The purification, crystallization, X-ray diffraction data acquisition and molecular-replacement results of royal palm tree (R. regia) peroxidase are described.
Keywords: royal palm tree peroxidase, Roystonea regia
Abstract
Royal palm tree peroxidase (RPTP), which was isolated from Roystonea regia leaves, has an unusually high stability that makes it a promising candidate for diverse applications in industry and analytical chemistry [Caramyshev et al. (2005 ▶), Biomacromolecules, 6, 1360–1366]. Here, the purification and crystallization of this plant peroxidase and its X-ray diffraction data collection are described. RPTP crystals were obtained by the hanging-drop vapour-diffusion method and diffraction data were collected to a resolution of 2.8 Å. The crystals belong to the trigonal space group P3121, with unit-cell parameters a = b = 116.83, c = 92.24 Å, and contain one protein molecule per asymmetric unit. The V M value and solvent content are 4.07 Å3 Da−1 and 69.8%, respectively.
1. Introduction
Peroxidases (EC 1.11.1.7) are enzymes that are widely distributed in the living world and are involved in many physiological processes, including abiotic and biotic stress responses. As a result of environmental stress, an increased production of oxygen intermediates (ROS) such as 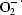 , H2O2 and OH˙ occurs. Effective metabolic conversion of
, H2O2 and OH˙ occurs. Effective metabolic conversion of 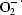 and H2O2 is crucial for organisms to overcome adverse environmental conditions. Thus, the primary function of peroxidases is conversion of H2O2 to H2O. In addition to this major role, they also participate in other processes, such as cell-wall formation, lignification and protection of tissues from pathogenic microorganisms (Dunford, 1991 ▶; Welinder, 1992 ▶). The peroxidase superfamily can be subdivided into peroxidases from mammals, catalases and the plant peroxidases. Moreover, according to their distinct structural features, members of the plant peroxidase family have been further classified as class I (intracellular prokaryotic enzymes), class II (extracellular fungal enzymes) and class III (secretory plant enzymes) peroxidases by Welinder (1992 ▶). The latter class, which share N-terminal signal peptides and four conserved disulfide bridges, includes the most studied peroxidases, such as that extracted from horseradish roots (Gajhede et al., 1997 ▶). In addition to its biological functions, this protein has various applications in industry and analytical chemistry, as exemplified, for example, by its widespread use in immunoenzymatic kits. Many peroxidases have been characterized to date, but their three-dimensional structures have only been unravelled in a few cases. The high percentage of glycosylation of the native enzymes makes them difficult to crystallize and their high stability means that their proteolytic digestion does not afford a sufficiently detailed peptide map. These characteristics, as well as the fact that the N-terminal end of many plant peroxidases is blocked by pyroglutamate and their poor homology with other proteins, are the prime reasons why primary sequences and three-dimensional structures have only been determined for a few class III peroxidases (Morita et al., 1993 ▶). The crystallographic structures of horseradish (Gajhede et al., 1997 ▶), barley (Henriksen et al., 1998 ▶) and peanut (Schuller et al., 1996 ▶) class III plant peroxidases revealed that they share the same helix-rich fold, with the central haem group sandwiched between a distal N-terminal domain and a proximal C-terminal domain. A total of ten α-helices of the structures are conserved in all known haem peroxidases (Henriksen et al., 1998 ▶). Royal palm tree (Roystonea regia) peroxidase (RPTP) is a member of the class III peroxidases and has a unique thermal stability as well pH stability in the pH range 4–11 (Sakharov et al., 2001 ▶). The unusually high stability of RPTP makes it a promising candidate for diverse applications in industry and analytical chemistry (Caramyshev et al., 2005 ▶). Here, we describe an improved purification procedure, crystallization and preliminary X-ray crystallographic analysis of RPTP.
