Crystallization and X-ray diffraction data collection of the Fab fragment of the monoclonal antibody WO2 in the absence or presence of amyloid β peptides associated with Alzheimer’s disease are reported.
Keywords: Aβ peptides, Alzheimer’s disease, antibody Fab fragments, immunotherapy
Abstract
The murine monoclonal antibody WO2 specifically binds the N-terminal region of the amyloid β peptide (Aβ) associated with Alzheimer’s disease. This region of Aβ has been shown to be the immunodominant B-cell epitope of the peptide and hence is considered to be a basis for the development of immunotherapeutic strategies against this prevalent cause of dementia. Structural studies have been undertaken in order to characterize the molecular basis for antibody recognition of this important epitope. Here, details of the crystallization and X-ray analysis of the Fab fragment of the unliganded WO2 antibody in two crystal forms and of the complexes that it forms with the truncated Aβ peptides Aβ1–16 and Aβ1–28 are presented. These crystals were all obtained using the hanging-drop vapour-diffusion method at 295 K. Crystals of WO2 Fab were grown in polyethylene glycol solutions containing ZnSO4; they belonged to the orthorhombic space group P212121 and diffracted to 1.6 Å resolution. The complexes of WO2 Fab with either Aβ1–16 or Aβ1–28 were cocrystallized from polyethylene glycol solutions. These two complex crystals grew in the same space group, P212121, and diffracted to 1.6 Å resolution. A second crystal form of WO2 Fab was grown in the presence of the sparingly soluble Aβ1–42 in PEG 550 MME. This second form belonged to space group P21 and diffracted to 1.9 Å resolution.
1. Introduction
Alzheimer’s disease (AD) is a progressive age-related neurodegenerative disorder and is the fourth leading cause of death in the developed world after heart disease, cancer and stroke. AD represents the most common form of dementia, accounting for between 50 and 70% of all cases. The disease is characterized by the presence of extracellular amyloid plaques in the brain (Selkoe, 2002 ▶). The major component of these plaques are aggregates of a 4 kDa peptide called amyloid β (Aβ). Aβ peptides are proteolytically derived from the membrane-bound amyloid precursor protein (APP; Kang et al., 1987 ▶) by sequential cleavage by β-site APP-cleaving enzyme (BACE) and the integral membrane-protein complex γ-secretase (Tabaton & Tamagno, 2007 ▶).
The Aβ peptide can exist as a soluble prefibrillar form that can compromise neuronal function and trigger cell death (Cappai & Barnham, 2007 ▶) and upon binding metal ions such as copper, zinc and iron can form either insoluble aggregates (Atwood et al., 1998 ▶) or oligomers that are capable of penetrating cell membranes (Lau et al., 2006 ▶). Aβ peptides (1–40 and 1–42) are sparingly soluble and prone to aggregation, particularly in the presence of metals such as copper and zinc. Aβ peptides are also pleiomorphic in that they can adopt different conformations depending on their environment. These features have complicated the study of the structural biology of Aβ. Nuclear magnetic resonance spectroscopists have used organic solvents and detergents to great effect to study membrane-bound forms of the peptide as well as truncated forms of Aβ in various media (Watson et al., 1998 ▶; Poulsen et al., 2000 ▶; D’Ursi et al., 2004 ▶). Aβ1–16 and Aβ1–28 are often studied as they are soluble and contain the metal-binding domain of Aβ and the residues involved in oligomeric assembly of Aβ.
The monoclonal antibody WO2 (Ida et al., 1996 ▶) recognizes the N-terminus of the Aβ peptide. There is great interest in this region as it constitutes the immunodominant B-cell epitope of Aβ and is thus a leading target for the development of anti-Aβ immunotherapies (Hamaguchi et al., 2006 ▶). Our approach has been to characterize WO2 binding to the immunodominant B-cell epitope and to exploit this Fab to crystallize longer forms of the peptide (residues 1–28 and 1–42).
