Preliminary neutron crystallographic data from the sweet protein thaumatin have been recorded using the LADI-III diffractometer at the Institut Laue Langevin (ILL). The results illustrate the feasibility of a full neutron structural analysis aimed at further understanding the molecular basis of the perception of sweet taste. Such an analysis will exploit the use of perdeuterated thaumatin.
Keywords: thaumatin, neutron diffraction, sweet proteins
Abstract
A preliminary neutron crystallographic study of the sweet protein thaumatin is presented. Large hydrogenated crystals were prepared in deuterated crystallization buffer using the gel-acupuncture method. Data were collected to a resolution of 2 Å on the LADI-III diffractometer at the Institut Laue Langevin (ILL). The results demonstrate the feasibility of a full neutron crystallographic analysis of this structure aimed at providing relevant information on the location of H atoms, the distribution of charge on the protein surface and localized water in the structure. This information will be of interest for understanding the specificity of thaumatin–receptor interactions and will contribute to further understanding of the molecular mechanisms underlying the perception of taste.
1. Introduction
Despite its importance in biology, medicine and biotechnology, the molecular basis of sweet taste is still poorly understood. There are many compounds that cause the sweet taste response, from low-molecular-weight synthetic products such as aspartamine to larger molecules such as thaumatin (de Vos et al., 1985 ▶). For a number of years, the search for the mechanism of sweet taste involved comparisons of the structural features of these sweet molecules and indirect mapping of the receptor-binding site (Temussi, 2006a ▶). The sweet receptor was only identified a few years ago (Bachmanov et al., 2001 ▶; Kitagawa et al., 2001 ▶; Li et al., 2001 ▶; Max et al., 2001 ▶; Montmayeur et al., 2001 ▶; Nelson et al., 2001 ▶; Sainz et al., 2001 ▶); the T1R2-T1R3 receptor (Li et al., 2002 ▶) was identified soon after. Studies by Tancredi et al. (2004 ▶) showed that low-molecular-weight sweeteners occupy small receptor cavities inside two subdomains of the receptor, whereas sweet proteins can interact with the sweet receptor according to a mechanism called the ‘wedge model’, in which they bind to a large external cavity (Temussi, 2006b ▶). It is likely that there are more active sites that have not yet been identified (Jiang et al., 2004 ▶).
Thaumatin, which has been commercialized as a sweetener since 1983, is 1600 times sweeter than sucrose on a weight basis (van der Wel & Loeve, 1972 ▶). Thaumatin can be obtained naturally from the arils of the tropical plant Thaumatococcus daniellii and consists of six closely related 22 kDa proteins (designated I, II, II, a, b and c) that are the sweetest substances known. Owing to the lack of sequence or three-dimensional structure homology between sweet proteins (Somoza et al., 1993 ▶), the mechanism of sweetness has remained obscure for a number of years. Thaumatin shows a striking homology to the PR5 proteins produced by plants to defend themselves against pathogen attacks (Selitrennikoff, 2001 ▶). However, these thaumatin-like proteins (TLPs) do not have sweet taste (Tattersall et al., 1997 ▶) and it has been suggested that the sweetness may arise from a pattern of lysine residues that are unique to thaumatin (Kim & Weickmann, 1994 ▶). This, together with the alkalinity of a number of sweet proteins, has led to studies of the role of positive charge in thaumatin derivatives containing five phosphopyridoxylated lysine residues (Kaneko & Kitabatake, 2001 ▶). These studies showed that there is a cleft-containing side of the protein that is important for sweetness.
Despite the increasing amount of information available, the structural basis of thaumatin sweetness remains poorly understood. The mechanism by which a single receptor is responsible for the response to both small molecules and sweet proteins has also not been fully characterized. The studies described here aim to investigate the protonation states and ordered solvent structure of thaumatin I. This neutron diffraction study aims towards the design of new sweetners and a better understanding of the mechanism of taste by providing information that can help characterize active sites in the protein under different pH conditions. Furthermore, we expect the neutron crystallography results to shed light onto the effect of pH on the structural stability of TLPs (Perri et al., 2008 ▶).
