Abstract
A human fibroblast cDNA expression library was screened for cDNA clones giving rise to flat colonies when transfected into v-Ki-ras-transformed NIH 3T3 cells. One such gene, RECK, encodes a membrane-anchored glycoprotein of about 110 kDa with multiple epidermal growth factor-like repeats and serine-protease inhibitor-like domains. While RECK mRNA is expressed in various human tissues and untransformed cells, it is undetectable in tumor-derived cell lines and oncogenically transformed cells. Restored expression of RECK in malignant cells resulted in suppression of invasive activity with concomitant decrease in the secretion of matrix metalloproteinase-9 (MMP-9), a key enzyme involved in tumor invasion and metastasis. Moreover, purified RECK protein was found to bind to, and inhibit the proteolytic activity of, MMP-9. Thus, RECK may link oncogenic signals to tumor invasion and metastasis.
Keywords: Ras signaling/transformation/metastasis/protease inhibitor
Mutations of ras protooncogenes are found in a large variety of human tumors (1). It has been well established that Ras proteins are essential components in various intracellular signaling pathways involved in regulating gene expression and several other aspects of cellular behavior (2). Therefore, it is now important to find targets for these signals relevant to the expression of the malignant phenotype to understand the mechanism of cell transformation and to develop means to cure or prevent cancers.
To this end, we have been isolating and characterizing genes that induce flat morphology (or “flat reversion”) when expressed in a v-Ki-ras-transformed NIH 3T3 cell line, DT (3). The Krev-1 gene (4), also known as rap1A, which encodes a Ras-related protein containing a region identical to the effector domain of Ras, was isolated in a previous study (5) by using a plasmid-based human fibroblast cDNA expression library. Using a similar approach, Cutler et al. (6) isolated another transformation suppressor gene, rsp-1, encoding a leucine-rich-repeat protein. Recently, we performed a similar screen of a human fibroblast cDNA expression library constructed with a new phagemid shuttle vector and obtained two cDNA clones exhibiting significant biological activities. One of these, clone CT124, was found to encode a truncated form of the MSX-2 homeobox protein, which induces flat reversion through a dominant-negative mechanism over the endogenous MSX-2 protein (7).
Here we describe some properties of the other reversion-inducing gene named RECK (reversion-inducing-cysteine-rich protein with Kazal motifs) and its product. This reversion-inducing gene is unique in that it encodes an extracellular protein with protease inhibitor-like domains and its expression is suppressed strongly in many tumors and cells transformed by various kinds of oncogenes. Restored expression of the RECK gene inhibits the invasive and metastatic activities of tumor cells. We also found that RECK negatively regulates matrix metalloproteinase-9 (MMP-9) (8) in two ways: suppression of MMP-9 secretion from the cells and direct inhibition of its enzymatic activity. These findings suggest that RECK may serve as a negative regulator for MMP-9 in normal cells, and its down-regulation by oncogenic signals therefore would facilitate tumor invasion and metastasis.
MATERIALS AND METHODS
Cell Lines.
The origins of NIH 3T3 derivatives transformed by v-src, v-fms, or v-fes have been described (3). Other transformants were newly generated by infecting with appropriate helper-free retroviruses. RZmet-2, a variant of the HT1080 cell line (9), was established by three cycles of subcutaneous inoculation and selection for high lymph node metastasis in nude mice (M.N., unpublished data). Transfection was carried out by using Lipofectamine (GIBCO/BRL). Soft agar assays and nude mouse assays were performed as described previously (7).
Antibody and Detection of RECK Protein.
The mouse mAbs 32C10A and 5B11D12 directed against bacterially expressed hRECK fragments were generated as described by Harlow and Lane (10). Isolation of cytosolic and membrane fractions was performed as described by Courtneidge et al. (11). For N-glycosidase treatment, membrane fractions were denatured in 0.5% SDS/1% 2-mercaptoethanol at 100°C for 10 min and then incubated in 50 mM sodium phosphate (pH 7.5) containing 1% Nonidet P-40 and 1 × 104 units/ml PNGaseF (New England Biolabs) at 37°C for 3 hr (12).
Invasion and Chemotaxis Assays.
