The crystallization strategy for the RNA polymerase I subcomplex A14/A43, which is based on iterative cycles of prediction, probing and removal of four flexible protein regions, is described.
Keywords: multiprotein crystals, mapping unstructured regions, protein engineering, crystal engineering, RNA polymerase I, Rpb4/7 subcomplex
Abstract
The removal of flexible protein regions is generally used to promote crystallization, but advanced strategies to quickly remove multiple flexible regions from proteins or protein complexes are lacking. Here, it is shown how a protein heterodimer with multiple flexibilities, the RNA polymerase I subcomplex A14/A43, could be crystallized with the use of an iterative procedure of predicting flexible regions, experimentally testing and improving these predictions and combining deletions of flexible regions in a stepwise manner. This strategy should enable the crystallization of other proteins and subcomplexes with multiple flexibilities, as required for hybrid structure solution of large macromolecular assemblies.
1. Introduction
Structure determination of large multi-component complexes often requires hybrid approaches that combine electron microscopy (EM) with X-ray crystallography. Whereas EM can establish the structure of the complex at medium resolution, the subunit architecture may be resolved by interpreting the EM density using high-resolution structures of subcomplexes obtained by X-ray crystallography. However, such subcomplexes often fail to crystallize because they contain flexible regions, some of which result from the removal of the subcomplex from the intact native complex. Recently, we reported the hybrid structure of yeast RNA polymerase (Pol) I, a 14-subunit 600 kDa complex (Kuhn et al., 2007 ▶). Hybrid analysis of the complex relied on the crystal structure of the heterodimeric polymerase subcomplex A14/A43, which contained several extended flexible regions. Here, we show how iterative cycles of structure prediction and experiments were used to identify and remove flexible protein regions, which led to the crystallization of the A14/A43 subcomplex. This approach can be applied to comparable crystallization challenges in the future.
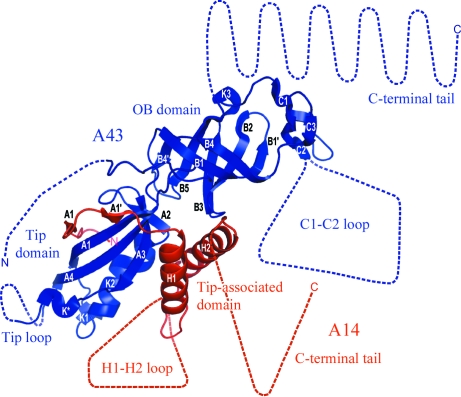
The recent crystal structure of A14/A43 revealed an overall similarity to its counterparts Rpb4/7 in Pol II, C17/25 in Pol III and RpoF/E in the archaeal Pol (Kuhn et al., 2007 ▶; Meka et al., 2003 ▶; Peyroche et al., 2002 ▶; Hu et al., 2002 ▶; Sadhale & Woychik, 1994 ▶; Siaut et al., 2003 ▶; Shpakovskii, 1999 ▶; Todone et al., 2001 ▶; Fig. 1 ▶; Table 1 ▶). A14 contains the ‘tip-associated domain’, which includes a flexible loop (H1-H2), but lacks the C-terminal helicase and RNAse D C-terminal (HRDC) domain present in all A14 counterparts, instead having a long flexible C-terminal tail. A43 consists of an N-terminal ‘tip domain’ and a C-terminal OB domain that is present in all its counterparts. The overall structural similarity of A14/A43 to its counterparts is surprising, as there is no sequence similarity between A14 and its counterparts and only 8% of the A43 residues are identical in Rpb7 (Table 1 ▶). Nevertheless, 78% of the Rpb7 residues have the same fold as in A43 (Kuhn et al., 2007 ▶; Table 1 ▶). A43 differs from Rpb7 by having flexible N- and C-terminal tails, an extended loop (C1-C2) within the C-terminal OB domain and a ten-residue insertion in the ‘tip loop’ (Kuhn et al., 2007 ▶). Thus, A14/A43 contains at least four very extended flexible regions (Fig. 1 ▶) which had to be identified and at least partially removed for crystallization.
Figure 1.
Crystal structure of yeast A14/A43. A43 and A14 are coloured blue and red, respectively. Disordered regions and regions which are not present in the protein variant are drawn to scale, assuming extended folds, as dashed lines. Adapted from Kuhn et al. (2007 ▶).