Here, we report the successful crystallization of unliganded WO2 Fab in two space groups and of complexes of WO2 Fab with Aβ1–16 and Aβ1–28, which have led to their successful structure determination as reported elsewhere (Miles et al., 2008 ▶).
2. Experimental procedures and results
2.1. Purification, Fab production and complex formation
22 harvests (25 ml each) from the WO2 hybridoma (Ida et al., 1996 ▶) were obtained from the Walter and Eliza Hall Institute Monoclonal Antibody Facility, Victoria, Australia. The cells were cultured in hybridoma serum-free media (Gibco, Australia) supplemented with interleukin-6 at 310 K in an atmosphere of 8%(v/v) CO2. Each harvest was centrifuged (4000g for 10 min) to remove cell debris and filtered (0.22 µm). WO2 IgG was purified using a Protein G-Sepharose (1.0 × 5 cm) column (GE Healthcare, Uppsala, Sweden) equilibrated in 20 mM sodium phosphate buffer pH 7.2 containing 150 mM NaCl (PBS). The column was washed with PBS, bound WO2 IgG was eluted with 100 mM glycine pH 3.0 and the fraction was neutralized in 1.0 M Tris–HCl pH 8.0. The purified IgG fractions were pooled, dialyzed against PBS (8000 Da molecular-weight cutoff dialysis membrane; SpectraPor, Gardena, California, USA) and stored at 253 K.
WO2 Fab fragments were prepared using the Immunopure Fab Preparation kit (Pierce, Rockford, Illinois, USA). Briefly, 40 mg WO2 IgG was digested with 2 ml immobilized papain slurry for 16 h at 293 K. The immobilized papain was removed by centrifugation (5000g for 15 min) and the supernatant containing the digest was loaded onto a Protein-A agarose (1.0 × 3 cm) column previously equilibrated with PBS. Fab fragments eluted in the column flowthrough fractions using PBS. Fractions containing the Fab fragments were pooled, concentrated using a Centriprep-10 centrifugal concentrator (10 000 Da molecular-weight cutoff; Millipore, Bedford, Massachusetts, USA) and further purified on a Superdex-200 HR (1.0 × 30 cm) gel-filtration column (GE Healthcare, Uppsala, Sweden) equilibrated in PBS. The purified Fab fragments were concentrated and buffer-exchanged into 20 mM HEPES pH 7.0. The digestion and purification steps were monitored by SDS–PAGE.
Aβ peptides corresponding to residues 1–16, 1–28 and 1–42 of the Aβ sequence (1-DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA-42) were synthesized using standard F-moc chemistry using side-chain and terminus-protected amino acids (AusPEP, Victoria, Australia). The peptides were cleaved from the resin, purified by reverse-phase HPLC and characterized by MALDI–TOF mass spectrometry, amino-acid analysis and HPLC.
Aβ peptides 1–16 and 1–28 were dissolved in PBS and incubated separately with WO2 Fab in a 2:1 molar excess of peptide overnight at 277 K. Excess peptide was removed from the Fab–peptide complex by either by gel-filtration chromatography (Superdex 75 HR 1.0 × 30 cm column equilibrated in PBS; GE Healthcare, Uppsala, Sweden) in the case of Aβ1–16 or dialysis for Aβ1–28. Both complexes were dialyzed against 10 mM MES pH 6.8, concentrated to 4 mg ml−1 and stored at 277 K until required for crystallization experiments. Complex formation was monitored by SDS–PAGE using Tricine gels.
Aβ1–42 was solubilized in 6 M urea, 20 mM Tris–HCl for 2 h at 277 K and then added to WO2 Fab solution in a 1:1 ratio and incubated overnight at 293 K. The resulting complex was extensively dialyzed against 10 mM HEPES pH 7.0 to remove urea and then concentrated to 5 mg ml−1 and stored at 277 K prior to setting up crystallization trials.
