The haem-binding protein HbpS from Streptomyces reticuli was crystallized and diffraction data were collected to a maximal resolution of 2.25 Å.
Keywords: Streptomyces reticuli, haem-binding protein, HbpS
Abstract
Streptomyces reticuli is a soil-growing Gram-positive bacteria that has been shown to secrete a novel haem-binding protein known as HbpS. Sequence analysis reveals that homologues of HbpS are found in a wide variety of bacteria, including different Actinobacteria and the Gram-negative Vibrio cholera and Klebsiella pneumoniae. The in vivo production of HbpS is greatly increased when S. reticuli is cultured in the presence of the natural antibiotic haemin (Fe3+ oxidized form of haem). Mutational analysis demonstrated that HbpS significantly increases the resistance of S. reticuli to toxic concentrations of haemin. Previous data show that the presence of the newly identified two-component sensor system SenS–SenR also considerably enhances the resistance of S. reticuli to haemin and the redox-cycling compound plumbagin, suggesting a role in the sensing of redox changes. Specific interaction between HbpS and SenS–SenR, which regulates the expression of the catalase–peroxidase CpeB, as well as HbpS, has been demonstrated in vitro. HbpS has been recombinantly overexpressed, purified and crystallized in space group P213, with a cell edge of 152.5 Å. Diffraction data were recorded to a maximal resolution of 2.25 Å and phases were obtained using the SAD method from crystals briefly soaked in high concentrations of sodium bromide.
1. Introduction
Streptomyces is the largest genus of Actinobacteria, with more than 500 species currently documented. The species are found worldwide and are characterized by a complex developmental life cycle and secondary metabolism. Indeed, Streptomyces spp. produce over two-thirds of naturally occurring antibiotics (e.g. neomycin and chloramphenicol). The Gram-positive soil bacterium S. reticuli has recently been shown to produce an extracellularly located novel haem-binding protein named HbpS (Ortiz de Orué Lucana et al., 2004 ▶) and a comparative sequence analysis (Fig. 1 ▶) shows that HbpS shares a high homology with proteins of unknown function from the Gram-positive bacteria S. coelicolor (Bentley et al., 2002 ▶), Rhodococcus sp. RHA1 (McLeod et al., 2006 ▶), Arthrobacter aurescens TC1 (Mongodin et al., 2006 ▶) as well as the pathogenic Gram-negative bacteria Vibrio cholera (Heidelberg et al., 2000 ▶) and Klebsiella pneumoniae (Arakawa et al., 1995 ▶).
Figure 1.
A sequence alignment of HbpS-like proteins colour-coded by sequence conservation (blue, lowest; red, highest). The predicted signal peptides (where appropriate) have been removed to improve the quality of the alignment. The following symbols are used in the ‘conserved’ alignment displayed at the bottom of the figure: ‘.’ weakly conserved, ‘:’ strongly conserved, ‘*’ absolutely conserved. This figure was generated from the sequence alignment performed by the package T-COFFEE (Notredame et al., 2000 ▶).
The production of HbpS is greatly increased in vivo during the cultivation of S. reticuli and S. lividans transformants in the presence of the naturally occurring antibiotic haemin and HbpS has been shown directly to bind haemin (Ortiz de Orué Lucana et al., 2004 ▶). Haemin, a natural porphyrin, is known to possess significant antibacterial activity when augmented by physiological concentrations of hydrogen peroxide or reducing agents (Stojiljkovic et al., 2001 ▶; Baker et al., 2003 ▶); it has therefore been suggested that HbpS-like proteins may play an important role in defence against haemin-based toxicity (Ortiz de Orué Lucana et al., 2004 ▶). Studies have also demonstrated that secretion of HbpS is carried out under the control of the twin-arginine translocation (TAT) pathway, through which folded and oligomeric proteins are translocated across the outer cell membrane (Chaddock et al., 1995 ▶; Berks, 1996 ▶). The native oligomeric state of HbpS is currently unclear as SDS–PAGE analysis of wild-type secreted HbpS indicates the presence of monomeric, dimeric and higher order oligomers (Ortiz de Orué Lucana et al., 2004 ▶).
