Good-quality crystals of selenomethionine-substituted Msmeg_3380 were obtained by the hanging-drop vapour-diffusion technique and diffracted to 1.2 Å using synchrotron radiation.
Keywords: pyridoxine 5′-phosphate oxidases, Mycobacterium smegmatis, FMN
Abstract
Pyridoxine 5′-phosphate oxidases (PNPOxs) are known to catalyse the terminal step in pyridoxal 5′-phosphate biosynthesis in a flavin mononucleotide-dependent manner in humans and Escherichia coli. Recent reports of a putative PNPOx from Mycobacterium tuberculosis, Rv1155, suggest that the cofactor or catalytic mechanism may differ in Mycobacterium species. To investigate this, a putative PNPOx from M. smegmatis, Msmeg_3380, has been cloned. This enzyme has been recombinantly expressed in E. coli and purified to homogeneity. Good-quality crystals of selenomethionine-substituted Msmeg_3380 were obtained by the hanging-drop vapour-diffusion technique and diffracted to 1.2 Å using synchrotron radiation.
1. Introduction
Pyridoxal 5′-phosphate (PLP; vitamin B6) is required as a cofactor for a number of enzymes involved in amino-acid metabolism (Christen & Mehta, 2001 ▶). In fact, over 1.5% of the proteins in prokaryotes are PLP-dependent and several of these are known drug targets (Amadasi et al., 2007 ▶). The wide distribution and physiological importance of PLP makes understanding the mechanism of PLP regeneration and the participation of pyridoxine 5′-oxidases (PNPOxs) in this process a priority. The X-ray structure of a PNPOx from Escherichia coli has been solved (Safo et al., 2000 ▶) and its catalytic mechanism has been characterized (di Salvo et al., 2003 ▶). Oxidation of the C-4 alcohol (pyridoxine 5′-phosphate) or amine (pyridoxamine 5′-phosphate) group to an aldehyde is catalysed in a flavin mononucleotide (FMN) cofactor-dependent manner, resulting in the formation of PLP. This is thought to occur via direct hydride transfer from the C-4 group to the bound FMN. Such FMN-catalyzed redox chemistry is potentially applicable to the degradation of aromatic compounds, as shown in the reduction of coumarin to 3,4-dihydrocoumarin by the structurally related xenobiotic reductase XenA (Griese et al., 2006 ▶). However, in contrast to the PNPOx reaction, the reduction of coumarin is thought to occur via direct hydride transfer from FMN to C-4 of coumarin.
Recently, the structure of a putative PNPOx from M. tuberculosis, Rv1155, has been solved (Canaan et al., 2005 ▶; Biswal et al., 2005 ▶). However, the identity and function of the cofactor is disputed. Canaan et al. (2005 ▶) demonstrated that up to a concentration of 15 µM, FMN did not bind to Rv1155. In contrast, Biswal et al. (2005 ▶) obtained structures of Rv1155 in complex with FMN as a result of soaking the crystals in 2 mM FMN overnight, although it should be noted that 2 mM is not a physiologically relevant concentration and the enzyme activity was not tested. Therefore, characterization of the cofactor for the PNPOx-like enzymes in Mycobacterium species will require further work before it is well understood. We intend to further investigate the identity of the cofactor, the catalytic mechanism and the potential application of these enzymes for xenobiotic breakdown. Here, we describe the cloning, recombinant expression, purification, crystallization and preliminary X-ray analysis of a putative PNPOx from M. smegmatis, Msmeg_3380, which shares 17% sequence identity with Rv1155 and is predicted to belong to the same family of FMN-dependent PNPOx enzymes.