and H2O2 is crucial for organisms to overcome adverse environmental conditions. Thus, the primary function of peroxidases is conversion of H2O2 to H2O. In addition to this major role, they also participate in other processes, such as cell-wall formation, lignification and protection of tissues from pathogenic microorganisms (Dunford, 1991 ▶; Welinder, 1992 ▶). The peroxidase superfamily can be subdivided into peroxidases from mammals, catalases and the plant peroxidases. Moreover, according to their distinct structural features, members of the plant peroxidase family have been further classified as class I (intracellular prokaryotic enzymes), class II (extracellular fungal enzymes) and class III (secretory plant enzymes) peroxidases by Welinder (1992 ▶). The latter class, which share N-terminal signal peptides and four conserved disulfide bridges, includes the most studied peroxidases, such as that extracted from horseradish roots (Gajhede et al., 1997 ▶). In addition to its biological functions, this protein has various applications in industry and analytical chemistry, as exemplified, for example, by its widespread use in immunoenzymatic kits. Many peroxidases have been characterized to date, but their three-dimensional structures have only been unravelled in a few cases. The high percentage of glycosylation of the native enzymes makes them difficult to crystallize and their high stability means that their proteolytic digestion does not afford a sufficiently detailed peptide map. These characteristics, as well as the fact that the N-terminal end of many plant peroxidases is blocked by pyroglutamate and their poor homology with other proteins, are the prime reasons why primary sequences and three-dimensional structures have only been determined for a few class III peroxidases (Morita et al., 1993 ▶). The crystallographic structures of horseradish (Gajhede et al., 1997 ▶), barley (Henriksen et al., 1998 ▶) and peanut (Schuller et al., 1996 ▶) class III plant peroxidases revealed that they share the same helix-rich fold, with the central haem group sandwiched between a distal N-terminal domain and a proximal C-terminal domain. A total of ten α-helices of the structures are conserved in all known haem peroxidases (Henriksen et al., 1998 ▶). Royal palm tree (Roystonea regia) peroxidase (RPTP) is a member of the class III peroxidases and has a unique thermal stability as well pH stability in the pH range 4–11 (Sakharov et al., 2001 ▶). The unusually high stability of RPTP makes it a promising candidate for diverse applications in industry and analytical chemistry (Caramyshev et al., 2005 ▶). Here, we describe an improved purification procedure, crystallization and preliminary X-ray crystallographic analysis of RPTP.
2. Materials and methods
2.1. Enzyme purification
RPTP, a 347-amino-acid plant enzyme with a molecular weight of 44.6 kDa, was purified from the leaves of the royal palm tree R. regia as described elsewhere (Sakharov et al., 2001 ▶) with essential modifications. Leaves (835 g) from seven-year-old Royal palm trees were milled and homogenized in 3.34 l distilled water for 22–24 h at room temperature. Excess material was removed by vacuum filtration and centrifugation (10 000g, 277–278 K for 15 min). Pigments were extracted by phase separation over 20–22 h at 277–278 K after the addition to the supernatant of solid PEG to 14%(w/v) and solid ammonium sulfate to 10%(w/v). Two phases were formed after the addition of ammonium sulfate: an upper polymer phase (dark red in colour) that contained pigments, phenols, polyphenols, oxidized phenols and PEG and a lower aqueous phase (yellow in colour) containing peroxidase. Each phase consisted of 50% of the initial volume. These phases were separated and the phase containing peroxidase activity was centrifuged. The clear supernatant containing peroxidase activity was titrated with ammonium sulfate to a conductivity value of 326 mS cm−1 and was applied onto a Phenyl-Sepharose column (1.5 × 35 cm) equilibrated with 100 mM phosphate buffer pH 6.5 with 1.7 M ammonium sulfate, which has the same conductivity as the sample. The enzyme was eluted with 100 mM phosphate buffer pH 6.5 plus 0.2 M ammonium sulfate at a flow rate of 1 ml min−1. 15 ml fractions were collected and those showing peroxidase activity were dialyzed against 5 mM Tris buffer pH 8.3 for 24 h with constant stirring at 277–278 K. These fractions were membrane-concentrated (Amicon, 10 kDa cutoff) to 15 ml and applied onto a DEAE-Toyopearl 650 column (1 × 30 cm) equilibrated with 5 mM Tris buffer pH 8.3. Elution was carried out with a linear 0–300 mM NaCl gradient in the same buffer at a flow rate of 1 ml min−1. The fractions with peroxidase activity were collected, membrane-concentrated (Amicon, 10 kDa cutoff) and applied onto a Superdex-200 column equilibrated with 5 mM Tris buffer pH 8.3. Elution was carried out in the same buffer at a flow rate of 1 ml min−1. Finally, the peroxidase was dialyzed against distilled water and freeze-dried.