The disruption of HbpS also induces a significant decrease in the in vivo production of CpeB, a mycelia-associated haem-containing enzyme (Zou et al., 1999 ▶). CpeB has been shown to utilize H2O2 to oxidize a number of substrates via an attached haem group (ferric protoporphyrin) or in a haem-independent reaction, which is coupled to manganese(II)/(III) peroxidation (Zou & Schrempf, 2000 ▶). As the haem-dependent catalase activity of CpeB leads to the dissociation of H2O2 to H2O and O2, it also plays an important part in the detoxification of H2O2.
CpeB shows a high degree of amino-acid identity (62% identity, 73% similarity) to the catalase–peroxidase KatG from Mycobacterium tuberculosis. The front-line anti-tuberculosis drug isoniazid (isonicotinic acid hydrazide) requires KatG activation before exerting a lethal effect (Zhang et al., 1992 ▶) and isoiazid-resistant strains of M. tuberculosis have been shown to possess no detectable levels of KatG, although they acquire a compensatory mutation resulting in an upregulation of expression of an alkyl hydroperoxide reductase protein, AhpC (Sherman et al., 1996 ▶). Sherman and co-workers suggested that KatG would confer protection against H2O2-mediated damage, even in the absence of adequate catalase and peroxidase activities, thus promoting survival of the organism in the environment of the phagocyte oxidative burst.
A specific interaction has been identified between HbpS and the sensor kinase SenS from the recently identified two-component sensor system, SenS–SenR, which negatively regulates the furS–cpeB operon and the hbpS gene (Ortiz de Orué Lucana et al., 2005 ▶; Bogel et al., 2007 ▶). Disruption of the SenS–SenR two-component system also results in an increased sensitivity of S. reticuli to haemin and hydrogen peroxide. The closest available annotated homologue of SenS is the sensor kinase ChrS of Corynebacterium diphtheriae (34% identity, 46% similarity), which has been shown to play a role in the utilization of haem and haemoglobin as iron sources (Schmitt, 1999 ▶).
This homology in HbpS and HbpS interaction partners implies a common functional role across a wide range of bacterial species, including a number of medically relevant bacteria. From the previous research reviewed above, the major role of HbpS-like proteins would appear to be the regulation of the expression of CpeB/KatG-like proteins in response to extracellular redox conditions (i.e. the presence of H2O2, haemin etc.) that are detrimental to bacterial survival. In order to gain further insight into the physiological role of HbpS-like proteins, we have overexpressed recombinant wild-type HbpS from S. reticuli in Escherichia coli for examination and in this report we detail the crystallization and X-ray diffraction characterization of the crystals obtained. The measurement of experimental phases, using a brief halide soak, is described and the current status of the three-dimensional model is given.
2. Materials and methods
2.1. Cleavage of DNA, ligation and agarose gel electrophoresis
The DNAs were cleaved with various restriction enzymes according to the suppliers’ instructions. Ligation was performed with T4 ligase (Promega). Gel electrophoresis was carried out in 0.8–2% agarose gels using Tris–borate–EDTA (TBE) buffer (89 mM Tris, 89 mM boric acid and 2 mM EDTA pH 8.0). E. coli was transformed with plasmid DNA by electroporation (Dower et al., 1988 ▶). Plasmids were isolated from E. coli with the aid of a mini plasmid kit (Qiagen).
2.2. Cloning of the hbpS gene into an E. coli expression plasmid
The hbpS-coding region (without the codons encoding for the signal peptide) was amplified by PCR from the previously described construct pWKS10 (Zou et al., 1999 ▶) using the primers HBP8 (5′-GAGACCATGGCCGACACCACGGAG-3′), consisting of an NcoI restriction site (bold) followed by the sequence encoding the N-terminal amino acids of the mature HbpS (without the signal peptide), and HBP9 (5′-GGGCAAGCTTGTGGCCGAGCACGG-3′) coding for the C-terminal amino acids of HbpS followed by the HindIII restriction site (bold). The PCR product was digested with NcoI and HindIII, ligated with NcoI/HindIII-cleaved pETM11 and subsequently transformed into E. coli XL1-blue. Sequencing of the resulting plasmid (pETHbpS) confirmed the presence of the hbpS gene in frame fusion with the His6 tag and the TEV-protease cleavage site.