Cells (2 × 105) suspended in DMEM containing 0.1% BSA were placed in the upper compartment of a BioCoat Matrigel Invasion Chamber (Collaborative Biomedical Products, Bedford, MA), the lower compartment was filled with DMEM containing fibronectin (25 μg/ml for B16-BL6, 10 μg/ml for HT1080), and, after incubation at 37°C under 5% CO2 (10 hr for B16-BL6, 20 hr for HT1080), the number of penetrated cells was counted (13). Chemotaxis assay was performed under similar conditions except that uncoated Transwell Chambers (Costar), instead of Matrigel Invasion Chambers, were used.
Metastasis Assays.
For experimental metastasis assays, 5 × 104 viable cells suspended in 0.2 ml of PBS were injected into the tail vein of a mouse (strain: ICR-nu/nu for HT1080 and C57BL/6 for B16-BL6). For spontaneous metastasis assays, 1 × 106 viable RZmet-2 transfectants in 0.2 ml PBS were injected s.c. into an ICR-nu/nu mouse. Twenty-one days after inoculation, the lung (after tail vein inoculation) and lymph nodes (after s.c. inoculation) of the animals were inspected for metastatic nodules (14).
Conditioned Medium and Gelatin Zymography.
Culture supernatants were prepared by incubating cells in DMEM containing 0.1% BSA (volume: 300 μl per 5 × 105 cells) at 37°C for 8 hr in the absence or presence of PI-PLC (Calbiochem, 1–100 milliunits/ml). Proteins in the conditioned media were separated, without prior boiling, by electrophoresis through SDS/polyacrylamide (10%) gels containing 1 mg/ml gelatin (Difco) under nonreducing conditions. The proteins in gel were renatured and stained as described previously (15).
Purification of hRECKΔC Protein.
Conditioned medium prepared from CHOd− cells transfected with pRC/RSV-hRECKΔC (vector: Invitrogen) was concentrated 20-fold by using a Pellicon tangential flow ultrafiltration device (Amicon) fitted with a 50-kDa molecular mass cutoff filter (Filtron) and diafiltered with 4 vol of 25 mM Tris, pH 8.45. The concentrate was loaded onto a Q-Sepharose HP column (Pharmacia) equilibrated with 25 mM Tris/20 mM NaCl, pH 8.45, and the column was eluted with a linear NaCl gradient from 20 to 275 mM. A pool of the hRECKΔC-containing fractions was bound to WGA agarose (EY Laboratories). After washing with 25 mM Tris/100 mM NaCl, pH 7.8, the bound hRECKΔC was eluted with 50 mM N-acetylglucosamine in the same buffer. The bound hRECKΔC (sample QW: more than 50% purity) was buffer-exchanged by using a YM-30 membrane with 20 mM Mes/25 mM NaCl, pH 5.8, and then loaded on a Resource S cation-exchange column (Pharmacia) equilibrated in the same buffer. The column was eluted with a linear NaCl gradient from 25 to 300 mM to obtain the protein sample QWS (more than 95% purity).
Solid-Phase Binding Assay.
ELISA plates (Falcon) were coated with 30 μl of hRECKΔC protein (sample QW, 1 μg/ml in PBS) and subsequently with 200 μl of BSA (5 mg/ml in PBS) as described (16). Plates were incubated with 30 μl of either pro-MMP-2 (5 units/ml) or pro-MMP-9 (10 milliunits/ml) (both from Boehringer Mannheim) at 37°C for 1 hr and washed four times with 200 μl of 0.05% Tween-20 in PBS and an additional four times with 200 μl of PBS. Bound MMPs were solubilized with 50 μl of 2% SDS in 60 mM Tris⋅HCl (pH 6.8) and subjected, together with the diluted input MMP samples, to gelatin zymography followed by densitometric quantitation of the corresponding bands.
Measurement of MMP-9 Activity.
Purified 92-kDa pro-MMP-9 protein (human) was activated following the procedure recommended by the provider (Boehringer Mannheim, cat. no. 1758896), and its activity was measured by using a 2,4-dinitrophenyl (Dnp)-labeled synthetic peptide, Dnp-Pro-Gln-Gly-Ile-Ala-Gly-Gln-d-Arg (Peptide Institute, Osaka), as a substrate following the procedure described by Masui et al. (17).
RESULTS
Structure and Subcellular Localization of RECK Protein.