Table 1. Similarity of A14 and A43 to homologous and orthologous polymerase subunits.
| Pol I subunit | Homologous subunit | Sequence identity† (%) | Conserved fold‡ (%) | Sequence similarity§ (%) |
|---|---|---|---|---|
| A14 | Rpb4 (Pol II) | 4.5 | 25.0 | |
| C17 (Pol III) | 6.6 | 52.9 | ||
| RpoF (archeal Pol) | 3.7 | 76.6 | ||
| A14 (C. glabrata) | 25.6 | 31.4 | ||
| A14 (A. gossypii) | 36.5 | 52.6 | ||
| A14 (K. lactis) | 42.3 | 55.5 | ||
| A43 | Rpb7 (Pol II) | 8.0 | 78.4 | |
| C25 (Pol III) | 8.6 | 64.2 | ||
| RpoE (archeal Pol) | 6.1 | 69.5 | ||
| A43 (C. glabrata) | 62.9 | 73.6 | ||
| A43 (A. gossypii) | 55.5 | 69.0 | ||
| A43 (K. lactis) | 56.4 | 69.0 |
Number of amino-acid residues in A14 or A43 that are identical in the homologous polymerase subunit divided by the total number of residues in A14 or A43. For orthologues, the number of amino-acid residues in S. cerevisiae A14 or A43 that are identical in the orthologous polymerase subunit divided by the total number of residues in S. cerevisiae A14 or A43.
Number of amino-acid residues in the homologous polymerase subunit that have the same fold in the A14/A43 structure divided by the total number of residues in the homologous polymerase subunit. For A14, the number of amino-acid residues in the homologous polymerase subunit that have the same fold in the A14 structure divided by the number of residues of the tip-associated domain of the homologous polymerase subunit (HRDC domain excluded).
The number of amino-acid residues in S. cerevisiae A14 or A43 that are similar (e.g. hydrophobic, acidic or basic) in the orthologous polymerase subunit divided by the total number of residues in S. cerevisiae A14 or A43.
Here, we present our approach to obtaining diffraction-quality crystals of the A14/A43 subcomplex and an evaluation of this approach in light of the structure that is now available. We show how repetitive cycles of prediction and experiments were used to obtain a minimal A14/A43 variant that lacks most of the flexible residues and forms well ordered crystals. The described approach is superior to standard sequence analysis and structure prediction. For example, there were conflicting predictions for the A14 structure, one of which suggested that an HRDC domain is present (Meka et al., 2003 ▶) and another that it is absent (Peyroche et al., 2002 ▶). Our approach revealed the absence of the HRDC domain, which was key to obtaining crystals. The strategy presented may be adapted to comparable crystallogenesis challenges that will be faced more frequently in the future.
2. Experimental methods
2.1. Bioinformatic analysis
ClustalW (Chenna et al., 2003 ▶) was used to align A14 (GeneID 851734) and A43 (GeneID 854518) with their counterparts Rpb4, C17, RpoF and Rpb7, C25, RpoE, respectively. Alignments were manually edited using SEAVIEW (Galtier et al., 1996 ▶). In addition, A14 (137 residues, molecular weight 14.6 kDa) and A43 (326 residues, molecular weight 36.2 kDa) were aligned with related yeast orthologues (Candida glabrata, Candida albicans, Yarrowia lipolytica, Debaryomyces hanseii, Ashbya gossypii, Kluyveromyces lactis and Pichia stipitis). The secondary structures of A14 and A43 were predicted with PredictProtein (Rost et al., 2004 ▶). The sequences of A14 and A43 were submitted to the HHpred server (Soding et al., 2005 ▶) using default settings.