2. Methods
2.1. Crystallization
Thaumatin I (Sigma–Aldrich) was used directly without further purification. Crystals were obtained at room temperature in deuterated buffer (to reduce hydrogen incoherent scattering) using both the hanging-drop and sitting-drop vapour-diffusion methods following published protocols (Tomčovà & Smatanovà, 2007 ▶). X-ray diffraction tests on the ID14-2 beamline at the European Synchrotron Radiation Facility (ESRF) showed that the crystals formed in space group P41212 (unit-cell parameters a = b = 57.8, c = 150.1 Å) and diffracted to 1.1 Å resolution when cryocooled. Larger samples were required for the neutron work, but as often happens when crystallizing with larger volumes the crystals fractured easily during sample preparation and mounting (Yadav et al., 2005 ▶). The final sample was grown in a quartz capillary using the gel-acupuncture method (López-Jaramillo et al., 2001 ▶). The solvent surrounding the D2O-soaked thaumatin crystal was removed and crystallization buffer was added to each side of the crystal in a quartz capillary prior to sealing with wax for neutron data collection.
2.2. Neutron diffraction
Neutron Laue diffraction data were collected at room temperature using the LADI-III instrument installed on cold neutron guide H142 at the Institut Laue Langevin (ILL). The LADI-III instrument (Blakeley et al., in preparation), which uses a large neutron image-plate detector that completely encircles the sample, is a recent replacement for the LADI-I instrument that was used to collect data for human aldose reductase (Blakeley et al., 2006 ▶, 2008 ▶), xylose isomerase (Meilleur et al., 2006 ▶), concanavalin A (Blakeley et al., 2004 ▶; Ahmed et al., 2007 ▶), rasburicase (Budayova-Spano et al., 2006 ▶) and endothiapepsin/hydroxyethylene (Coates et al., 2006 ▶) from perdeuterated crystals as small as 0.15 mm3. The improved detector design of LADI-III provides a twofold to threefold gain in neutron detection (Wilkinson et al., 2007 ▶), while the larger radius of the drum aids the signal-to-noise ratio of recorded reflections and decreases spatial overlap. A nickel/titanium multi-layer bandpass filter was used to select a restricted neutron wavelength range (Δλ/λ ≃ 25%) centred at 3.7 Å and extending from 3.2 to 4.2 Å. Data were recorded in a series of seven contiguous Laue images using 12 h exposures with a step separation of 9° about the vertical rotation (ϕ) axis of the detector. Neutron Laue data were indexed and integrated using the LAUEGEN software suite (Helliwell et al., 1989 ▶; Campbell, 1995 ▶), modified for the cylindrical geometry of the LADI-III detector (Campbell et al., 1998 ▶). The LSCALE program (Arzt et al., 1999 ▶) was used to derive the wavelength-normalization curve using the intensities of symmetry-equivalent reflections measured at different wavelengths. No account was made for crystal damage since neutrons do not induce detectable radiation damage and no explicit absorption corrections were applied. SCALA (Collaborative Computational Project, Number 4, 1994 ▶) was used to combine and merge the 26 595 observed reflections. Fourier maps were calculated using the observed structure-factor amplitudes (F o) derived from these data in conjunction with neutron amplitudes (F c) and phases (αc) calculated from the high-resolution X-ray structure of thaumatin using neutron coherent scattering lengths. The maps were displayed and studied using the program O (Jones et al., 1991 ▶; Jones, 2004 ▶).
