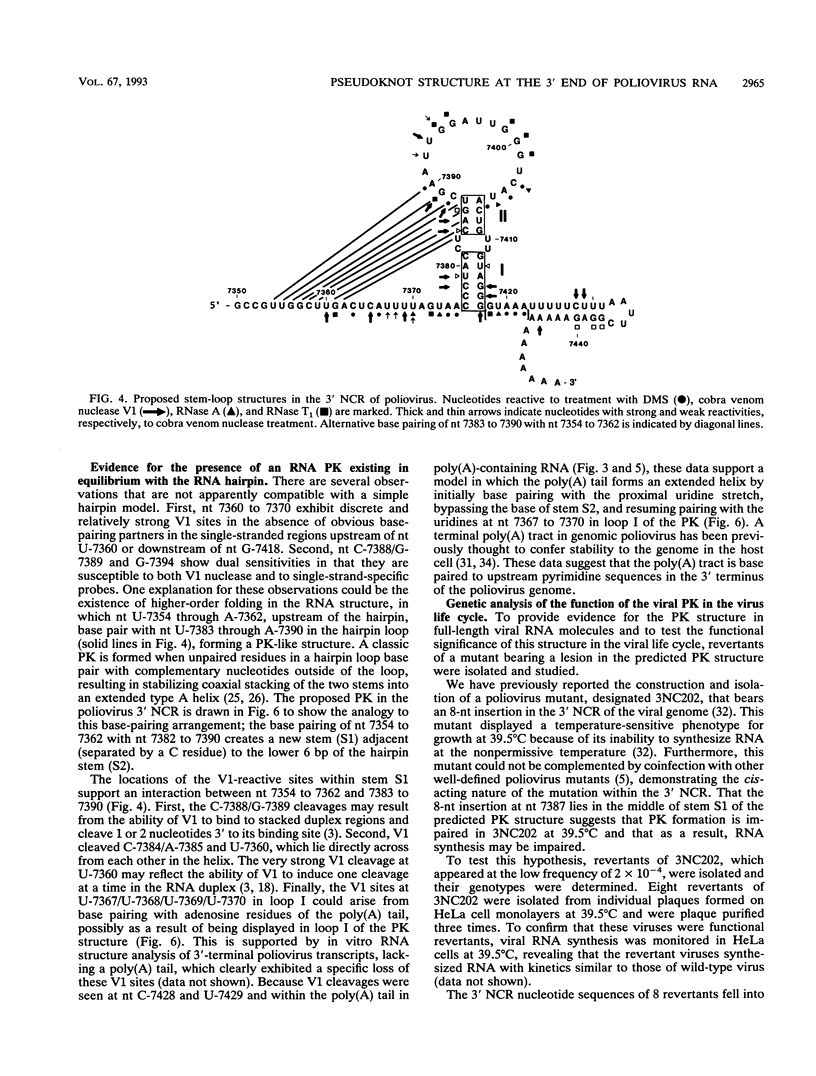
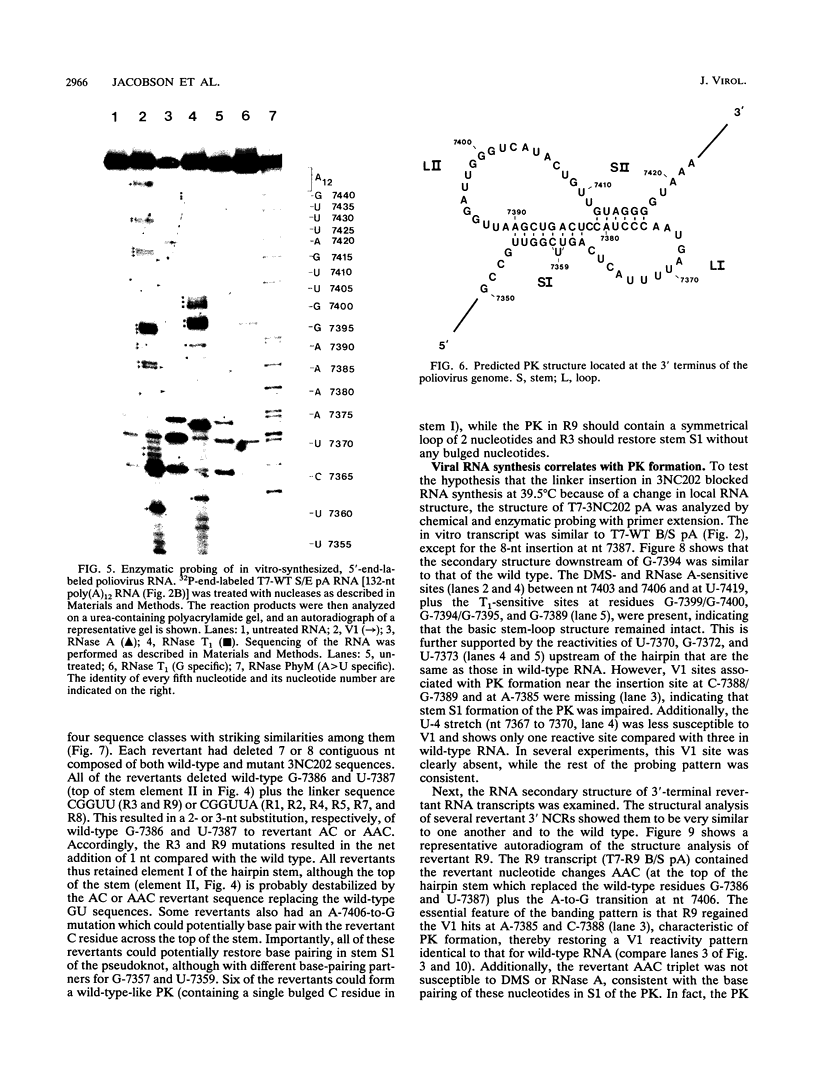
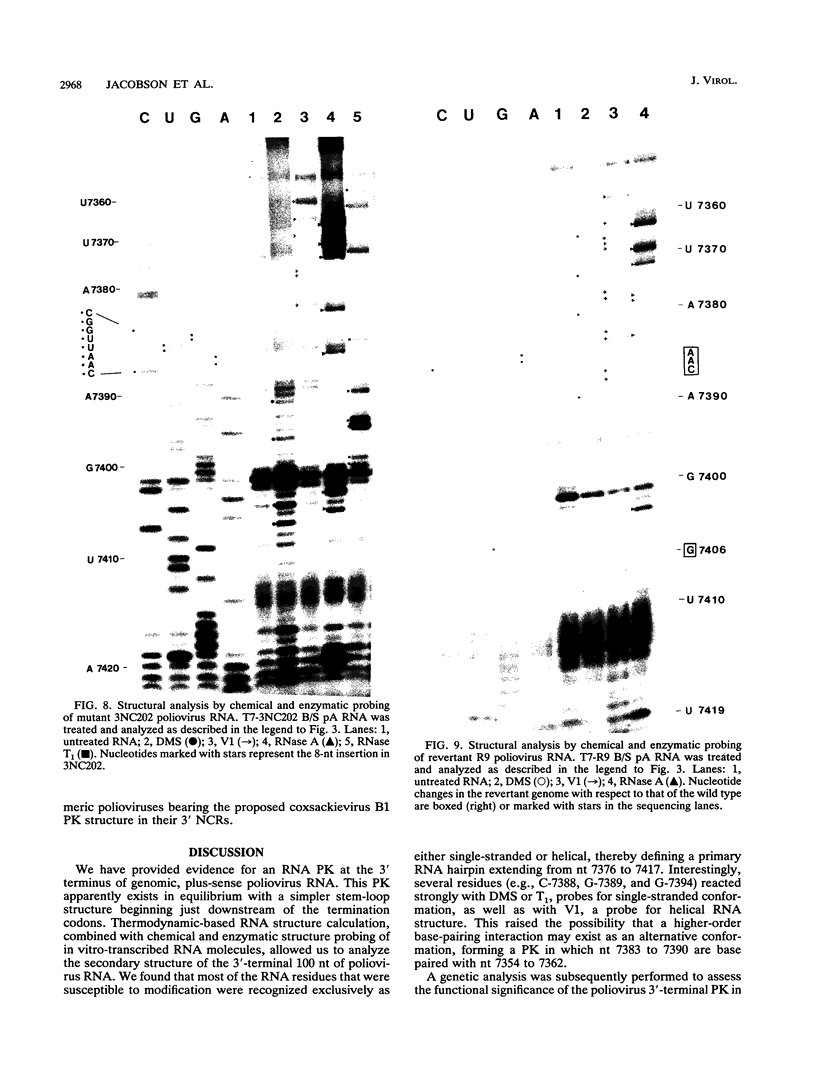
The sequences in the plus-stranded poliovirus RNA genome that dictate the specific amplification of viral RNA in infected cells remain unknown. We have analyzed the structure of the 3' noncoding region of the viral genome by thermodynamic-based structure calculation and by chemical and enzymatic probing of in vitro-synthesized RNAs and provide evidence for the existence of an RNA pseudoknot structure in this region. To explore the functional significance of this structure, revertants of a mutant bearing a lesion in the proposed pseudoknot and exhibiting a temperature-sensitive defect in viral RNA synthesis were isolated and mapped. The results of this genetic analysis established a correlation between the structure of the 3' terminus of the viral RNA and its function in vivo in RNA amplification. Furthermore, phylogenetic analysis indicated that a similar structure could be formed in coxsackievirus B1, a related enterovirus, which further supports a role for the pseudoknot structure in viral RNA amplification in infected cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed Y. F., Gilmartin G. M., Hanly S. M., Nevins J. R., Greene W. C. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991 Feb 22;64(4):727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Auron P. E., Weber L. D., Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982 Sep 14;21(19):4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Sarnow P., Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986 Dec;60(3):1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., Carmichael G. G. RNA replication: function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Structural elements in RNA. Prog Nucleic Acid Res Mol Biol. 1991;41:131–177. doi: 10.1016/S0079-6603(08)60008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Cohen L. W., Riggs A. D., Morin C., Itakura K., Richards J. H. Oligonucleotide-directed mutagenesis as a general and powerful method for studies of protein function. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6409–6413. doi: 10.1073/pnas.79.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Zabel P., Baltimore D. Dependence of the activity of the poliovirus replicase on the host cell protein. Cell. 1980 Feb;19(2):423–429. doi: 10.1016/0092-8674(80)90516-4. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Van Dyke T. A. Isolation of a soluble and template-dependent poliovirus RNA polymerase that copies virion RNA in vitro. J Virol. 1979 Oct;32(1):155–161. doi: 10.1128/jvi.32.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991 Nov;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Iizuka N., Kuge S., Nomoto A. Complete nucleotide sequence of the genome of coxsackievirus B1. Virology. 1987 Jan;156(1):64–73. doi: 10.1016/0042-6822(87)90436-3. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- McPheeters D. S., Stormo G. D., Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J Mol Biol. 1988 Jun 5;201(3):517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988 Jul 28;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991 Apr 4;350(6317):434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Maslova S. V., Sinyakov A. N., Agol V. I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992 Apr 11;20(7):1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr A pseudoknotted RNA oligonucleotide. Nature. 