2.2. Cloning, expression, purification and limited proteolysis of protein variants
Cloning of all A14/A43 variants (Fig. 2 ▶) was carried out as described previously (Kuhn et al., 2007 ▶), except that genes were cloned sequentially into vector pET21b, resulting in a C-terminal hexahistidine tag on A43. Additional residues from the polylinker and tag sequence which remain on A43 were AAALEHHHHHH. All A14/A43 variants were expressed recombinantly (Maniatis et al., 1982 ▶) and purified as described by Kuhn et al. (2007 ▶), except for the thrombin-cleavage step and some individual changes, as follows. Full-length A14/A43 and variants A14/A43-1, A14/A43-2, A14/A43-4, A14/A43-5 and A14/A43-6 were purified by immobilized metal-affinity chromatography (IMAC) using as buffer 150 mM NaCl, 50 mM Tris pH 7.5, 5% glycerol, 10 mM β-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, 200 µM pepstatin and 60 µM leupeptin. Full-length A14/A43, A14/A43-2 and A14/A43-6 were further purified on a Mono Q 10/100 GL anion-exchange column (GE Healthcare), using as buffer 150 mM NaCl, 50 mM Tris pH 7.5, 5 mM DTT, and were subsequently applied onto various gel-filtration columns (Superose 6 HR 10/300, Superose 12 HR 10/300 and Superdex 75 FPLC; GE Healthcare). For limited proteolysis (Hubbard, 1998 ▶), the protein was diluted to a concentration of 1 mg ml−1. 100 µl protein solution was incubated with 1 µl chymotrypsin or trypsin (1 mg ml−1) and incubated for 1 h at 310 K. 15 µl samples were taken at different time points ranging from 1 to 60 min. The reaction was stopped by adding 5 µl of 4× SDS sample buffer (50 mM Tris pH 7.0, 14% 1,4-dithiothreitol, 10% glycerol, 1% β-mercaptoethanol, 0.1% bromophenol blue, 0.1% sodium lauryl sulfate) and incubated for 5 min at 358 K. Samples were analyzed by SDS–PAGE.
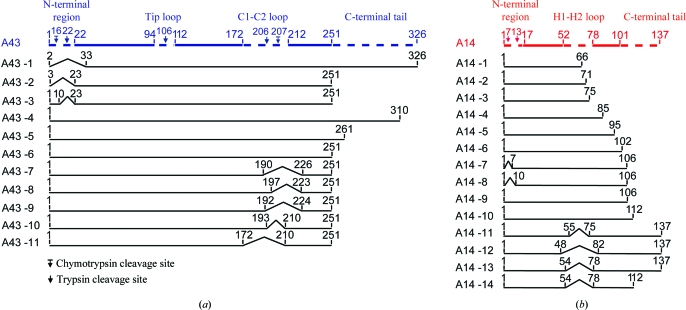
Figure 2.
Schematic representation of yeast A43 and A14 variants. The full-length proteins A14 and A43 are shown as coloured bars (red and blue, respectively). Disordered regions that are not present in the crystal structure are depicted as dashed lines. Arrows indicate proteolytic cleavage sites. Variants are numbered A43-1 to A43-11 for A43 and A14-1 to A14-14 for A14. All residue numbers correspond to residues present in the variant. The minimal A43 and A14 variants used for final crystallization are shown at the bottom (variants A43-11 and A14-14).
2.3. Crystallization and crystal screening
Initial crystallization experiments of all A14/A43 variants were performed at 293 K in sitting drops using the Hydra II Plus One crystallization robot (Thermo Fisher Scientific). Commercially available screening kits were used for initial screens (Qiagen, Jena Bioscience, Hampton Research). The A14/A43 variants were concentrated to 10–30 mg ml−1. Details of the crystallization of native and selenomethionine-derivatized A14-14/A43-11 can be found in Kuhn et al. (2007 ▶).