The purity of the RPTP was determined by SDS–PAGE as described by Fairbanks et al. (1971 ▶) on a Bio-Rad minigel device using a flat block with a 15% polyacrylamide concentration, by gel filtration, which was performed using a Superdex 200 10/30 HR column in an FPLC Amersham Akta system, and by UV–visible spectrophotometry (RZ = A 403/A 280 = 2.8–3.0) and MALDI–TOF mass spectrometry.
2.2. Crystallization
Initial attempts to crystallize royal palm tree peroxidase were performed by hanging-drop vapour diffusion using the sparse-matrix screening method with Crystal Screens 1 and 2 and ammonium sulfate over a broad pH range (Hampton Research). Hanging drops containing 1 µl protein at 4.8 mg ml−1 in 5 mM Tris buffer pH 7.4 were mixed with equal amounts of reservoir solution and equilibrated against 500 µl reservoir solution at 277 K. Small RPTP crystals grew in hanging drops with a reservoir solution containing 2.4–3.2 M ammonium sulfate over a broad pH range (pH 6.0–8.0).
2.3. Data collection and processing
A single RPTP crystal was harvested using a nylon loop and transferred to a cryoprotectant solution consisting of 100% mineral oil for a few seconds. The crystal was then flash-cooled to 100 K in a nitrogen stream and used for data acquisition. X-ray diffraction experiments were performed with a MAR Research MAR345dtb image-plate detector mounted on a Rigaku UltraX-18 rotating-anode X-ray generator providing Cu Kα radiation (1.5418 Å) operated at 50 kV and 100 mA and equipped with Osmic confocal Max-Flux optics. A data set of 96 images was collected using the oscillation method, with an angular range of 1° and an exposure time of 20 min per image. The data set was reduced and merged using the MOSFLM (Leslie, 1992 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶) programs.
3. Results and discussion
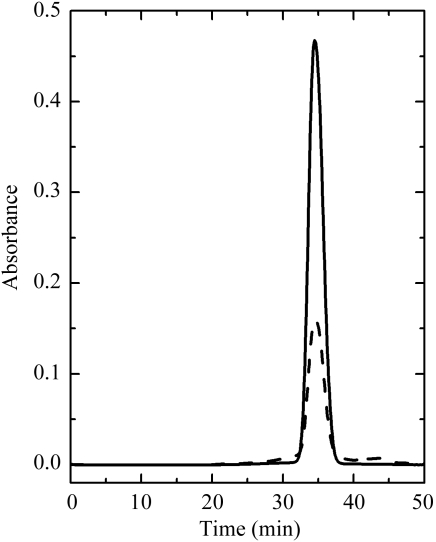
RPTP was purified to homogeneity with a high yield from royal palm tree (R. regia) leaves. The purification steps and their efficiencies are summarized in Table 1 ▶. Purified peroxidase migrated in SDS–PAGE as a single band corresponding to a molecular weight of 51 kDa (Fig. 1 ▶). The retention time of the elution of the protein from the size-exclusion column indicates that the protein forms dimers in solution with an approximate molecular weight of 90 kDa (Fig. 2 ▶). The experimentally observed high molecular weight of the native enzyme is a consequence of its extensive glycosylation. The glycosylation sites and the chemical nature of the carbohydrate moieties are currently being determined.