2.2. Protein crystallization
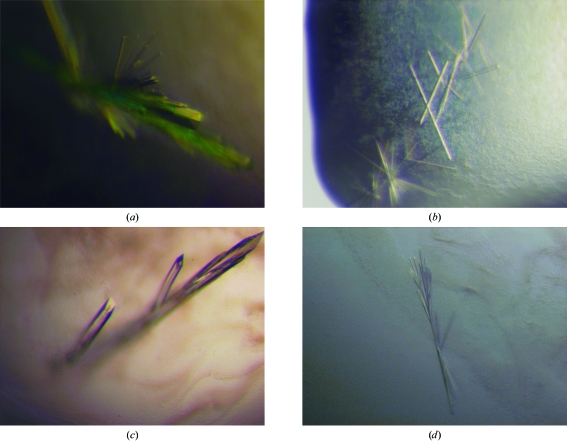
Crystallization trials of the WO2 Fab and of the WO2 Fab in complex with either Aβ1–16, Aβ1–28 or Aβ1–42 were performed at 295 K using the hanging-drop vapour-diffusion method by mixing 1–2 µl protein solution with an equal volume of reservoir solution and equilibrating against 0.5–1.0 ml reservoir solution. Initial crystallization conditions were established using the screens described by Jancarik & Kim (1991 ▶) and Cudney et al. (1994 ▶). Initial screening resulted in small needle-like clusters of crystals for the WO2 Fab and all the Fab–peptide complexes using different molecular-weight PEGs as the precipitants at a range of pH values, which were subsequently optimized by a combination of streak-seeding and grid screens around the pH and PEG concentrations (Table 1 ▶). Some of this work involved use of the Bio21 Collaborative Crystallization Centre (http://www.csiro.au/c3/). Crystals of WO2 Fab were grown in 100 mM MES pH 6.7, 20%(v/v) PEG 550 MME and 10 mM ZnSO4 (Fig. 1 ▶ a). The inclusion of ZnSO4 was essential for crystallization. Crystals of the WO2 Fab complexed with Aβ1–16 were grown in 100 mM MES pH 6.5 containing 25%(v/v) PEG 400 (Fig. 1 ▶ b). In contrast, crystals of the WO2 Fab complexed with Aβ1–28 were grown in 100 mM bicine pH 9.0 containing 20%(w/v) PEG 8000 and 200 mM MgCl2 (Fig. 1 ▶ c). Crystals of the latter complex were dissolved and analyzed by mass spectrometry on an Agilent 6510 Q-TOF LC/MS (Bio21 Institute, Melbourne). MS/MS confirmed that the observed ions of the Aβ1–28 peptide were of the expected length with no sign of degradation.
Table 1. Crystal data and X-ray diffraction data-collection statistics.
Values in parentheses are for the highest resolution bin (approximate interval of 0.1 Å).