3. Results and discussion
3.1. Prediction of unstructured regions
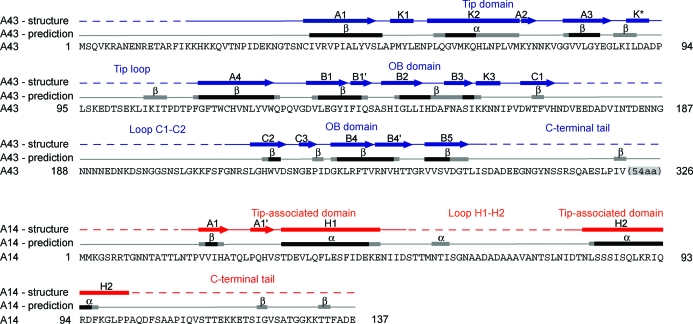
Recombinantly expressed full-length A14/A43 (§2) did not crystallize, indicating the presence of unstructured flexible regions within the complex. To aid in the design of protein variants that were suitable for crystallization, we used bioinformatics tools to predict the structured and unstructured regions in both subunits. Firstly, we predicted the secondary-structure elements in both subunit sequences using PredictProtein (Rost et al., 2004 ▶). Secondly, we prepared a sequence alignment with closely related yeast orthologues detected by BLAST (Altschul et al., 1997 ▶). This alignment was used to judge the reliability of the secondary-structure predictions, assuming that structured elements must be conserved among closely related species. Thirdly, a preliminary alignment of the A14 and A43 sequences was obtained by matching the predicted secondary-structure elements with the precisely located secondary-structure elements present in the crystal structures of the A14/A43 counterparts in other RNA polymerases. Since sequence homology was extremely weak (A43) or essentially absent (A14), the obtained alignments obviously contained errors, but they still allowed an initial prediction of the location of structured and unstructured regions. The final alignment of the secondary-structure elements observed in the A14/A43 crystal structure with predicted secondary-structure elements is shown in Fig. 4 ▶. The preliminary alignment between A43 and its counterparts in the Rpb7 family suggested that A43 contains a long flexible C-terminal tail and a disordered internal loop. However, the precise length of the tail and the exact position of the loop were unclear owing to a lack of conservation and possible misalignment in the C-terminal region. For A14, two generally possible alignments were obtained. The first comprised a C-terminal HRDC domain, consistent with one published model (Meka et al., 2003 ▶), yet not all of the helices required to form this domain were predicted. The second model postulated that the HRDC domain was absent and A14 contained extended flexible regions, consistent with another model (Peyroche et al., 2002 ▶).
Figure 4.
Comparison of secondary-structure elements observed in the A14/A43 crystal structure with predicted secondary-structure elements. Predicted secondary-structure elements (α-helices and β-strands) are shown above the amino-acid sequences of A43 and A14 as black or grey bars for elements predicted with high (9–5) or low (4–1) reliability index (§2). Secondary-structure elements taken from the crystal structure (Kuhn et al., 2007 ▶) are shown above the predicted secondary-structure elements (bars, α-helices; arrows, β-strands; lines, loops; dashed lines, disordered regions).
3.2. Identification of unstructured regions in A43
We tested the predictions of flexible regions in the A14/A43 heterodimer by preparing deletion variants of A14 and A43, examining their interaction after coexpression in Escherichia coli and assessing the solubility of the obtained subcomplexes. We first investigated the nature of a possible N-terminal tail in A43, since previously published data suggested that 32 N-terminal residues of A43 could be removed from the protein (Meka et al., 2003 ▶). However, our variant A14/A43-1, which also lacks 32 N-terminal residues, could only be expressed as a substoichiometric complex (Figs. 2 ▶ a and 3 ▶). The A14/A43 structure subsequently showed that residues 24–27 of A43 interact with residues 28–30 of A14. Limited proteolysis of full-length A14/A43 revealed two cleavage sites in the N-terminal region of A43 after residues 16 and 22 (Fig. 2 ▶ a). Variants A14/A43-2 and A14/A43-3, containing two different deletions before residue 23 at the N-terminus, could be purified as stable stoichiometric subcomplexes (Figs. 2 ▶ a and 3 ▶). Thus, the first 22 residues at the N-terminus can be removed without affecting the stability of the complex, while residues 23–32 were important for stable binding of A14 to A43. Owing to the remaining uncertainties we decided not to remove any residues at the N-terminus and instead focused on more extended loops in A43.
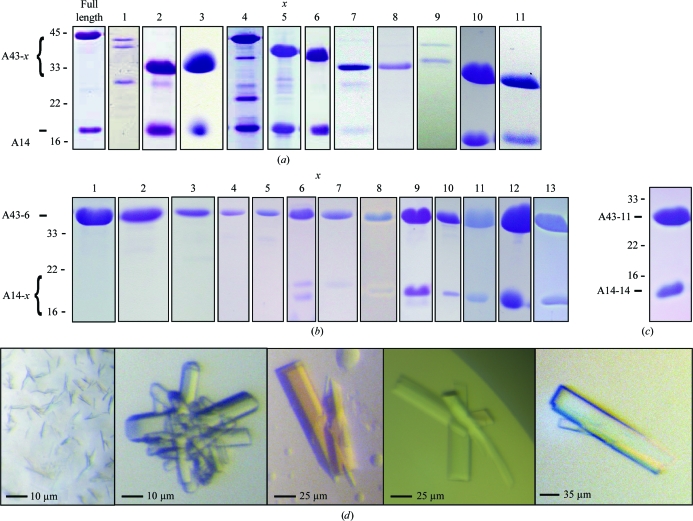
Figure 3.