A reversion-inducing cDNA, CT192, was isolated initially by screening a human fibroblast cDNA expression library, λHK-1-MRC-5, for reversion-inducing clones in DT cells (7). We subsequently isolated a mouse cDNA by hybridization screening and confirmed its reversion-inducing activity (data not shown). Sequence analyses revealed that both the human and mouse cDNAs encode proteins of 971 amino acid residues sharing 93.0% identity with each other (Fig. 1A). These proteins are rich in cysteine (9%) and contain hydrophobic regions at both the NH2- and COOH-terminal ends (Fig. 1B). Peptide sequencing of the mature human protein expressed in mammalian cells indicated that the NH2-terminal hydrophobic region (26 residues) serves as a signal peptide (18). The COOH-terminal hydrophobic region (ca. 29 residues) appears to serve as a signal for glycosylphosphatidylinositol anchoring (19). The middle portion of the protein contains three serine-protease inhibitor-like domains (Fig. 1B): the first domain (residues 635–654) completely matches the Kazal motif (C-X7-C-X6-Y-X3-C-X2,3-C) (20), whereas the second (residues 716–735) and the third (residues 754–772) define incomplete Kazal motifs. We also detected two regions (residues 493–523 and 676–709) of weak homology to the epidermal growth factor-like repeat. The NH2-terminal one-third of the protein contains five repeats of a putative cysteine knot motif (6-Cys repeat: C2-X7–8-C-X3-C-X12–22-C-X9–12-C; residues 37–84, 104–141, 151–197, 216–263, and 292–338) and five potential glycosylation sites (asparagines at positions 39, 86, 200, 297, and 352). Based on its activity and these structural features, we named this gene RECK for reversion-inducing cysteine-rich protein with Kazal motifs (hRECK for the human and mRECK for the mouse gene).
Figure 1.
Structure of the RECK protein. (A) Primary translation products predicted from the human and mouse RECK cDNA sequences. Identical amino acids and conservative substitutions are indicated by asterisks and dots, respectively. (B) Structural organization of the predicted hRECK protein. EGF, epidermal growth factor; SPI, serine protease inhibitor.
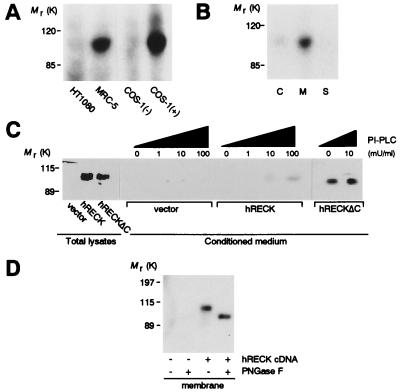
Two mAbs were generated by using bacterially expressed COOH-terminal fragments of the hRECK protein (residues 621–832 for antibody 32C10A and residues 801–971 for antibody 5B11D12). Both antibodies detected a specific band of about 110 kDa in immunoblots by using total lysates of MRC-5 human embryo fibroblasts, but no specific bands were detectable in immunoblots of HT1080 human fibrosarcoma cells (9). Subsequent to, but not before, transfection with the hRECK-expression vector, these antibodies also detected a specific 110-kDa band in COS-1 cells (Fig. 2A). As predicted from its structure, the hRECK protein was found primarily in the membrane fraction of MRC-5 cells (Fig. 2B) and released into the culture supernatant when the cells were treated with phosphatidylinositol-specific phospholipase C (PI-PLC), which cleaves off glycosylphosphatidylinositol anchors (19), or when the COOH-terminal 23 aa were deleted from the protein (hRECKΔC) (Fig. 2C). N-glycosylation of the hRECK protein was demonstrated by the downward shift of the hRECK protein band after treatment of the membrane fraction with N-glycosidase F (PNGaseF) (Fig. 2D).
Figure 2.
Immunoblot detection of the hRECK protein using the 32C10A antibody. (A) Total lysates of indicated cells or COS-1 cells transfected with either pCXN2neo (37) (−) or pCXN2-hRECK expression vector (+) were analyzed. (B) The cytosolic (C) and membrane (M) fractions or the culture supernatant (S) prepared from equivalent numbers of MRC-5 cells were analyzed. (C) Total cell lysates and culture supernatants (conditioned medium), prepared in the presence of various concentrations of PI-PLC, from HT1080 cells transfected with either the pCXN2neo, pCXN2-hRECK, or pCXN2-hRECKΔC were analyzed. The amount of sample applied per lane corresponds to 5 × 104 cells for total lysates and 1 × 105 cells (8-hr harvest) for conditioned medium. (D) Membrane fractions prepared from pCXN2neo (−) or pCXN2-hRECK-transfected HT1080 cells (+), either untreated (−) or treated with PNGaseF (+), were analyzed.
Expression of RECK Gene in Normal Tissues and Cultured Cells.