| Crystal | WO2 Fab, form A | WO2 Fab–Aβ1–16 | WO2 Fab–Aβ1–28 | WO2 Fab, form B |
|---|---|---|---|---|
| Crystallization conditions | 100 mM MES pH 6.7, 20%(v/v) PEG 550 MME, 10 mM ZnSO4 | 100 mM MES pH 6.5, 25%(v/v) PEG 400 | 100 mM bicine pH 9.0, 20%(w/v) PEG 8000, 200 mM MgCl2 | 100 mM MES pH 6.5, 25%(v/v) PEG 550 MME, Aβ1–42 (equimolar) |
| Cryoprotectant | 100 mM MES pH 6.7, 30%(v/v) PEG 550 MME, 10 mM ZnSO4 | Not required | 100 mM bicine pH 9.0, 25%(v/v) PEG 8000, 200 mM MgCl2 | 100 mM MES pH 6.5, 25%(v/v) PEG 550 MME, 20%(v/v) glycerol |
| X-ray source | 14-BMC, APS | 14-BM-C, APS | Rotating anode | Rotating anode |
| X-ray wavelength (Å) | 0.99988 | 0.99988 | Cu Kα | Cu Kα |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Space group | P212121 | P212121 | P212121 | P21 |
| Unit-cell parameters | ||||
| a (Å) | 52.4 | 51.7 | 51.7 | 40.4 |
| b (Å) | 90.5 | 66.6 | 66.4 | 111.2 |
| c (Å) | 123.3 | 115.2 | 115.5 | 53.1 |
| β (°) | 103.8 | |||
| Maximum resolution (Å) | 1.6 | 1.6 | 1.6 | 1.9 |
| Total observations | 328859 | 273773 | 248683 | 100646 |
| Unique reflections used | 76778 | 53462 | 53254 | 33358 |
| Redundancy | 4.3 (3.2) | 5.1 (3.2) | 4.7 (3.1) | 3.0 (2.0) |
| Data completeness (%) | 98.0 (87.8) | 99.2 (92.2) | 99.7 (100) | 93.3 (68.1) |
| Rmerge† (%) | 10.1 (28.2) | 11.8 (26.3) | 7.3 (30.0) | 5.8 (16.4) |
| 〈I/σ(I)〉 | 16.6 (2.3) | 14.3 (2.7) | 11.4 (3.8) | 14.4 (4.8) |
R
merge = 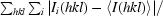
 , where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
, where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
Figure 1.
Crystals of WO2 Fab (a) by itself, (b) grown in the presence of the Aβ1–16 peptide, (c) grown in the presence of the Aβ1–28 peptide and (d) grown in the presence of the Aβ1–42 peptide.
A second crystal form of the WO2 Fab was grown in the presence of the sparingly soluble Aβ1–42 in 100 mM MES pH 6.5 containing 25%(v/v) PEG 550 MME (Fig. 1 ▶ d). Subsequent crystal structure determination showed no evidence of the Aβ1–42 peptide bound to the protein (Miles et al., 2008 ▶). Hence, we refer to this crystal form as WO2 Fab form B in order to distinguish it from the initial WO2 Fab crystals, which we call form A (Table 1 ▶).
2.3. Data collection and preliminary X-ray analysis
Complete X-ray diffraction data sets were collected from single crystals of the WO2 Fab and of the WO2 Fab in complex with each of the Aβ peptides flash-cooled to 100 K. Synchrotron data were indexed with DENZO and integrated with SCALEPACK (Otwinowski & Minor, 1997 ▶), while all other data were indexed with MOSFLM (Leslie, 1992 ▶) and integrated using SCALA (Collaborative Computational Project, Number 4, 1994 ▶). Data-collection statistics are shown in Table 1 ▶.
Diffraction data from crystals of the WO2 Fab and of the WO2 Fab complexed with Aβ1–16 were collected on the BioCARS beamline 14-BM-C (Advanced Photon Source, Chicago, USA) using a MAR Research CCD 165 detector. The data set from the WO2 Fab crystals extended to 1.6 Å resolution and the crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 52.4, b = 90.5, c = 123.3 Å (form A in Table 1 ▶). Crystals of the WO2 Fab–Aβ1–16 complex also diffracted to 1.6 Å resolution and belonged to the same space group P212121, but had different unit-cell parameters of a = 51.7, b = 66.6, c = 115.21 Å. Diffraction data from crystals of the complexes of the WO2 Fab with Aβ1–28 and with Aβ1–42 were collected in-house on an R-AXIS IV++ image-plate detector using Cu Kα X-rays from a Rigaku MicroMax-007 HF rotating-anode X-ray generator. The crystals of the WO2 Fab–Aβ1–28 complex diffracted to 1.6 Å and belonged to the same space group P212121, with almost identical unit-cell parameters to those of the WO2 Fab–Aβ1–16 complex crystals: a = 51.7, b = 66.4, c = 115.3 Å. Crystals of WO2 Fab grown in the presence of the Aβ1–42 peptide were found to belong to a different space group to the other crystal forms, P21, with unit-cell parameters a = 40.4, b = 111.2, c = 53.1 Å, β = 103.8 °. These crystals diffracted to 1.9 Å resolution (form B in Table 1 ▶).