Purification and crystallization of A14/A43 variants. (a) SDS–PAGE analysis (Coomassie staining) of A14 purification after coexpression of different A43 deletion variants A43-x (compare Fig. 2 ▶ a). (b) SDS–PAGE analysis (Coomassie staining) of A43-6 purification after coexpression of different A14 deletion variants A14-x (compare Fig. 2 ▶ b). (c) SDS–PAGE analysis (Coomassie staining) of the A14-14/A43-11 variant. (d) Crystals obtained of the A14-14/A43-11 variant. The protein was crystallized using as protein buffer 100 mM NaCl, 20 mM Tris–HCl pH 8.0 and 5 mM DTT. Reservoir solutions: small microcrystals, 200 mM sodium acetate, 30%(w/v) PEG 4000, 100 mM Tris–HCl pH 8.5 (left panel); intergrown crystals, 50 mM potassium phosphate, 20%(w/v) PEG 8000 (second panel from the left); larger crystals, 200 mM potassium acetate, 20%(w/v) PEG 3350 (third panel from the left); clusters of single crystals, 220 mM potassium acetate, 26%(w/v) PEG 3350 (fourth panel from the left); crystal obtained after microseeding, 300 mM potassium acetate, 20%(w/v) PEG 3350 (right panel).
Limited proteolysis of the A14/A43 subcomplex showed an additional cleavage site within the predicted ‘tip-loop’ region (Fig. 2 ▶ a). However, we did not create a deletion variant lacking the tip loop because it is involved in a key interaction with the core of Pol I. During purification of the A14/A43-1 variant, an additional protein variant was detected by SDS–PAGE. Edman sequencing revealed a truncated A43 variant starting at residue 81, preceded by an additional methionine residue. Analysis of the A43 nucleotide sequence revealed a Shine–Dalgarno sequence within the A43 open reading frame. To prevent expression of the truncated variant, two silent point mutations (nucleotide changes A228G and G243T) were inserted and were retained in all subsequent A14/A43 variants.
Consistent with the prediction that A43 contains a long flexible C-terminal tail, a 46-amino-acid C-terminal deletion variant (Peyroche et al., 2002 ▶) could be expressed without affecting the solubility or stoichiometry of the complex. Screening of variants that were shortened from the C-terminus demonstrated that up to 75 residues of A43 could be removed without affecting the solubility or stability of the subcomplex (variants A14/A43-4, A14/A43-5 and A14/A43-6; Figs. 2 ▶ a and 3 ▶). After the C-terminal tail had been successfully delineated and removed, variant A14/A43-6 was subjected to limited proteolysis to identify the location of the major loop. Comparison with Rpb7 revealed approximately 35 additional amino acids that were predicted to form a major loop in A43. In order to locate the loop, limited proteolysis experiments were performed, which revealed two cleavage sites after residues Lys206 and Phe207 of A43 (Fig. 2 ▶ a). However, variants A14/A43-7, A14/A43-8 and A14/A43-9, which contained different A43 loop deletions including the proteolytic cleavage sites, showed strongly impaired solubility (Figs. 2 ▶ a and 3 ▶), indicating that the loop was smaller. Indeed, a A43 variant lacking residues 194–209 (variant A14/A43-10) could be expressed as a soluble stochiometric heterodimer (Figs. 2 ▶ a and 3 ▶). The loop deletion could be extended to residues 173–209 (A14/A43-11; Figs. 2 ▶ a and 3 ▶). These results suggested that the loop was located between β-strands C1 and C2 within the OB domain (Figs. 1 ▶ and 2 ▶ a), as β-strand C1 was predicted to lie between residues 165 and 170, immediately before the loop deletion. Combining this loop deletion with the C-terminal truncation resulted in a variant that lacks residues 173–209 and 252–326 and was well expressed and soluble (variant A14/A43-11; Figs. 2 ▶ a and 3 ▶).
