A periplasmic membrane-fusion protein MacA from Actinobacillus actinomycetemcomitans, an essential component of the multidrug efflux pump in Gram-negative bacteria, was crystallized.
Keywords: membrane-fusion proteins, multidrug efflux pumps, ABC-type transporters, Gram-negative bacteria
Abstract
Periplasmic membrane-fusion proteins (MFPs) are an essential component of the multidrug efflux pump in Gram-negative bacteria. They play a crucial role in bridging the outer membrane porin TolC and two distinct types of inner membrane transporters. The MFP MacA bridges the inner membrane ABC-type multidrug transporter MacB and the outer membrane porin TolC. MacA from the pathogenic bacterium Actinobacillus actinomycetemcomitans was expressed in Escherichia coli B834 (DE3) and the recombinant protein was purified using Ni–NTA affinity, Q anion-exchange and gel-filtration chromatography. The purified MacA protein was crystallized using the vapour-diffusion method. A MAD diffraction data set was collected to a resolution of 3.0 Å at 100 K. The crystal belongs to space group P622, with unit-cell parameters a = b = 109.2, c = 255.4 Å, α = β = 90, γ = 120°, and contains one molecule in the asymmetric unit.
1. Introduction
The multidrug efflux pumps of Gram-negative bacteria cause intrinsic drug tolerance by expelling a wide range of completely unrelated chemotherapeutic drugs (Lewis, 2000 ▶). These pumps consist of an inner membrane transporter, a multifunctional outer membrane porin (TolC) and a periplasmic membrane-fusion protein (MFP) (Lewis, 2000 ▶). The MFPs play a crucial role in bridging the outer membrane TolC and the inner membrane transporter. The inner membrane transporters consist of two structurally unrelated families (Zgurskaya & Nikaido, 1999 ▶): ATP-binding-cassette-type (ABC-type) transporters and resistance-nodulation-division-type (RND-type) transporters. Since ABC-type transporters differ structurally from RND-type transporters, the MFPs associated with ABC-type transporters should show unique features in their three-dimensional structures. Whereas Escherichia coli MacA is associated with the ABC-type transporter MacB (Tikhonova et al., 2007 ▶), E. coli AcrA and Pseudomonas aeruginosa MexA are linked with the RND-type transporters AcrB and MexB, respectively (Zgurskaya & Nikaido, 1999 ▶). To date, crystal structures of MFPs associated with RND-type transporters have been determined, revealing that the MFPs consist of three linearly arranged domains (Mikolosko et al., 2006 ▶; Higgins et al., 2004 ▶; Akama et al., 2004 ▶), but no crystal structure of an MFP associated with an ABC-type transporter has been determined. Additionally, the structure of the active form of MFPs remains unknown; all the available crystal structures failed to provide the functionally relevant forms as MFPs operate in vivo as oligomers (Eswaran et al., 2004 ▶).
Actinobacillus actinomycetemcomitans is a pathogenic bacterium associated with periodontitis, namely acute juvenile periodontitis and chronic adult periodontitis (Meyer & Fives-Taylor, 1997 ▶; Zambon, 1985 ▶). It was recently discovered that the MacA–MacB–TdeA complex is used to pump out macrolide antibiotics in A. actinomycetemcomitans, in which TdeA, a homologue of E. coli TolC, plays the role of the outer membrane porin (Crosby & Kachlany, 2007 ▶). Here, we report the crystallization and preliminary X-ray analysis of MacA from A. actinomycetemcomitans.
2. Materials and methods
2.1. DNA construction and protein expression
A DNA fragment encoding A. actinomycetemcomitans MacA (residues 29–394) was amplified from the genomic DNA library of A. actinomycetemcomitans using the polymerase chain reaction. The DNA fragment was inserted into the NcoI and XhoI sites of the pPROEX-HTA (Invitrogen, USA) vector, which contains a hexahistidine tag (Met-Ser-Tyr-Tyr-His-His-His-His-His-His), a spacer region (Asp-Tyr-Asp-Ile-Pro-Thr-Thr) and a TEV protease cleavage site (Glu-Asn-Leu-Tyr-Phe-Gln) at the N-terminus. The resulting protein contained three additional amino acids (Gly-His-Met) between the TEV protease cleavage site and the mature protein as a cloning artifact. The recombinant MacA protein was expressed in E. coli strain B834 (DE3) using M9 medium supplemented with an amino-acid mixture containing l-(+)-selenomethinonine and 50 µg ml−1 ampicillin at 310 K until the OD600 reached 0.5. Protein expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation at 5000g for 15 min at 277 K.