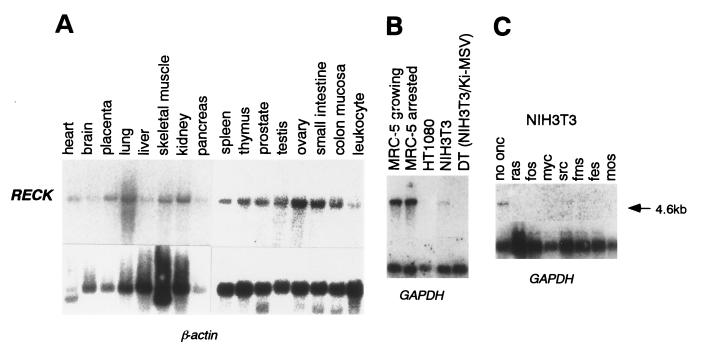
The hRECK gene was mapped to human chromosome 9p12–13 (S.T., C.T., and M.N., unpublished data). Its expression could be detected by Northern blot hybridization in a wide variety of normal human tissues (Fig. 3A) and in MRC-5 cells cultured under both growing and serum-starved conditions (Fig. 3B). Consistent with the immunoblot data (Fig. 2A), RECK mRNA was undetectable in the human fibrosarcoma cell line HT1080 (Fig. 3B). Moreover, RECK mRNA was undetectable in all malignant cell lines examined, including 4 rodent (PCC4, N18, B16, and PC12) and 19 human (GOTO, SK-N-SH, SK-N-AS, SW48, SW480, Colo320, T24, MCF7, A253, A375, A431, A549, A673, HeLa, HT1080, Y79, HL60, Raji, and HepG2) cell lines derived from various types of tumors (data not shown). The endogenous mRECK mRNA was detectable in untransformed mouse NIH 3T3 cells, but was down-regulated in ras-transformed cells (Fig. 3B). This down-regulation seems to occur at the transcription level, since we could demonstrate, using the dual luciferase reporter system, the repression of the RECK promoter by activated ras oncogenes in a cotransfection assay as well as in cells harboring an inducible ras gene (R.M.S., C.T., and M.N., unpublished data). Down-regulation of RECK mRNA also was observed in NIH 3T3 cells transformed by a variety of other oncogenes, such as v-fos, c-myc, v-src, v-fms, v-fes, and v-mos (Fig. 3C). Our results suggest that the RECK gene is a common negative target for oncogenic signals.
Figure 3.
Detection of RECK mRNA by Northern blot hybridization. (A) Human multiple tissue blots (CLONTECH) were probed with hRECK and β-actin (control) cDNAs. (B) Poly(A)+ RNAs prepared from either MRC-5 normal human fibroblasts [cultured in medium containing 10% FCS (growing) or in medium containing 1% FCS for 48 hr (arrested)] or indicated cells were probed with hRECK and glyceraldehyde-3-phosphate dehydrogenase (GAPDH: control) cDNAs. (C) Poly(A)+ RNAs prepared from NIH 3T3 cell line or its derivatives expressing activated human myc (exons 2 and 3) or other viral oncogenes (3) were probed with hRECK and GAPDH (control) cDNAs.
Effects of Restored RECK Gene Expression on Tumor Invasion and Metastasis.
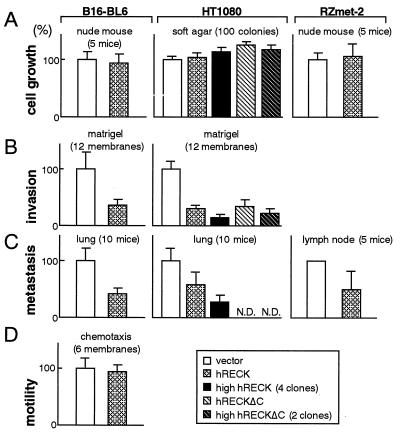
To assess the importance of RECK down-regulation in the manifestation of the malignant properties of tumor cells, we established stable transfectants of hRECK-expression vectors from various tumor-derived cell lines, including HT1080, B16, SW48, A549, HeLa, and A673. We could not detect any prominent effects of the restored expression of hRECK on the growth of these cells in vitro or in vivo (Fig. 4A, and data not shown), although flattening of some cell lines was noted, consistent with the manner in which this gene was first isolated. Interestingly, however, matrix invasion and the metastatic activity of the B16-BL6 mouse melanoma (21) and HT1080 fibrosarcoma (10) or its highly metastatic variant RZmet-2 cells (M.N., T. Matsuzaki, and S. Kumar, unpublished data) were found to be significantly suppressed in hRECK-transfectants (Fig. 4 B and C). These results cannot be explained by mere suppression of cell viability or motility, since we could not detect any differences in colony-forming efficiency (data not shown) or chemotactic activity (Fig. 4D) between hRECK-expressing cells and the vector-transfected control cells. Importantly, the solubilized form of RECK, encoded by the hRECKΔC mutant, retained the ability to suppress tumor invasion (Fig. 4B, striped bars, and see below).