3. Results
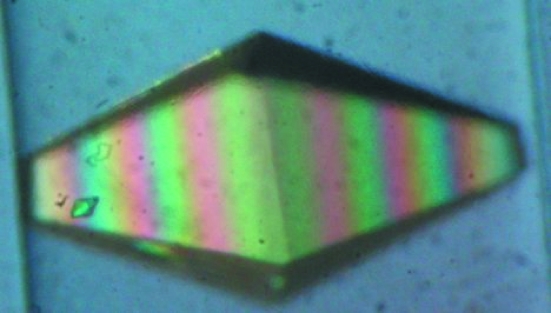
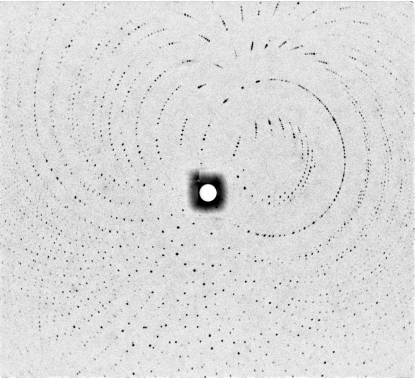
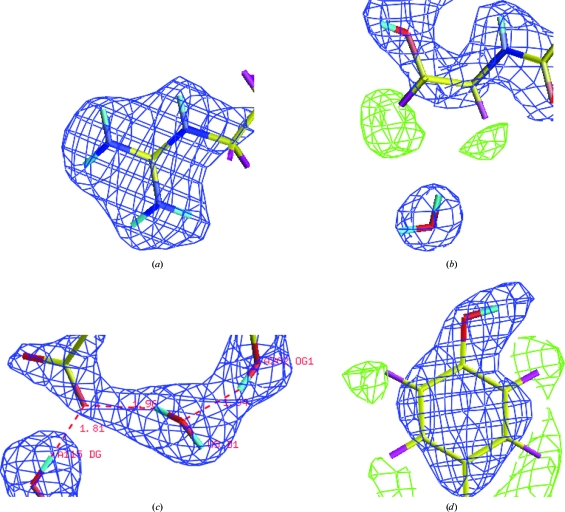
Fig. 1 ▶ shows the thaumatin crystal used in this study. The crystal dimensions were 2.0 × 1.0 × 0.7 mm. Diffraction peaks were observed to 1.8 Å resolution and data were processed to 2.0 Å resolution. A representative neutron Laue diffraction pattern is shown in Fig. 2 ▶. While the final stages of neutron data collection have not yet been carried out, the relevant statistics for the data so far collected are shown in Table 1 ▶. Fig. 3 ▶ shows four regions of density in the thaumatin neutron (2F o − F c) maps. In Fig. 3 ▶(a), the effect of deuterium replacement of labile H atoms in arginine is very clear, whereas in Thr54 (Fig. 3 ▶ b), hydrogen attached to C atoms appears as negative (green) peaks in the density map, with a D2O molecule located within hydrogen-bonding distance. Fig. 3 ▶(c) shows evidence of a water molecule located between Asp113 and Thr202 and Fig. 3 ▶(d) shows negative (green) density around Tyr169 corresponding to ring H atoms. Note that in Figs. 3 ▶(b) and 3 ▶(d) the density map clearly shows the orientation of two O—D groups.
Figure 1.
The thaumatin crystal used for this neutron diffraction study on LADI-III. The crystal volume was 1.4 mm3.
Figure 2.
Neutron Laue diffraction pattern recorded from the hydrogenated thaumatin crystal shown in Fig. 1 ▶ using the LADI-III instrument at the ILL. The exposure time was 12 h.
Table 1. Summary of 2 Å neutron Laue diffraction data from thaumatin.
| dmin (Å) | Nmeas | Nref | Rmerge (%) | I/σ(I) | Completeness (%) | Multiplicity |
|---|---|---|---|---|---|---|
| 6.32 | 1394 | 547 | 5.6 | 18.1 | 86.0 | 2.5 |
| 4.47 | 3400 | 964 | 7.5 | 18.3 | 91.2 | 3.5 |
| 3.65 | 3906 | 1178 | 9.3 | 14.7 | 88.8 | 3.3 |
| 3.16 | 3762 | 1201 | 11.4 | 10.8 | 79.5 | 3.1 |
| 2.83 | 3180 | 1119 | 14.7 | 6.7 | 65.7 | 2.8 |
| 2.58 | 2722 | 1073 | 16.8 | 4.3 | 57.8 | 2.5 |
| 3.39 | 2316 | 977 | 17.8 | 3.4 | 48.7 | 2.4 |
| 2.24 | 2364 | 1020 | 18.2 | 2.6 | 46.4 | 2.3 |
| 2.11 | 2045 | 946 | 18.5 | 2.2 | 40.6 | 2.2 |
| 2.00 | 1506 | 779 | 18.6 | 2.0 | 32.0 | 1.9 |
| All | 26595 | 9804 | 11.0 | 8.1 | 56.9 | 2.7 |
Figure 3.