2.3. Production and purification of HbpS
The plasmid pETHbpS was transformed into E. coli strain BL21 (DE3) pLysS. Protein expression was induced at OD600 = 0.6 at 310 K with 1 mM IPTG for 4 h. Cell pellets were resuspended in buffer W (150 mM NaCl, 20 mM Tris–HCl pH 8) containing DNaseI (1 µg ml−1) and then lysed by ultrasonication (Branson sonifier, 5 × 10 s, with 10 s intervals). After centrifugation, the supernatant was incubated with Ni-NTA agarose (Qiagen) in the presence of an additional 25 mM imidazole. The resin was washed with buffer W and the protein was then eluted with buffer W containing an additional 250 mM imidazole. The eluted protein was incubated overnight in the presence of 5 mM DTT and His6-tag-TEV protease for cleavage of the His6 tag. Imidazole and DTT were removed from the protein solution using PD10 columns (Amersham) previously equilibrated in buffer W. A second Ni-NTA affinity chromatography was performed to remove remaining His6 tags, His6-tagged TEV protease and uncleaved His6-tag-HbpS. The solution containing unbound proteins (His6-tag-free HbpS) was then desalted into buffer A (20 mM Tris–HCl pH 7) using PD10 columns (Amersham). HbpS was then further purified by anion-exchange chromatography over a DEAE-Sepharose column previously equilibrated in 20 mM Tris–HCl pH 7. Interestingly, under such conditions HbpS (pI 5.9) does not interact with the matrix and remains in the flowthrough in high purity (over 99% as judged from SDS–PAGE analysis; Fig. 2 ▶). The protein sequence was confirmed by mass-spectrometric analysis (ESI LC-MS) and the protein was concentrated to 14 mg ml−1 for crystallization, as judged from its absorbance at 280 nm (∊o = 8250 cm−1 M −1; MW = 15 498 Da).
Figure 2.
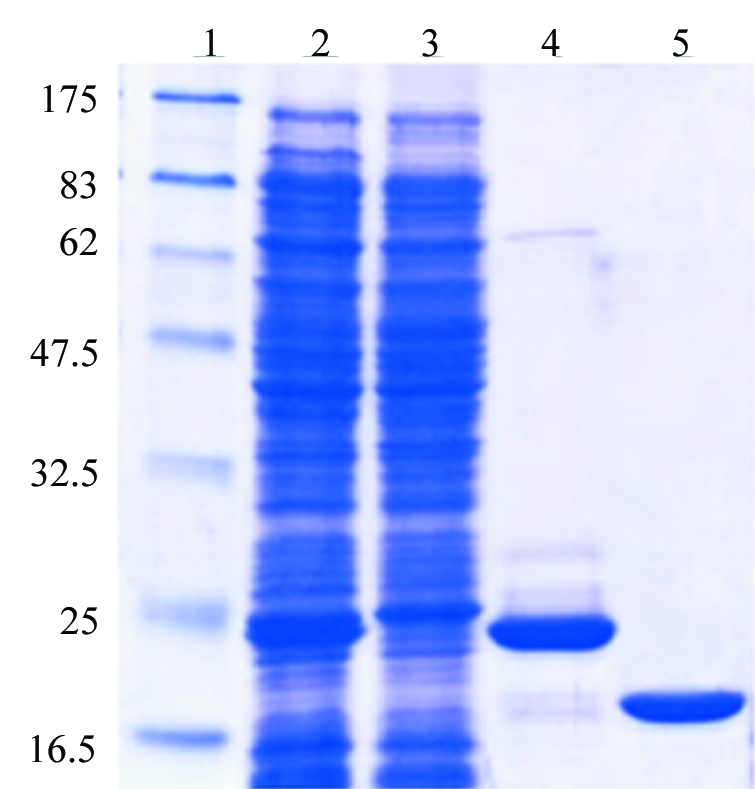
An SDS–PAGE analysis of the purified HbpS. The E. coli BL21 (DE3) pLysS strain containing the plasmid pETHbpS was grown and induced as described in §2. The cytoplasmic fraction containing His6-tag-HbpS (lane 2) was incubated with Ni-NTA agarose. Unbound proteins remained in the flowthrough (lane 3). His6-tag-HbpS was eluted in the presence of 250 mM imidazole (lane 4). The His6 tag was then cleaved using TEV-protease; after a second Ni-NTA affinity chromatography, HbpS was then further purified by anion-exchange chromatography through DEAE-Sepharose (lane 5). The molecular weight (in kDa) of each of the protein markers is indicated (lane 1). A total of 5 mg purified His6-tag-free HbpS was obtained from 1 l of bacterial culture.