Figure 4.
Effects of restored expression of hRECK on the growth (A), invasion (B), metastasis (C), and motility (D) of B16 melanoma and HT1080 fibrosarcoma cells. (Left) B16-BL6 cells stably transfected with either the pSRαneo (38) or pSRα-hRECK expression vector were pooled and their biological activities were assessed. (Center) HT1080 transfectants carrying either pCXN2neo, pCXN2-hRECK, or the pCXN2-hRECKΔC were analyzed. The data with all clones and with selected clones expressing higher levels of hRECK protein (number of tested clones are indicated) are both presented. (Right) The lymph node metastatic activities and growth of RZmet-2 cells transfected with either pBabe-puro (39) or the pBabe-hRECK were assessed by using nude mice. All the data are presented as ratios to the data with vector-transfected cells. Error bars represent the SE from the number of experiments or animals (n).
Regulation of MMP-9 Secretion by RECK.
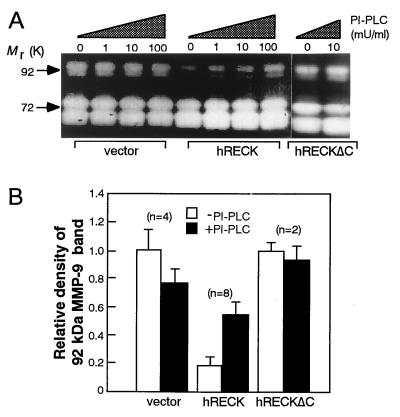
Analysis of the conditioned media by gelatin zymography revealed that the release of the 92-kDa MMP-9 (8) was decreased consistently in hRECK transfectants as compared with the vector-transfected cells (Fig. 5A); note that during this assay, total secreted MMP-9 was activated and detected, and therefore the results reflect the total amount of MMP-9, rather than only the amount of “active” MMP-9, secreted. This decrease must be a result of posttranscriptional event(s), since no difference in the levels of MMP-9 mRNA was detected by Northern hybridization between the two groups of cells (data not shown). Moreover, concomitant increases in the release of MMP-9 and cleaved hRECK protein into culture supernatants were observed when the hRECK transfectants were treated with PI-PLC or when hRECKΔC-transfectants were used (compare Figs. 2C and 5A). This phenomenon was confirmed by testing multiple clones (Fig. 5B). Thus, membrane association of RECK protein is necessary for RECK-mediated suppression of MMP-9 release from the cells. Our observations are consistent with the earlier findings by others that pharmacological inhibition of MMP-9 gene expression in HT1080 cells resulted in suppression of matrix invasion (22–23) and support the proposition that MMP-9 is able to influence the malignant behavior of this cell line.
Figure 5.
Effects of restored hRECK expression on the release of MMP-9 from HT1080 cells. (A) Release of the 92-kDa MMP-9 from the typical HT1080 transfectant clone carrying either the pCXN2neo, pCXN2-hRECK, or pCXN2-hRECKΔC. Culture supernatants prepared in the presence of different concentrations of PI-PLC were subjected to gelatin zymography. (B) Summary of the relative levels of the 92-kDa MMP-9 released from several independent HT1080 transfectant clones. Conditioned medium prepared in the absence (open bar) or presence (solid bar) of PI-PLC (10 milliunits/ml) from the indicated number (n) of transfectant clones were analyzed by gelatin zymography, and the relative levels of 92-kDa MMP-9 release were determined by densitometry. Error bars represent the SE among clones.
Interaction Between MMP-9 and RECK Protein.