Four separate regions of the (2F o − F c) neutron Fourier map calculated for the thaumatin LADI-III data. The density map is contoured at +1.5 r.m.s. (blue) and at −1.5 r.m.s. (green). (a) Arg53, with labile H atoms replaced by deuterium. (b) Thr54, showing the H atoms attached to C atoms as negative peaks (green). (c) A D2O molecule located between Asp113 and Thr202. (d) Tyr169, showing the ring H atoms as well as the position and orientation of an O—D group.
4. Discussion
This work clearly demonstrates the feasibility of a detailed study of thaumatin using neutron crystallography. It is anticipated that data collection will be completed during the forthcoming ILL scheduling period, yielding a data set with high completeness to a resolution of 1.8 Å. The final structure refinement will be carried out using both X-ray and neutron data recorded at the same temperature, using the nCNS program (Langan & Mustyakimov, 2008 ▶), which combines global X-ray data with neutron data and energy refinement for the first time and allows cross-validated and maximum-likelihood simulated annealing. Such a study will provide important information on H-atom positions and hydration and will be used to attempt to relate structural aspects of the protein to its physiological properties. Central to this programme will be structural studies of a range of closely related protein structures in which subtle differences in structure have marked consequences for taste. A study of perdeuterated thaumatin is under way and will be used to improve data quality and analysis as well as adding to the growing base of data that provide detailed comparison of the structural effects of protein deuteration (Artero et al., 2005 ▶).
More generally, this study emphasizes a number of key issues that are linked to the development of neutron crystallography and its exploitation for the study of biological systems. Firstly, while it is clear from the data presented here that neutron crystallography can deliver high-resolution results from hydrogenated protein in deuterated buffer, major gains are possible through the use of perdeuterated protein, in which hydrogen incoherent scattering is eliminated and the coherent scattering power of deuterium is fully exploited. In practical terms, this yields up to a factor of 10 in efficiency, reducing the crystal sizes for these experiments and enhancing the quality of the data and its interpretation. On the Grenoble campus where this work is being carried out, a Deuteration Laboratory was first proposed (Forsyth et al., 2002 ▶) and subsequently built with the purpose of making the provision of deuterated proteins more routine for crystallography, fibre diffraction, solution scattering (Callow et al., 2007 ▶) and reflectometry (Lu et al., 2000 ▶; Grage et al., 2008 ▶) as well as dynamics studies (Wood et al., 2007 ▶) and NMR work (Varga et al., 2007 ▶). The laboratory, which now exists within the Grenoble Partnership for Structural Biology (PSB), offers a peer-review user access programme, has an active in-house programme and is committed to methodological developments (see, for example, Laux et al., 2008 ▶) that will greatly widen the access of neutron scattering approaches to structural biologists. Such developments come at a particularly important time given the major developments that are occurring around the world for neutron scattering (Teixeira et al., 2008 ▶).
Acknowledgments
We acknowledge the ILL for provision of beamtime under proposal TEST-1429, the ESRF for test time on beamline ID14-2 and the EMBL for partial support of the LADI-III diffractometer. We thank the staff of the ILL-EMBL Deuteration Laboratory for helpful discussion (in particular Shona Gillespie) and Joanne McCarthy for help with ESRF instrumentation. SCMT and VTF acknowledge support from the EPSRC under grant EP/C015452/1 and from the EU under contract RII3-CT-2003-505925.