2. Experimental procedures and results
2.1. Cloning, expression and purification of Msmeg_3380
2.1.1. Cloning
The open reading frame encoding Msmeg_3380 (GenBank accession No. ABK72884) was amplified from M. smegmatis mc2 155 genomic DNA using the following primers: forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAAACC TGTATTTTCAGGGA ATGGTCGCCGTGCCCGAG; reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT CTACTGCTTGCTGAACG. The primers were designed to incorporate Gateway (Invitrogen) recombination sites (bold), a TEV protease-cleavage site (underlined) and start and stop codons (italicized). The amplified product was purified using a Qiagen gel-purification kit and recombined into pDONR201 by Lysogenic (BP) Gateway recombination. After sequence confirmation (Micromon, Australia), Msmeg_3380 was recombined into the pDEST17 expression vector by Lytic (LR) Gateway recombination. Recombination into the pDEST17 vector resulted in the addition of an N-terminal hexahistidine tag to the protein.
2.1.2. Expression of native and selenomethionine-substituted Msmeg_3380
The Msmeg_3380 pDEST 17 expression construct was transformed into E. coli BL21-AI cells (Invitrogen) and plated onto LB media supplemented with 100 mg l−1 ampicillin (Sigma). A single colony was picked from a plate and used to inoculate a 50 ml LB seed culture supplemented with 100 mg l−1 ampicillin. The seed culture was incubated with shaking at 310 K for 16 h and diluted 1:20 to inoculate a 1 l expression culture. The expression culture was incubated with shaking at 310 K and protein expression was induced at a cell OD of 600 by the addition of 0.2% arabinose. After 4 h of further growth, cells were harvested by centrifugation. For the preparation of selenomethionine-substituted (SeMet) Msmeg_3380, cells were grown in minimal media as described by Van Duyne et al. (1993 ▶).
2.1.3. Purification of native and SeMet Msmeg_3380
Both native and SeMet Msmeg_3380 were purified using immobilized metal-ion affinity chromatography (IMAC) followed by anion-exchange chromatography. Cells were harvested by centrifugation (8000g for 15 min at 277 K) and pellets were resuspended in IMAC equilibration/wash buffer (20 mM HEPES pH 7.4, 300 mM NaCl) containing 1% BugBuster (Novagen) for cell lysis. The resuspended cell pellet was incubated at room temperature for 30 min. The soluble fraction was separated from insoluble material by centrifugation (38 000g for 45 min at 277 K) and loaded onto a pre-equilibrated 5 ml Ni–NTA column (Qiagen). Captured Msmeg_3380 was eluted with an imidazole gradient of 0–500 mM. Fractions containing Msmeg_3380 were pooled and exchanged into TEV protease-cleavage buffer using a desalting column (GE Biosciences; Fig. 1 ▶). The proteins were incubated at 287 K for 3 d. After TEV cleavage, one glycine residue remained before the initial methionine. The proteins were then exchanged into low-salt buffer (20 mM Tris pH 8.0) for anion exchange using a desalting column before being passed over a 5 ml DEAE Sepharose column (GE Biosciences). Msmeg_3380 was eluted with a gradient of 0–1 M NaCl. Fractions containing cleaved Msmeg_3380 were pooled and exchanged into IMAC equilibration buffer using a desalting column. The protein was loaded onto a 5 ml Ni–NTA column and the flowthrough containing cleaved Msmeg_3380 was collected. Finally, Msmeg_3380 was exchanged into crystallization buffer (10 mM Tris pH 8.0, 100 mM NaCl) using a desalting column and concentrated to 10 mg ml−1. Protein concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) by measurement of the absorbance at 280 nm using an extinction coefficient of 20 340 M −1 cm−1.
Figure 1.