3.3. Identification of unstructured regions in A14
Next, we tested experimentally the two models for the A14 structure that differed by the presence or absence of a C-terminal HRDC domain. We checked for the presence of a HRDC domain by coexpressing A43 with A14 variants with various C-terminal deletions and assessed the solubility and stoichiometry of the A14/A43 heterodimer. Since in the counterpart structures subunit interaction does not rely on the HRDC domain, we expected that the HRDC domain could be removed without impairing the solubility of the subcomplex, as observed for the C17/25 subcomplex (Jasiak et al., 2006 ▶). Multiple C-terminal deletion variants were designed, lacking 71, 66 and 62 residues, respectively (variants A14-1/A43-6, A14-2/A43-6 and A14-3/A43-6; Figs. 2 ▶ b and 3 ▶), consistent with the previously predicted HRDC domain (Meka et al., 2003 ▶). Purification of the hexahistidine-tagged A43 did not result in copurification of the truncated A14 protein, indicating that the deletion variant lacked parts of the A43-binding domain. We therefore reduced the length of the A14 C-terminal deletion until a minimal stoichiometric A14/A43 subcomplex was obtained (A14-4/A43-6 to A14-10/A43-6; Figs. 2 ▶ b and 3 ▶). The A14-10/A43-6 complex could be purified at stoichiometric levels. This result argued against the presence of a HRDC domain in A14. Instead, the insolubility of the shorter A14 variants suggested that these contained an incomplete tip-associated domain.
We therefore tested the second model for the A14 structure, which suggested that a predicted α-helix between residues 82 and 96 corresponds to helix H2 of the tip-associated domain (Peyroche et al., 2002 ▶). This model proposed a major loop in A14 between helices H1 and H2, ranging approximately from residues 50 to 80. Consistent with this model, coexpression of several A14 variants with various loop deletions (variants A14-11/A43-6 to A14-13/A43-6; Figs. 2 ▶ b and 3 ▶) led to a minimal A14 variant which forms a soluble and stochiometric complex with the A43 variant (variant A14-14/A43-11; Figs. 2 ▶ and 3 ▶ c).
3.4. Additional changes to obtain diffraction-quality crystals
By combining a total of four precise deletions, two each in A14 and A43, we obtained a minimal A14/A43 variant (A14-14/A43-11; Figs. 2 ▶ and 3 ▶ c). This variant was purified and subjected to various crystallization screens, varying the protein concentration, protein buffer concentration and pH, reducing agents and additives. Although the variant was highly soluble, not sensitive to limited proteolysis and apparently contained stoichiometric amounts of subunits, only very small microcrystals and clusters of small crystals were obtained under varying conditions and using different crystallization methods (Fig. 3 ▶ d, left panel). We therefore changed the position of the affinity-purification tag from the C-terminus of A43 to the N-terminus of A14 and removed it by thrombin cleavage after affinity purification, finally leading to the formation of single protein crystals. However, over-nucleation resulted in crystals of medium size that were often clustered and intergrown (Fig. 3 ▶ d, middle three panels). Only very few crystals showed diffraction beyond 4 Å resolution. With the use of microseeding (Bergfors, 2003 ▶), we could reduce nucleation and obtain well shaped large single crystals when using 300 mM potassium acetate and 20%(w/v) PEG 3350 as reservoir solution. These crystals were suitable for data collection to high resolution and enabled structure determination (Fig. 3 ▶ d, right panel).
4. Conclusions
The removal of flexible protein regions has emerged as a general tool to achieve crystallization of proteins and their complexes. In the future, the number of crystallization projects hampered by multiple protein flexibilities will increase, in particular since there is a need to crystallize subcomplexes of larger assemblies that are subjected to hybrid analysis, combining EM of the entire assembly with X-ray crystallography of its subcomplexes. However, if more than one flexible region needs to be removed from a protein or a protein complex, the number of variants that have to be cloned, expressed, purified and subjected to crystallization trials becomes very large, as several variants for each deletion must be tested in all combinations. If four extended flexible regions exist, as in the example described here, only ten variants for each flexible region will lead to a total of 10 000 variants with different combinations of deletions. In addition, if only one flexible region is not predicted correctly, all variants will be insoluble and useless for the identification of a suitable variant. Furthermore, high-throughput techniques can be applied to delineate flexible terminal tails, but are not suited to finding and removing flexible internal loops, which is often crucial for crystallogenesis.