2.2. Protein purification
The harvested cells were suspended in a lysis buffer consisting of 20 mM Tris pH 8.0 and 150 mM NaCl and disrupted by sonication. The lysate was centrifuged at 45 000g for 30 min at 277 K. The resulting supernatant was loaded onto Ni–NTA agarose resin pre-equilibrated with lysis buffer. The resin was washed with lysis buffer supplemented with 20 mM imidazole and then eluted with lysis buffer supplemented with 200 mM imidazole. The fractions containing the MacA protein were pooled and β-mercaptoethanol was added to a final concentration of 10 mM. This solution was incubated with recombinant TEV protease overnight at 277 K to remove the hexahistidine tag. The reaction mixture was subsequently loaded onto a Q anion-exchange column (HiTrap-Q; GE Healthcare, USA) for further purification and proteins were eluted from the column using a 0–1 M NaCl gradient in 20 mM Tris buffer pH 8.0. The collected fractions containing the MacA protein were pooled, concentrated and separated on a HiLoad Superdex 200 gel-filtration column (GE Healthcare, USA) pre-equilibrated with lysis buffer. During purification, the presence of the protein was confirmed by SDS–PAGE. The purified protein was concentrated to 20 mg ml−1 in 20 mM Tris buffer pH 8.0 containing 150 mM NaCl and stored frozen at 193 K until use.
2.3. Crystallization
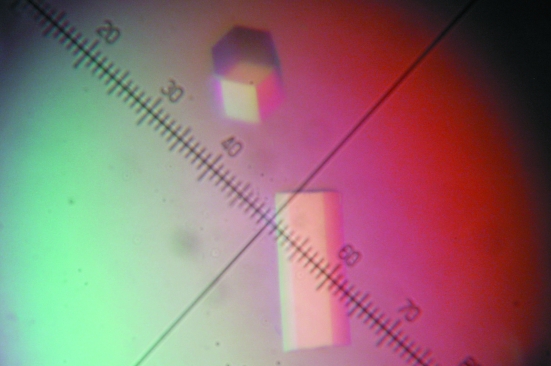
Crystals of recombinant MacA protein labelled with selenomethionine were obtained by the vapour-diffusion technique at 277 K. Initial crystallization screening was performed using the sitting-drop method with Crystal Screen HT, a high-throughput sparse-matrix screening kit (Hampton Research, USA). The crystallization conditions were optimized to produce shiny single crystals (Fig. 1 ▶) in droplets consisting of 1 µl protein solution (20 mg ml−1 protein in 20 mM Tris pH 8.0 buffer containing 150 mM NaCl) and 1 µl of a precipitant solution consisting of 0.01 M nickel chloride, 0.1 M Tris–HCl pH 8.5 and 1 M lithium sulfate. The droplets were equilibrated by the hanging-drop vapour-diffusion method against 1 ml precipitant solution at 277 K for one month.
Figure 1.
Crystals of selenomethionine-substituted MacA. Approximate dimensions are 0.3 × 0.1 × 0.1 mm. Eight divisions of the ruler represent 0.1 mm.
2.4. Crystallographic data collection
For X-ray data collection, a single crystal was gradually transferred into a cryoprotectant solution consisting of 0.01 M nickel chloride, 0.1 M MES pH 6.5, saturated lithium sulfate and 25% glycerol and then incubated overnight at 277 K. A multiwavelength anomalous dispersion (MAD) data set was collected on beamline 6C at Pohang Accelerator Laboratory using a Quantum 210 CCD detector (ADSC) at 100 K. Three sets of 180 images (0–180°) were obtained using a 1° oscillation width and 30 s exposure time at the selenium peak (0.9794 Å), edge (0.9796 Å) and remote (1.1271 Å) wavelengths without an inverse-beam experiment. The diffraction data were processed and scaled with the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
MacA is known to anchor to the bacterial inner membrane via the noncleavable N-terminal signal sequence (residues 1–28; Tikhonova et al., 2007 ▶). The signal sequence was removed from the full-length protein during the creation of the DNA construct for protein expression, which enabled soluble expression of the protein.
The crystal initially diffracted X-rays to only 7–8 Å resolution, although the crystals were large and shiny. We found that diffraction improved significantly when the crystals were incubated in a cryoprotectant solution saturated with the major precipitant at a different pH. From the diffraction data, the crystal belongs to space group P622, with unit-cell parameters a = b = 109.2, c = 255.4 Å. In particular, analysis of diffraction along the l axis clearly demonstrates that the c axis of the crystal is a rotational axis and not a screw axis. The diffraction data set had a resolution range of 50–3.0 Å with 91.9% completeness and an R merge of 0.06 at the remote wavelength (Table 1 ▶).