3. Results and discussion
3.1. Crystallization of HbpS
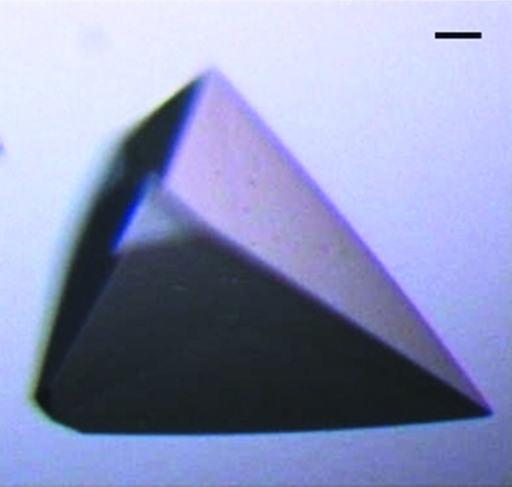
Purified HbpS was submitted to the high-throughput crystallization facility at the EMBL Hamburg (Mueller-Dieckmann, 2006 ▶) at the concentration described above. Sitting-drop sizes were 200 + 200 nl, equilibrated over 50 µl screen reagent. A large number of conditions (32 of 576 conditions screened) resulted in crystals and were screened for diffraction properties on beamline X13, EMBL-Hamburg. After initial diffraction screening, the crystals grown from the cryocondition sparse-matrix screen [40%(v/v) PEG 400, 100 mM MES pH 6.0, 5%(w/v) PEG 3000; Emerald Biostructures Wizard Cryo Screens I and II; deCODE Biostructures] were identified as those providing the highest initial diffraction quality. Subsequently, manual hanging-drop optimization screens were performed around these conditions, varying buffer pH and PEG concentrations, in order to reproduce and improve the crystal size. Optimized crystals exhibited pyramidal morphology and grew to maximum dimensions of 200 × 150 × 150 µm (Fig. 3 ▶). Final optimized conditions were in the range 39–42%(v/v) PEG 400, 100 mM MES pH 5.8–6.5, 5%(v/v) PEG 3000. Owing to the components of the crystallization experiment, no further cryoprotection was required for diffraction experiments at 100 K.
Figure 3.
An image of a typical HbpS crystal grown as described in §3.1. The bar in the upper right corner corresponds to 10 µm.
3.2. Diffraction experiments using native HbpS crystals
A native data set was collected from optimized HbpS crystals on beamline BW7A, EMBL-Hamburg, in dose mode. The collection of initial frames was performed to allow crystal characterization and space-group assignment using the software POINTLESS (Evans, 2006 ▶). Subsequently, the data-collection strategy software BEST (Popov & Bourenkov, 2003 ▶) was used to design a data-collection experiment suitable for the time available on the beamline which would provide a signal-to-noise ratio of over 3 at the edge of the detector used (Fig. 4 ▶; MarCCD-165 mm). A 2.2 Å native data set was collected, with statistics as detailed in Table 1 ▶. Data were processed using XDS and merged in XSCALE (Kabsch, 1988 ▶). A sequence search of the Protein Data Bank (Berman et al., 2000 ▶) using BLAST (Altschul et al., 1990 ▶) revealed that no structures of significant sequence homology were available for use as molecular-replacement models, requiring the collection of experimental phases. Native HbpS crystals were briefly soaked (15 s) in the crystallization well solution containing an additional 1 M sodium bromide (Dauter et al., 2000 ▶). Immediately following the soaking experiments, the crystals were mounted on beamline X12, EMBL-Hamburg, and screened for diffraction. The data-collection strategy software BEST (Popov & Bourenkov, 2003 ▶) was again used to design an SAD data-collection experiment which would provide a signal-to-noise ratio of 4 at the edge of the detector used (MarCCD-225 mm). A 3.4 Å data set was collected at a wavelength above the bromine edge (0.9197 Å), with statistics as detailed in Table 1 ▶. As described above for the native data, derivative data were processed using XDS and merged in XSCALE (Kabsch, 1988 ▶). The solvent content (Matthews, 1968 ▶) was predicted for a number of different oligomeric states, with between six and eight molecules predicted to occupy the asymmetric unit.