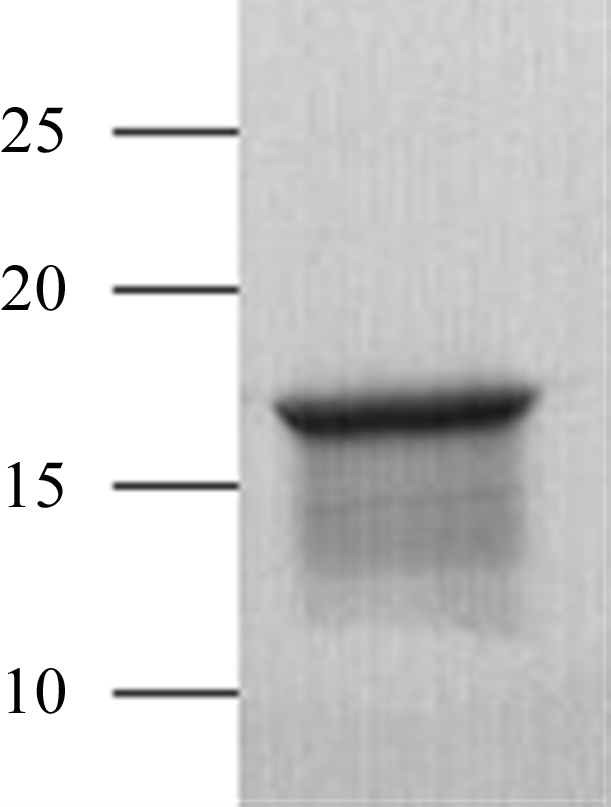
10–20% SDS–PAGE showing the purity of Msmeg_3380. The positions of molecular-weight markers are labelled (kDa).
Incorporation of SeMet into Msmeg_3380 was confirmed by liquid-chromatography electrospray ionization time-of-flight (LC-ESI-TOF) mass spectrometry on a 1100 LC-MSD-TOF mass spectrometer (Agilent). The calculated masses were 18 009.6 and 18 150.5 for native and SeMet Msmeg_3380, respectively; a mass difference of 140.7. This equates to the complete substitution of all three Met residues in Msmeg_3380 with SeMet. Additionally, mass spectrometry suggested that no FMN was bound to the purified enzyme.
2.2. Crystallization
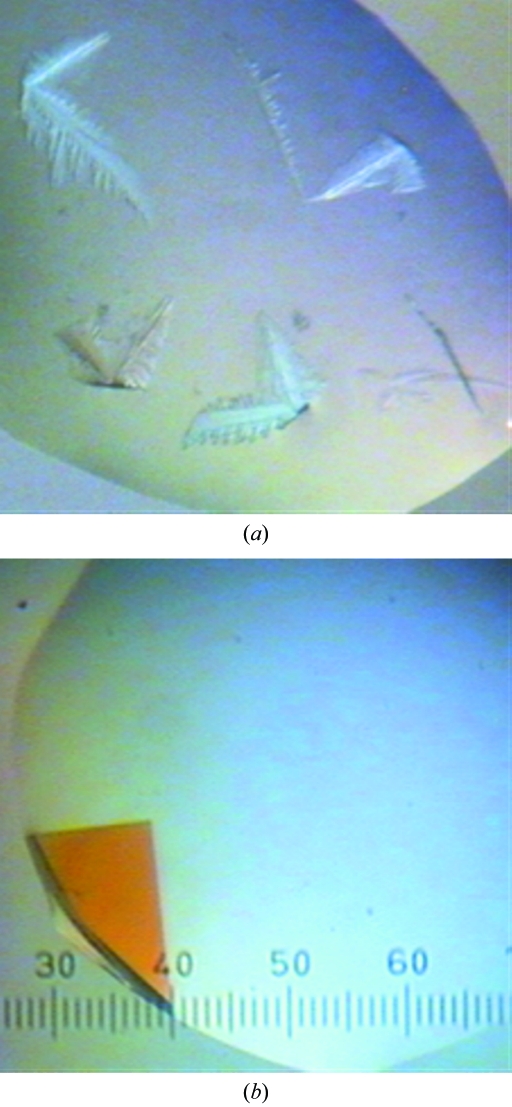
Initial crystallization experiments with native Msmeg_3380 were performed using a Cartesian Honeybee nanolitre-dispensing robot and the Qiagen PEG Suite, JCSG+ Suite and pHclear Suite crystallization screens. Equal volumes (0.3 µl) of protein and precipitant solution were mixed in a sitting-drop vapour-diffusion experiment using 96-well Intelliplates at 291 K (Robbins Scientific Equipment) with a reservoir volume of 100 µl. Dendritic crystals (Fig. 2 ▶ a) formed in several conditions overnight. A grid screen, using the hanging-drop vapour-diffusion technique and a reservoir volume of 1 ml, was performed around the most promising condition (20% PEG 10K, 0.1 mM Tris–HCl pH 8.5). The ratio of protein solution to precipitant solution was varied (0.5:2.5–2.5:0.5 µl), producing large single crystals (Fig. 2 ▶ b). The crystallization conditions used for SeMet Msmeg_3380 were identical. These crystals were too fragile to be transferred to cryosolvent (35% PEG 10K, 0.1 M Tris–HCl pH 8.5). Instead, the mother liquor was gradually replaced with cryosolvent by pipetting. Crystals were flash-cooled in a stream of nitrogen gas (100 K).