Table 1. Purification steps of RPTP.
| Procedure | Volume (ml) | Protein (mg) | Total activity (U) | Specific activity (U mg−1) | Purification | Yield (%) | |
|---|---|---|---|---|---|---|---|
| 1 | Homogenate | 9980 | 68862 | 1297422 | 18.84 | 1 | 100 |
| 2 | PEG + (NH4)2SO4 | 5400 | 11502 | 1293635 | 112.47 | 6 | 99.7 |
| 3 | Phenyl-Sepharose | 383 | 498 | 845825 | 1698.78 | 90 | 65.2 |
| 4 | DEAE-Toyopearl | 99.5 | 158 | 581665 | 3676.65 | 195 | 44.8 |
| 5 | Superdex 200 | 42 | 67 | 438626 | 6568.22 | 350 | 33.8 |
Figure 1.

SDS–PAGE analysis of RPTP (lane 1). Lane 2 contains molecular-weight markers: phosphorylase b from rabbit muscle (97 kDa), albumin from bovine serum (66 kDa), ovalbumin from chicken egg (45 kDa), carbonic anhydrase from bovine erythrocytes (29 kDa) and trypsin inhibitor from soybean (20 kDa).
Figure 2.
High-performance gel filtration of RPTP on Superdex 200 HR 10/30 at a flow rate of 0.4 ml min−1. The solid line is the absorbance at 403 nm and the dashed line is that at 280 nm. The molecular weight of RPTP estimated by this method is ∼90 kDa.
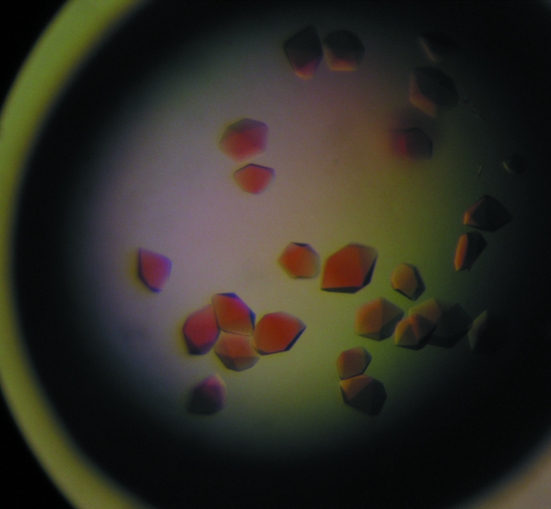
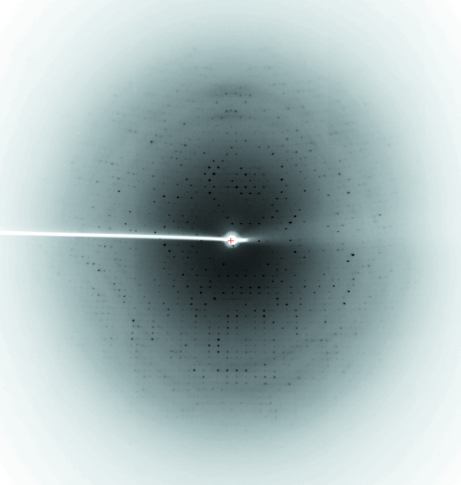
Optimization of the initial conditions with ammonium sulfate resulted in crystals suitable for X-ray diffraction data collection, which grew in about 3–4 d in hanging drops with reservoir solution containing 2.6 M ammonium sulfate and 0.1 M MES pH 6.5 at 291 K (Fig. 3 ▶). The resolution of the diffraction data collected at the home X-ray source extends to 2.8 Å (Fig. 4 ▶). Data-acquisition statistics are given in Table 2 ▶. Initial analysis of the crystal solvent content using the Matthews coefficient (Matthews, 1968 ▶) suggested that the asymmetric unit contained either one molecule with a 69.8% solvent content (V M = 4.07 Å3 Da−1) or two molecules with V M = 2.04 Å3 Da−1 and a solvent content of 39.6%. The X-ray structure of horseradish peroxidase (PDB code 1h57; 36% amino-acid sequence identity) was used as a search model for the molecular-replacement procedure. Prior to the rotation search, the search model was improved using CHAINSAW (Collaborative Computational Project, Number 4, 1994 ▶), which compares the chosen model and the given protein sequence and prunes nonconserved residues to the last common atom and retains conserved residues. This improved new model was used for molecular replacement with MOLREP (Vagin & Teplyakov, 1997 ▶).