Here, we present an alternative approach to engineering a protein with multiple extended flexible regions such that it will form well ordered crystals. The novelty of this approach is that it successfully integrates structure prediction and structure-guided variant design and that it involves cycles of predicting flexible regions and experimental verification or falsification of the predictions. In particular, flexible regions are detected stepwise and also combined in a stepwise manner, rather than in the parallel high-throughput approach that is often followed in structural proteomics. This stepwise detection and removal of flexible regions dramatically reduces the number of variants to be made and tested. The key to success of this procedure is to iteratively improve the prediction of unstructured regions and then use the improved prediction to design new variants for additional experiments. As more and more protein structures become available, the likelihood of finding a structure of a protein that is distantly related to the target protein to be crystallized increases. As a consequence, the number of crystallization projects that are able to use this approach will increase in the future.
Acknowledgments
We thank Michela Bertero and other members of the Cramer laboratory for help. Part of this work was performed at the Swiss Light Source (SLS) at the Paul Scherrer Institut, Villigen, Switzerland and at beamline 14.3 of the Protein Structure Factory (PSF) at BESSY, Berlin, Germany. We thank Claude Pradervand and Clemens Schulze-Briese at SLS and Uwe Müller at BESSY. This research was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich SFB646, the EU Research Grant Network 3D Repertoire, The Nanoinitiative Munich NIM, the Elitenetzwerk Bayern IDK-NanoBioTechnology and the Fonds der Chemischen Industrie.
References
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res.25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Bergfors, T. (2003). J. Struct. Biol.142, 66–76. [DOI] [PubMed]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G. & Thompson, J. D. (2003). Nucleic Acids Res.31, 3497–3500. [DOI] [PMC free article] [PubMed]
- Galtier, N., Gouy, M. & Gautier, C. (1996). Comput. Appl. Biosci.12, 543–548. [DOI] [PubMed]
- Hu, P., Wu, S., Sun, Y., Yuan, C. C., Kobayashi, R., Myers, M. P. & Hernandez, N. (2002). Mol. Cell. Biol.22, 8044–8055. [DOI] [PMC free article] [PubMed]
- Hubbard, S. J. (1998). Biochim. Biophys. Acta, 1382, 191–206. [DOI] [PubMed]
- Jasiak, A. J., Armache, K. J., Martens, B., Jansen, R. P. & Cramer, P. (2006). Mol. Cell, 23, 71–81. [DOI] [PubMed]
- Kuhn, C. D., Geiger, S. R., Baumli, S., Gartmann, M., Gerber, J., Jennebach, S., Mielke, T., Tschochner, H., Beckmann, R. & Cramer, P. (2007). Cell, 131, 1260–1272. [DOI] [PubMed]
- Maniatis, T., Fritsch, E. F. & Sambrook, J. (1982). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
- Meka, H., Daoust, G., Arnvig, K. B., Werner, F., Brick, P. & Onesti, S. (2003). Nucleic Acids Res.31, 4391–4400. [DOI] [PMC free article] [PubMed]
- Peyroche, G., Levillain, E., Siaut, M., Callebaut, I., Schultz, P., Sentenac, A., Riva, M. & Carles, C. (2002). Proc. Natl Acad. Sci. USA, 99, 14670–14675. [DOI] [PMC free article] [PubMed]
- Rost, B., Yachdav, G. & Liu, J. (2004). Nucleic Acids Res.32, W321–W326. [DOI] [PMC free article] [PubMed]
- Sadhale, P. P. & Woychik, N. A. (1994). Mol. Cell. Biol.14, 6164–6170. [DOI] [PMC free article] [PubMed]
- Shpakovskii, G. V. & Shematorova, E. K. (1999). Bioorg. Khim.25, 791–796. [PubMed]
- Siaut, M., Zaros, C., Levivier, E., Ferri, M. L., Court, M., Werner, M., Callebaut, I., Thuriaux, P., Sentenac, A. & Conesa, C. (2003). Mol. Cell. Biol.23, 195–205. [DOI] [PMC free article] [PubMed]
- Soding, J., Biegert, A. & Lupas, A. N. (2005). Nucleic Acids Res.33, W244–W248. [DOI] [PMC free article] [PubMed]
- Todone, F., Brick, P., Werner, F., Weinzierl, R. O. & Onesti, S. (2001). Mol. Cell, 8, 1137–1143. [DOI] [PubMed]