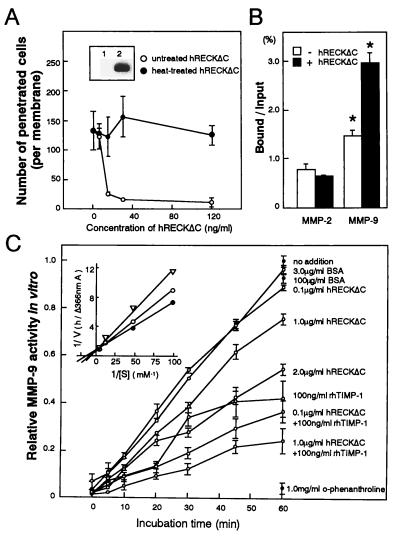
Although the hRECKΔC mutant failed to inhibit MMP-9 release (Fig. 5B), it retained the ability to suppress invasion when transfected into HT1080 cells (Fig. 4B). These results raised the possibility that this mutant protein might suppress invasion even when added exogenously to the cells. To test this hypothesis, we partially purified the hRECKΔC protein (sample QW, above 50% purity) from the conditioned medium of a CHO cell line expressing the hRECKΔC gene. This sample contained neither collagenase activity nor tissue inhibitor of metalloproteinase (TIMP)-like activity (24) as detected by gelatin zymography and reverse gelatin zymography, respectively, and it did not inhibit MMP-9 release from HT1080 cells when added to the medium (data not shown). This sample, however, strongly inhibited the invasion of HT1080 cells in vitro (Fig. 6A).
Figure 6.
Biological and biochemical activities of purified hRECKΔC protein. (A) Matrigel invasion activity of HT1080 cells in the presence of varied concentrations of untreated or heat-treated (90°C, 5 min) hRECKΔC protein. Error bars represent SE from three experiments. Inset shows an immunoblot detection of hRECKΔC protein in the original conditioned medium (lane 1) and the purified protein sample (lane 2). (B) Solid-phase binding assay with immobilized hRECKΔC protein. A solution of either pro-MMP-2 or pro-MMP-9 was incubated in ELISA plate wells coated with either hRECKΔC (sample QW) protein plus BSA (solid bars) or BSA alone (open bars), and after extensive washing, the bound MMP was solubilized. The proportion of bound proform MMPs was estimated by subjecting these samples and diluted input MMP samples to gelatin zymography, followed by densitometric quantitation. Error bars represent SE from two experiments. ∗, P = 0.001 (Student’s t test). (C) Inhibition of MMP-9 activity by purified hRECKΔC protein. Proteolytic activity of MMP-9 was measured in the presence of the indicated concentration of hRECKΔC protein. BSA was used as a negative control and recombinant human TIMP-1 (rhTIMP-1; FYK, Toyama, Japan) as well as o-phenanthroline (Sigma) were used as positive controls. Error bars represent the SE from two experiments. Inset shows the Lineweaver–Burke plot (36) of the proteolytic activity of MMP-9 incubated with 10–200 μM synthetic substrate in the absence (solid circle) or presence (open circle) of 20 nM hRECKΔC or 2 nM hTIMP-1 (triangle) for 1 hr. The Ki value of hRECKΔC for hydrolysis of the Dnp-peptide by activated MMP-9 was estimated as approximately 78 nM by using the equation 1/V = 1/Vmax + Km(1 + [I]/Ki)/Vmax[S].
We next examined the biochemical interaction between hRECK and MMP-9 in two ways. First, we assessed the binding of purified pro-MMP-2 and pro-MMP-9 to immobilized hRECKΔC protein (sample QW) and found that the hRECKΔC protein is able to retain a significant amount of pro-MMP-9 but not pro-MMP-2 (Fig. 6B). Second, we examined the effects of highly purified hRECKΔC protein (sample QWS, above 95% purity) on the proteolytic activity of MMP-9 and detected a significant MMP-9-inhibiting activity associated with hRECKΔC (IC50 ≈ 2 μg/ml) (Fig. 6C). An additive effect was observed when hRECKΔC and TIMP-1 were added together (Fig. 6C). Kinetic analysis of MMP-9 activity in the absence or presence of hRECKΔC also was performed. hRECKΔC weakly inhibited activated MMP-9 in a competitive manner with an inhibition constant (Ki) of around 78 nM (Fig. 6C). The Ki value for TIMP-1 determined under identical conditions was 2.9 nM. Taken together, these results suggest that RECK protein may directly interact with, and inhibit the activity of, MMP-9.