Figure 4.
A typical diffraction image obtained from the HbpS native crystals. This image was collected on beamline BW7A, EMBL-Hamburg, under cryoconditions (100 K) from a single crystal mounted directly from the crystallization conditions described. Exposure times were approximately 300 s, dependent upon the storage-ring intensity, for an oscillation range of 0.2°. The incident wavelength was 0.98 Å and the resolution at the edge of the detector was 2.2 Å. Further details of the data-collection parameters are given in Table 1 ▶.
Table 1. Data-collection parameters for the native and sodium-bromide-soaked crystals of HbpS.
Values in parentheses refer to the high-resolution shell of the respective data set.
| Native HbpS | 15 s soak in 1 M NaBr | |
|---|---|---|
| Experimental conditions | ||
| Beamline | BW7A (EMBL Hamburg) | X12 (EMBL-Hamburg) |
| Wavelength (Å) | 0.9778 | 0.9197 |
| Temperature (K) | 100 | 100 |
| Detector | Mar-CCD 165 mm | Mar-CCD 225 mm |
| Crystal parameters | ||
| Space group | P213 | P213 |
| Unit-cell parameters (Å, °) | a = b = c = 152.5, α = β = γ = 90 | a = b = c = 151.9, α = β = γ = 90 |
| Data-collection parameters | ||
| Resolution range (Å) | 20–2.20 (2.40–2.20) | 20–3.4 (3.7–3.4) |
| Mosaicity (°) | 0.18 | 0.33 |
| Crystal-to-detector distance (mm) | 174 | 295 |
| Oscillation range (°) | 0.2 | 0.3 |
| No. of frames | 450 | 140 |
| Data-processing parameters | ||
| No. of measured reflections | 247495 (59161) | 70896 (10638) |
| No. of unique reflections | 37888 (12979) | 29023 (5005) |
| Multiplicity | 6.53 (4.56) | 2.44 (2.12) |
| Completeness (%) | 98.5 (94.9) | 93.2 (76.1) |
| I/σ(I) | 12.63 (3.56) | 9.77 (3.87) |
| Rmerge† (%) | 7.3 (32.1) | 9.2 (26.5) |
| Rmeas‡ (%) | 8.1 (38.6) | 11.4 (33.5) |
| Mean anomalous difference§ | n/a | 1.127 (0.854) |
Merging R factor, R
merge = 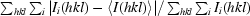 .
.
Redundancy-independent R factor, R
meas = 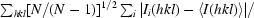
 (Diederichs & Karplus, 1997 ▶).
(Diederichs & Karplus, 1997 ▶).
Mean anomalous difference in units of its estimated standard deviation [|F(+) − F(−)|/σ]. F(+), F(−) are structure-factor estimates obtained from the merged intensity observations in each parity class.
3.3. Phase determination
Initial analysis of the anomalous signal from the bromide-soaked crystals indicated the presence of bound bromine ions, with significant peaks available in the Harker sections of the anomalous Patterson maps (Fig. 5 ▶). Subsequently, the package SOLVE (Terwilliger & Berendzen, 1999 ▶) was used to identify 20 independent bromine sites with an overall figure of merit of 0.21. Refinement of the experimental phases and automated building of the sequence into the resulting electron-density maps were performed using RESOLVE (Terwilliger, 2000 ▶, 2003 ▶). The initial figure of merit of phasing was 0.42 and the final figure of merit after automated building of 777 amino-acid residues was 0.60. Examination of the resulting electron-density maps indicated that HbpS is present as an octomer in the asymmetric unit, indicating that approximately 62% of the structure (777 out of 1248 residues) was built by RESOLVE. Further model building and refinement of the full structure are currently in progress.
Figure 5.