Figure 3.
Crystals of royal palm tree peroxidase. Typical dimensions are approximately 0.3 × 0.4 × 0.3 mm.
Figure 4.
Diffraction pattern of the RPTP crystal collected on a rotating-anode X-ray generator. The maximum resolution of the data extends to 2.8 Å.
Table 2. X-ray data-acquisition statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 68.1–2.8 (2.9–2.8) |
| Space group | P3121 |
| Unit-cell parameters (Å) | a = b = 116.83, c = 92.24 |
| Completeness (%) | 99.8 (99.9) |
| Redundancy | 6.5 (6.1) |
| Rmerge† (%) | 10.7 (67.4) |
| Average I/σ(I) | 13.7 (2.3) |
| Total reflections | 119107 |
| Unique reflections | 18423 |
R
merge = 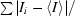
 , where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity of the reflection.
, where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity of the reflection.
The rotation search, which was conducted within the resolution range 68.1–3.0 Å, yielded a clear top solution with a value of 8.87σ (in contrast to 3.96σ for the second solution), which confirmed the presence of one protein molecule per asymmetric unit. The best solution after the translation function, which was conducted using the same resolution range, had a score of 0.436 and an R factor of 53.4%. The obtained molecular-replacement solution was visually examined using program Coot (Emsley & Cowtan, 2004 ▶) and showed no steric clashes or overlaps with the symmetry-related molecules. After ten cycles of refinement using REFMAC5 (Murshudov et al., 1997 ▶) against the collected X-ray diffraction data in the full resolution range, the R and R free factors fell to 38.2% and 42.4%, respectively, and the figure of merit was 58.9%. Complete model building and refinement of the structure to 2.8 Å resolution is in progress.
Acknowledgments
This work was supported in part by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) via grants 06/0153-5, 06/60860-0, 04/08070-9, 06/00182-8 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), as well as by Programa de Acciones Integradas de Investigación Cientifica y Tecnológica (2006–2007).
References
- Caramyshev, A. V., Evtushenko, E. G., Ivanov, V. F., Ros Barceló, A., Roig, M. G., Shnyrov, V. L., van Huystee, R. B., Kurochkin, I. N., Vorobiev, A. K. & Sakharov, I. Y. (2005). Biomacromolecules, 6, 1360–1366. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dunford, H. B. (1991). Peroxidases in Chemistry and Biology, edited by J. Everse, K. E. Everse & M. B. Grisham, Vol. II, pp. 1–24. Boca Raton, FL, USA: CRC Press.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fairbanks, G., Steck, T. & Wallach, D. F. N. (1971) Biochemistry, 10, 2606–2617. [DOI] [PubMed]
- Gajhede, M., Schuller, D. J., Henriksen, A., Smith, A. T. & Poulos, T. L. (1997). Nature Struct. Biol.4, 1032–1038. [DOI] [PubMed]
- Henriksen, A., Welinder, K. G. & Gajhede, M. (1998). J. Biol. Chem.273, 2241–2248. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Morita, Y., Funatsu, J. & Mikami, B. (1993). Plant Peroxidases: Biochemistry and Physiology, edited by K. G. Welinder, S. K. Rasmussen, C. Penel & H. Greppin, pp. 1–4. Geneva: University of Geneva.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Sakharov, I. Y., Vesga, M. K., Galaev, I. Y., Sakharova, I. V. & Pletjushkina, O. Y. (2001). Plant Sci.161, 853–860.
- Schuller, D. J., Ban, N., van Huystee, R. B., McPherson, A. & Poulos, T. (1996). Structure, 4, 311–321. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Welinder, K. G. (1992) Curr. Opin. Struct. Biol.2, 388–393.