Table 1. Diffraction statistics.
(a).
Data collection. Values in parentheses refer to the highest resolution shell.
| Data set | Peak | Edge | Remote |
|---|---|---|---|
| X-ray source | Beamline 6C, Pohang Accelerator Laboratory | ||
| Wavelength (Å) | 0.9794 | 0.9796 | 1.1271 |
| Resolution (Å) | 50–3.2 (3.31–3.2) | 50–3.2 (3.31–3.2) | 50–3.0 (3.11–3.0) |
| Unit-cell parameters (Å, °) | a = b = 109.2, c = 255.4, α = β = 90, γ = 120 | ||
| Completeness (%) | 92.0 (71.5) | 91.8 (73.0) | 91.9 (77.3) |
| Rmerge† | 0.077 (0.265) | 0.075 (0.267) | 0.061 (0.183) |
| Redundancy | 7.0 (4.2) | 7.0 (4.3) | 7.7 (4.6) |
| Average I/σ(I) | 15.8 (2.4) | 16.4 (2.5) | 29.4 (3.6) |
(b).
Cell content analysis.
| No. of molecules per ASU | 1 | 2 | 3 |
|---|---|---|---|
| VM (Å3 Da−1) | 5.22 | 2.62 | 1.75 |
| Solvent content (%) | 76 | 53 | 30 |
R
merge = 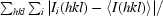
 , where I
i(hkl) and 〈I(hkl)〉 are the observed intensity and the mean intensity of related reflections, respectively.
, where I
i(hkl) and 〈I(hkl)〉 are the observed intensity and the mean intensity of related reflections, respectively.
In order to solve the structure, molecular replacement was attempted with the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶) using the E. coli AcrA (25% sequence identity) and P. aeruginosa MexA (24% sequence identity) structures as search models, but this method was not successful. Therefore, we attempted to use the MAD method to solve the phase problem. We could find three selenium sites using the program SOLVE (Terwilliger & Berendzen, 1999 ▶) using only 20–4.3 Å resolution data. Subsequently, solvent flattening was performed using the program RESOLVE (Terwilliger & Berendzen, 1999 ▶), but the experimental map produced low-resolution electron density that was insufficient for manual tracing. Candidates for the number of molecules per asymmetric unit and the corresponding Matthews coefficients (Matthews, 1968 ▶) and solvent contents were calculated and are listed in Table 1 ▶. However, the electron-density map indicated that one molecule is present in the asymmetric unit of the crystal, yielding an unusually high solvent content (76%). Attempts to determine the structure of the MacA protein from this data set are in progress; at the same time, we are optimizing the crystallization conditions and cryoprotectant solution in an attempt to obtain a higher resolution data set.
Acknowledgments
We thank Kihyun Nam at Korea University (Seoul, Korea) and Kyung-Jin Kim at Pohang Accelerator Laboratory (Pohang, Korea) for assistance during data collection. This study made use of beamline 6C at Pohang Accelerator Laboratory (Pohang, Korea). This work was supported for two years by a Pusan National University Research Grant.
References
- Akama, H., Matsuura, T., Kashiwagi, S., Yoneyama, H., Narita, S., Tsukihara, T., Nakagawa, A. & Nakae, T. (2004). J. Biol. Chem.279, 25939–25942. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Crosby, J. A. & Kachlany, S. C. (2007). Gene, 388, 83–92. [DOI] [PMC free article] [PubMed]
- Eswaran, J., Koronakis, E., Higgins, M. K., Hughes, C. & Koronakis, V. (2004). Curr. Opin. Struct. Biol.14, 741–747. [DOI] [PubMed]
- Higgins, M. K., Bokma, E., Koronakis, E., Hughes, C. & Koronakis, V. (2004). Proc. Natl Acad. Sci. USA, 101, 9994–9999. [DOI] [PMC free article] [PubMed]
- Lewis, K. (2000). Curr. Biol.10, R678–R681. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Meyer, D. H. & Fives-Taylor, P. M. (1997). Trends Microbiol.5, 224–228. [DOI] [PubMed]
- Mikolosko, J., Bobyk, K., Zgurskaya, H. I. & Ghosh, P. (2006). Structure, 14, 577–587. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Tikhonova, E. B., Devroy, V. K., Lau, S. Y. & Zgurskaya, H. I. (2007). Mol. Microbiol.63, 895–910. [DOI] [PubMed]
- Zambon, J. J. (1985). J. Clin. Periodontol.12, 1–20. [DOI] [PubMed]
- Zgurskaya, H. I. & Nikaido, H. (1999). J. Mol. Biol.285, 409–420. [DOI] [PubMed]