An anomalous Patterson map from data collected on the sodium-bromide-soaked crystals. The map corresponds to the Harker section (Z = 1/2) of the space group P213. The map is contoured from 1σ in 0.5σ steps. Signals originating from multiple bound bromine ions can be distinguished. 20 individual bromide ions were finally used in the phasing protocol of SOLVE (Terwilliger & Berendzen, 1999 ▶). See §3.3 for further details.
Acknowledgments
The authors acknowledge the support of EMBL-Hamburg, in particular that of the scientists responsible for the beamlines BW7A (Dr Andrea Schmidt) and X12 (Dr Manfred S. Weiss). We thank Professor Dr Hildgund Schrempf for stimulating discussions. This work was also supported in part by grant OR 224/1-1 of the Deutsche Forschungsgemeinschaft (DFG).
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol.215, 403–410. [DOI] [PubMed]
- Arakawa, Y., Wacharotayankun, R., Nagatsuka, T., Ito, H., Kato, N. & Ohta, M. (1995). J. Bacteriol.177, 1788–1796. [DOI] [PMC free article] [PubMed]
- Baker, H. M., Anderson, B. F. & Baker, E. N. (2003). Proc. Natl Acad. Sci. USA, 100, 3579–3583.
- Bentley, S. D. et al. (2002). Nature (London), 417, 141–147.
- Berks, B. C. (1996). Mol. Microbiol.22, 393–404. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Bogel, G., Schrempf, H. & Ortiz de Orué Lucana, D. (2007). FEBS J.274, 3900–3913. [DOI] [PubMed]
- Chaddock, A. M., Mant, A., Karnauchov, I., Brink, S., Herrmann, R. G., Klösgen, R. B. & Robinson, C. (1995). EMBO J.14, 2715–2722. [DOI] [PMC free article] [PubMed]
- Dauter, Z., Dauter, M. & Rajashankar, K. R. (2000). Acta Cryst. D56, 232–237. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Dower, W. J., Miller, J. F. & Ragsdale, C. W. (1988). Nucleic Acids Res.16, 6127–6145. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Heidelberg, J. F. et al. (2000). Nature (London), 406, 477–483. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1988). J. Appl. Cryst.21, 916–924.
- McLeod, M. P. et al. (2006). Proc. Natl Acad. Sci. USA, 103, 15582–15587.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mongodin, E. F. et al. (2006). PloS Genet.2, e214. [DOI] [PMC free article] [PubMed]
- Mueller-Dieckmann, J. (2006). Acta Cryst. D62, 1446–1452. [DOI] [PubMed]
- Notredame, C., Higgins, D. & Heringa, J. (2000). J. Mol. Biol.302, 205–217. [DOI] [PubMed]
- Ortiz de Orué Lucana, D., Schaa, T. & Schrempf, H. (2004). Microbiology, 150, 2575–2585. [DOI] [PubMed]
- Ortiz de Orué Lucana, D., Zou, P., Nierhaus, M. & Schrempf, H. (2005). Microbiology, 151, 3603–3614. [DOI] [PubMed]
- Popov, A. N. & Bourenkov, G. P. (2003). Acta Cryst. D59, 1145–1153. [DOI] [PubMed]
- Schmitt, M. P. (1999). J. Bacteriol.181, 5330–5340. [DOI] [PMC free article] [PubMed]
- Sherman, D. R., Mdluli, K., Hickey, M. J., Arain, T. M., Morris, S. L., Barry III, C. E. & Stover, C. K. (1996). Science, 272, 1641–1643. [DOI] [PubMed]
- Stojiljkovic, I., Evavold, B. D. & Kumar, V. (2001). Expert Opin. Investig. Drugs, 10, 309–320. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2003). Acta Cryst. D59, 38–44. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Zhang, Y., Heym, B., Allen, B., Young, D. & Cole, S. (1992). Nature (London), 358, 591–593. [DOI] [PubMed]
- Zou, P., Borovok, I., Ortiz de Orué Lucana, D., Muller, D. & Schrempf, H. (1999). Microbiology, 145, 549–559. [DOI] [PubMed]
- Zou, P. & Schrempf, H. (2000). Eur. J. Biochem.267, 2840–2849. [DOI] [PubMed]