References
- Ahmed, H. U., Blakeley, M. P., Cianci, M., Cruickshank, D. W. J., Hubbard, J. A. & Helliwell, J. R. (2007). Acta Cryst. D63, 906–922. [DOI] [PubMed]
- Artero, J.-B., Härtlein, M., McSweeney, S. & Timmins, P. (2005). Acta Cryst. D61, 1541–1549. [DOI] [PubMed]
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst.32, 554–562.
- Bachmanov, A. A., Li, X., Reed, D. R., Ohmen, J. D., Li, S., Chen, Z., Tordoff, M. G., de Jong, P. J., Wu, C., West, D. B., Chatterjee, A., Ross, D. A. & Beauchamp, G. K. (2001). Chem. Senses, 26, 925–933. [DOI] [PMC free article] [PubMed]
- Blakeley, M., Kalb, A., Helliwell, J. & Myles, D. (2004). Proc. Natl Acad. Sci. USA, 101, 16405–16410. [DOI] [PMC free article] [PubMed]
- Blakeley, M., Mitschler, A., Hazemann, I., Meilleur, F., Myles, D. & Podjarny, A. (2006). Eur. Biophys. J.35, 577–583. [DOI] [PubMed]
- Blakeley, M., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Budayova-Spano, M., Bonneté, F., Ferté, N., El Hajji, M., Meilleur, F., Blakeley, M. P. & Castro, B. (2006). Acta Cryst. F62, 306–309. [DOI] [PMC free article] [PubMed]
- Callow, P., Sukhodub, A., Taylor, J. E. & Kneale, G. G. (2007). J. Mol. Biol.369, 177–185. [DOI] [PMC free article] [PubMed]
- Campbell, J. W. (1995). J. Appl. Cryst.28, 228–236.
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst.31, 496–502.
- Coates, L., Erskine, P. T., Mall, S., Gill, R., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2006). Eur. Biophys. J.35, 559–566. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Forsyth, V. T., Myles, D., Timmins, P. & Haertlein, M. (2002). Opportunities for Neutron Scattering in the 3rd Millennium, pp. 47–54. Grenoble: Institut Laue Langevin.
- Grage, S., May, R., Holt, S., Haertlein, M., de Planque, M., Mendes, G., Contera, S., Turdzeladze, T., Burck, J., Martinac, B., Forsyth, V. T., Watts, A. & Ulrich, A. (2008). In preparation.
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst.22, 483–497.
- Jiang, P., Ji, Q., Liu, Z., Snyder, L. A., Benard, L. M. J., Margolskee, R. F. & Max, M. (2004). J. Biol. Chem.279, 45068–45075. [DOI] [PubMed]
- Jones, T. A. (2004). Acta Cryst. D60, 2115–2125. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kaneko, R. & Kitabatake, N. (2001). Biosci. Biotechnol. Biochem.65, 409–413. [DOI] [PubMed]
- Kim, S. & Weickmann, J. (1994). Thaumatin, edited by M. Witty & J. D. Higginbotham, pp. 135–149. Boca Raton: CRC Press.
- Kitagawa, M., Kusakabe, Y., Miura, H., Ninomiya, Y. & Hino, A. (2001). Biochem. Biophys. Res. Commun.283, 236–242. [DOI] [PubMed]
- Langan, P. & Mustyakimov, P. (2008). In preparation.
- Laux, V., Callow, P., Svergun, D., Timmins, P., Forsyth, V. T. & Haertlein, M. (2008). Eur. Biophys. J. doi:10.1007/s00249-008-0280-5. [DOI] [PubMed]
- Li, X., Inoue, M., Reed, D. R., Huque, T., Puchalski, R. B., Tordoff, M. G., Ninomiya, Y., Beauchamp, G. K. & Bachmanov, A. A. (2001). Mamm. Genome, 12, 13–16. [DOI] [PMC free article] [PubMed]
- Li, X., Staszewski, L., Xu, H., Durick, K., Zoller, M. & Adler, E. (2002). Proc. Natl Acad. Sci. USA, 99, 4692–4696. [DOI] [PMC free article] [PubMed]
- López-Jaramillo, F. J., García-Ruiz, J. M., Gavira, J. A. & Otálora, F. (2001). J. Appl. Cryst.34, 365–370.