DISCUSSION
Our findings are consistent with a model in which the release of the 92-kDa MMP-9 is somehow gated by the membrane-anchored hRECK protein on the plasma membrane. When an oncogenic signal is turned on, the RECK gene is down-regulated, resulting in increased secretion of MMP-9, and this contributes to morphological transformation as well as to the invasive behavior of the cells. This malignant behavior can be reversed to some extent by restored expression of the hRECK gene or by the addition of the solubilized form of the hRECK protein (hRECKΔC) into the medium. It has been well documented that the MMP-9 gene is up-regulated by activated ras genes (25–29); therefore, MMP-9 is positively regulated by Ras signaling through multiple mechanisms, i.e., transcriptional activation of MMP-9 itself and down-regulation of a negative regulator of MMP-9, RECK.
Emerging evidences suggest that membrane-anchored proteases play important roles during growth, morphogenesis, tissue repair, and pathogenesis. It has been proposed that, by concentrating at or near the cell surface, the proteolytic activities of these enzymes can be effective even in the presence of high concentrations of (diffusable) inhibitors (30–33). RECK is a protease inhibitor-like molecule that has been found to be anchored to the membrane. Although the exact molecular mechanism is yet to be explored, RECK clearly regulates the secretion of MMP-9 (Fig. 5). The purified hRECKΔC protein was found to bind to the purified pro-MMP-9 and to inhibit the proteolytic activity of this enzyme in vitro, albeit with apparently lower potency than TIMP-1 (Fig. 6). Although this relatively low potency merely may reflect the effects of the artificial COOH-terminal truncation and/or inactivation during purification, even this weak activity may be influential when concentrated on the cell surface. Alternatively, the strong influence of RECK on MMP-9 secretion may suggest that other cooperating molecules on the cell surface or some indirect mechanism besides direct interaction may be involved in RECK-mediated regulation of MMP-9. Nevertheless, it is interesting that a molecule structurally related to the inhibitors of one class of proteases (i.e., serine proteases) regulates a member of another class of proteases (i.e., MMPs). This is not, however, unprecedented: several serine protease inhibitors, including α-macroglobulins (34), the Streptomyces subtilisin family of inhibitors (35), and the amyloid precursor protein (36), have been reported to inhibit metalloproteinases. Further studies on the interaction between RECK and MMP-9 may yield new insights into how such cross-class protease–protease inhibitor interactions occur. It is also important to establish the specificity of the interaction between RECK and MMP-9. This will require an extensive survey of proteases, as well as other types of proteins, which have the potential to interact physically and/or functionally with RECK protein. Analysis of the phenotypes of RECK-deficient mice may also provide some clues to the understanding of the physiological functions of this molecule.
Acknowledgments
We thank Drs. I. Fidler, R. Bassin, and T. Tsuruo for cell lines, Y. Takebe, N. Arai, H. Okayama, N. Satake, J. Miyazaki, H. Sato, M. Seiki, and Y. Ito for plasmids, H. Kobayashi and R. Takahashi for technical advice, D. Alexander for critical reading of the manuscript, and H. Sugano for encouragement. This work was supported by grants from the Japanese Foundation for Cancer Research; the Ministry of Education, Science, Sports, and Culture; the Science and Technology Agency of Japanese Government; and Amgen Inc.
ABBREVIATIONS
- RECK
reversion-inducing cysteine-rich protein with Kazal motif
- MMP-9
matrix metalloproteinase-9
- PI-PLC
phosphatidylinositol-specific phospholipase C
Footnotes
References
- 1. Bos J L. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 3.Noda M, Selinger Z, Scolnick E, Bassin R. Proc Natl Acad Sci USA. 1983;80:5602–5606. doi: 10.1073/pnas.80.18.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 5.Noda M, Kitayama H, Sugimoto Y, Okayama H, Bassin R H, Ikawa Y. Proc Natl Acad Sci USA. 1989;86:162–166. doi: 10.1073/pnas.86.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutler M L, Bassin R H, Zanoni L, Talbot N. Mol Cell Biol. 1992;12:3750–3756. doi: 10.1128/mcb.12.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi C, Akiyama N, Matsuzaki T, Takai S, Kitayama H, Noda M. Oncogene. 1996;12:2137–2146. [PubMed] [Google Scholar]