The structures of the WO2 Fab and of its complexes with the Aβ peptides have been determined using molecular replacement from the data reported here. Details have been published elsewhere (Miles et al., 2008 ▶).
Acknowledgments
This work was supported in part by a grant from the National Health and Medical Research Council of Australia (NHMRC) to MWP. This work, including the use of the BioCARS sector, was also supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Facilities Program. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Sciences, Office of Energy Research. We thank Dr Harry Tong and the staff at BioCARS as well as Dr Julian Adams for their help with data collection during our visit to the Advanced Photon Source. DBA is an Australian Postgraduate Award Scholar and a recipient of a St Vincent’s Institute Foundation Scholarship sponsored by Colin North and Major Engineering, RC and KJB are NHMRC Senior Research Fellows, MWP is an Australian Research Council Federation Fellow and NHMRC Honorary Fellow and WJM was a NHMRC Industry Fellow.
References
- Atwood, C. S., Moir, R. D., Huang, X., Scarpa, R. C., Bacarra, N. M., Romano, D. M., Hartshorn, M. A., Tanzi, R. E. & Bush, A. I. (1998). J. Biol. Chem.273, 12817–12826. [DOI] [PubMed]
- Cappai, R. & Barnham, K. J. (2007). Future Neurol.2, 397–409.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cudney, R., Patel, S., Weisgraber, K., Newhouse, Y. & McPherson, A. (1994). Acta Cryst. D50, 414–423. [DOI] [PubMed]
- D’Ursi, A. M., Armenante, M. R., Guerrini, R., Salvadori, S., Sorrentino, G. & Picone, D. (2004). J. Med. Chem.47, 4231–4238. [DOI] [PubMed]
- Hamaguchi, T., Ono, K. & Yamada, M. (2006). Cell. Mol. Life Sci.63, 1538–1552. [DOI] [PMC free article] [PubMed]
- Kang, J., Lemaire, H. G., Unterbeck, A., Salbaum, J. M., Masters, C. L., Grzeschik, K.-H., Multhaup, G., Beyreuther, K. & Muller-Hill, B. (1987). Nature (London), 325, 733–736. [DOI] [PubMed]
- Ida, N., Hartmann, T., Pantel, J., Schroder, J., Zerfass, R., Forstl, H., Sandbrink, R., Masters, C. L. & Beyreuther, K. (1996). J. Biol. Chem.271, 22908–22914. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411.
- Lau, T. L., Ambroggio, E. E., Tew, D. J., Cappai, R., Masters, C. L., Fidelio, G. D., Barnham, K. J. & Separovic, F. (2006). J. Mol. Biol.356, 759–770. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Miles, L. A., Wun, K. S., Crespi, G. A. N., Fodero-Tavoletti, M. T., Galatis, D., Bagley, C. J., Beyreuther, K., Masters, C. L., Cappai, R., McKinstry, W. J., Barnham, K. J. & Parker, M. W. (2008). J. Mol. Biol.377, 181–192. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Poulsen, S. A., Watson, A. A., Fairlie, D. P. & Craik, D. J. (2000). J. Struct. Biol.130, 142–152. [DOI] [PubMed]
- Selkoe, D. J. (2002). Science, 298, 789–791. [DOI] [PubMed]
- Tabaton, M. & Tamagno, E. (2007). Cell. Mol. Life Sci.64, 2211–2218. [DOI] [PMC free article] [PubMed]
- Watson, A. A., Fairlie, D. P. & Craik, D. J. (1998). Biochemistry, 37, 12700–12706. [DOI] [PubMed]