- Lu, J., Thomas, R. & Penfold, J. (2000). Adv. Colloid Interface Sci.84, 143–304. [DOI] [PubMed]
- Max, M., Shanker, Y. G., Huang, L., Rong, M., Liu, Z., Campagne, F., Weinstein, H., Damak, S. & Margolskee, R. F. (2001). Nature Genet.28, 58–63. [DOI] [PubMed]
- Meilleur, F., Snell, E., van der Woerd, M., Judge, R. & Myles, D. (2006). Eur. Biophys. J.35, 601–609. [DOI] [PubMed]
- Montmayeur, J. P., Liberles, S. D., Matsunami, H. & Buck, L. B. (2001). Nature Neurosci.4, 492–498. [DOI] [PubMed]
- Nelson, G., Hoon, M. A., Chandrashekar, J., Zhang, Y., Ryba, N. J. & Zuker, C. S. (2001). Cell, 106, 381–390. [DOI] [PubMed]
- Perri, F., Romitelli, F., Rufini, F., Secundo, F., Stasio, E. D., Giardina, B. & Vitali, A. (2008). Protein J.27, 13–20. [DOI] [PubMed]
- Sainz, E., Korley, J. N., Battey, J. F. & Sullivan, S. L. (2001). J. Neurochem.77, 896–903. [DOI] [PubMed]
- Selitrennikoff, C. P. (2001). Appl. Environ. Microbiol.67, 2883– 2894. [DOI] [PMC free article] [PubMed]
- Somoza, J. R., Jiang, F., Tong, L., Kang, C. H., Cho, J. M. & Kim, S.-H. (1993). J. Mol. Biol.234, 390–404. [DOI] [PubMed]
- Tancredi, T., Pastore, A., Salvadori, S., Esposito, V. & Temussi, P. A. (2004). Eur. J. Biochem.271, 2231–2240. [DOI] [PubMed]
- Tattersall, D. B., van Heeswijck, R. & Høj, P. B. (1997). Plant Physiol.114, 759–769. [DOI] [PMC free article] [PubMed]
- Teixeira, S. et al. (2008). Chem. Phys. doi:10.1016/j.chemphys.2008.02.030.
- Temussi, P. (2006a). J. Mol. Recognit.19, 188–199. [DOI] [PubMed]
- Temussi, P. A. (2006b). Cell. Mol. Life Sci.63, 1876–1888. [DOI] [PMC free article] [PubMed]
- Tomčovà, I. & Smatanovà, I. (2007). J. Cryst. Growth, 306, 383–389.
- Varga, K., Aslimovska, L., Parrot, I., Dauvergne, M.-T., Haertlein, M., Forsyth, V. T. & Watts, A. (2007). Biochim. Biophys. Acta, 1768, 3029–3035. [DOI] [PubMed]
- Vos, A. M. de, Hatada, M., van der Wel, H., Krabbendam, H., Peerdeman, A. F. & Kim, S.-H. (1985). Proc. Natl Acad. Sci. USA, 82, 1406–1409. [DOI] [PMC free article] [PubMed]
- Wel, H. van der & Loeve, K. (1972). Eur. J. Biochem.31, 221–225. [DOI] [PubMed]
- Wilkinson, C., Blakeley, M. & Dauvergne, F. (2007). ILL Internal Report, p. ILL07WI02T. Grenoble: Institut Laue Langevin.
- Wood, K., Plazanet, M., Gabel, F., Kessler, B., Oesterhelt, D., Tobias, D., Zaccai, G. & Weik, M. (2007). Proc. Natl Acad. Sci. USA, 104, 18049–18054. [DOI] [PMC free article] [PubMed]
- Yadav, M. K., Gerdts, C. J., Sanishvili, R., Smith, W. W., Roach, L. S., Ismagilov, R. F., Kuhn, P. & Stevens, R. C. (2005). J. Appl. Cryst.38, 900–905. [DOI] [PMC free article] [PubMed]