1988 Jan 21;331(6153):283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O. C., Ehrenfeld E. Poliovirus RNA replication. Curr Top Microbiol Immunol. 1990;161:89–119. doi: 10.1007/978-3-642-75602-3_4. [DOI] [PubMed] [Google Scholar]
- Rietveld K., Pleij C. W., Bosch L. Three-dimensional models of the tRNA-like 3' termini of some plant viral RNAs. EMBO J. 1983;2(7):1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sarnow P., Bernstein H. D., Baltimore D. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc Natl Acad Sci U S A. 1986 Feb;83(3):571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P. Role of 3'-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989 Jan;63(1):467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes E. A., Sarnow P. An RNA hairpin at the extreme 5' end of the poliovirus RNA genome modulates viral translation in human cells. J Virol. 1991 Feb;65(2):913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. H., Baltimore D. Requirement of 3'-terminal poly(adenylic acid) for the infectivity of poliovirus RNA. Proc Natl Acad Sci U S A. 1974 Aug;71(8):2983–2987. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Tobin G. J., Young D. C., Flanegan J. B. Self-catalyzed linkage of poliovirus terminal protein VPg to poliovirus RNA. Cell. 1989 Nov 3;59(3):511–519. doi: 10.1016/0092-8674(89)90034-2. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Kohara M., Kataoka Y., Suganuma T., Omata T., Imura N., Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984 Apr 25;174(4):561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Maizels N. tRNA-like structures tag the 3' ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]