- 8.Wilhelm S M, Collier I E, Marmer B L, Eisen A Z, Gerant G A, Goldberg G I. J Biol Chem. 1989;264:17213–17211. [PubMed] [Google Scholar]
- 9.Rasheed S, Nelson-Rees W A, Teth E M, Arnstein P, Gardner M B. Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 139–283. [Google Scholar]
- 11.Courtneidge S A, Levinson A D, Bishop J M. Proc Natl Acad Sci USA. 1980;77:3183–3787. doi: 10.1073/pnas.77.7.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarentino A L, Quinones G, Trumble A, Changchien L M, Duceman B, Maley F, Plummer T H., Jr J Biol Chem. 1990;265:6961–6966. [PubMed] [Google Scholar]
- 13.Kobayashi H, Ohi H, Sugimura M, Shinohara H, Fujii T, Terao T. Cancer Res. 1992;52:3610–3614. [PubMed] [Google Scholar]
- 14.Poste G, Fidler I J. Nature (London) 1980;283:139–145. doi: 10.1038/283139a0. [DOI] [PubMed] [Google Scholar]
- 15.Welch D R, Fabra A, Nakajima M. Proc Natl Acad Sci USA. 1990;87:7678–7682. doi: 10.1073/pnas.87.19.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks P C, Sillet S, von Schalscha T L, Friedlander M, Cheresh D A. Cell. 1998;92:391–400. doi: 10.1016/s0092-8674(00)80931-9. [DOI] [PubMed] [Google Scholar]
- 17.Masui Y, Takemoto T, Sakakibara S, Hori H, Nagai Y. Biochem Med. 1977;17:215–221. doi: 10.1016/0006-2944(77)90026-6. [DOI] [PubMed] [Google Scholar]
- 18.Neurath H. Trends Biochem Sci. 1989;14:268–271. doi: 10.1016/0968-0004(89)90061-3. [DOI] [PubMed] [Google Scholar]
- 19.Low M G. Biochim Biophys Acta. 1989;988:427–454. doi: 10.1016/0304-4157(89)90014-2. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski M, Kato I. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 21.Hart I R. Am J Pathol. 1979;97:587–600. [PMC free article] [PubMed] [Google Scholar]
- 22.Cha H, Bae S, Lee H, Lee O, Sato H, Seiki M, Park B C, Kim K. Cancer Res. 1996;56:2281–2284. [PubMed] [Google Scholar]
- 23.McMillan J I, Weeks R, West J W, Bursten S, Rice G C, Lovett D H. Int J Cancer. 1996;67:523–531. doi: 10.1002/(SICI)1097-0215(19960807)67:4<523::AID-IJC11>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Docherty A J P, Lyons A, Smith B J, Wright E M, Stephens P E, Harris T J R, Murphy G, Reynolds J J. Nature (London) 1985;318:66–69. doi: 10.1038/318066a0. [DOI] [PubMed] [Google Scholar]
- 25.Stetler-Stevenson W G, Hewitt R, Corcoran M. Semin Cancer Biol. 1996;7:147–154. doi: 10.1006/scbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 26.Liotta L A, Steeg P S, Stetler-Stevenson W G. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 27.Ballin M, Gomez D E, Sinha C C, Thorgeirsson U P. Biochem Biophys Res Commun. 1988;154:832–838. doi: 10.1016/0006-291x(88)90215-x. [DOI] [PubMed] [Google Scholar]
- 28.Himelstein B P, Lee E J, Sato H, Seiki M, Muschel R J. Oncogene. 1997;14:1995–1998. doi: 10.1038/sj.onc.1201012. [DOI] [PubMed] [Google Scholar]
- 29.Steeg P S. Semin Cancer Biol. 1991;2:105–110. [PubMed] [Google Scholar]
- 30.Brooks P C, Strömblad S, Sanders L C, von Schalscha W G, Aimes R T, Stetler-Stevenson W G, Quigley J P, Cheresh D A. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- 31.Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. Nature (London) 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 32.Blasi F, Verde P. Semin Cancer Biol. 1990;1:117–126. [PubMed] [Google Scholar]
- 33.Werb Z. Cell. 1997;91:439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 34.Barret A J, Starkey P M. Biochem J. 1973;133:709–724. doi: 10.1042/bj1330709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajiwara K, Fujita A, Tsuyuki H, Kumazaki T, Ishii S. J Biochem. 1991;110:350–354. doi: 10.1093/oxfordjournals.jbchem.a123584. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki K, Hasegawa M, Funahashi K, Umeda M. Nature (London) 1993;362:839–841. doi: 10.1038/362839a0. [DOI] [PubMed] [Google Scholar]
- 37.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 38.Takebe Y, Seki M, Fujisawa J, Hoy P, Yokota Y, Arai K, Yoshida M, Arai N. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]