Figure 2.
Crystal forms of Msmeg_3380 in 20% PEG 10K, 0.1 mM Tris–HCl pH 8.5. (a) Dendrites, (b) a single crystal. The scale is 0.025 mm per gradation.
2.3. Data collection and processing
X-ray diffraction data were collected from crystals of SeMet Msmeg_3380 at the Australian Synchrotron (Melbourne, Australia). One data set consisting of 360 images was collected (10 s exposures) on a MAR SX-165 CCD detector (MAR-USA). Intensity data were collected at 100 K using a wavelength corresponding to the SeMet edge (0.9793 Å) and a 1.0° oscillation angle per image. Diffraction data were integrated and scaled using the programs MOSFLM (Leslie, 1992 ▶) and SCALA (Evans, 1997 ▶). Details of the data collection are given in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | Synchrotron |
| Space group | P212121 |
| Unit-cell parameters (Å) | |
| a | 56.90 |
| b | 65.35 |
| c | 69.75 |
| Matthews coefficient (Å3 Da−1) | 2.22 |
| Solvent content (%) | 45.3 |
| Data collection | |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.9793 |
| Resolution (Å) | 22.03–1.20 (1.26–1.20) |
| Total reflections | 1033563 (143602) |
| Unique reflections | 80647 (11419) |
| Multiplicity | 12.8 (12.6) |
| 〈I/σ(I)〉 | 21.6 (4.4) |
| Completeness (%) | 98.7 (97.0) |
| Rmerge† (%) | 6.7 (56.1) |
| Anomalous completeness | 98.0 (95.9) |
| Anomalous multiplicity | 6.7 (6.5) |
R
merge = 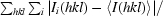
 , where the summations are over i observations of each reflection and all hkl. 〈I(hkl)〉 is the average intensity of the i observations.
, where the summations are over i observations of each reflection and all hkl. 〈I(hkl)〉 is the average intensity of the i observations.
3. Results and discussion
We have obtained crystals of Msmeg_3380 and collected a diffraction data set to 1.2 Å resolution (Table 1 ▶). Based on systematic absences, the crystals were assigned to the orthorhombic space group P212121. Cell-content analysis indicated a Matthews coefficient of 2.23 Å3 Da−1 and a solvent content of 45.3% assuming the presence of a dimer in the asymmetric unit. This is consistent with other dimeric PNPOx structures (Canaan et al., 2005 ▶; Biswal et al., 2005 ▶) and was confirmed by the observation of a large non-origin peak in the self-rotation function of the orthorhombic crystals on the κ = 180° plane, representing a NCS twofold axis at (θ, ϕ) = (103.2, 170.5°) in polar angles. The large anomalous signal obtained from SeMet labelling makes it probable that this structure could be solved using SAD phasing. Owing to the low sequence identity to the currently known structures of this family (<22%), it is unlikely that molecular replacement will be possible. Phasing efforts are currently under way and the structure will be reported elsewhere.
Acknowledgments
We thank the Australian Synchrotron for beam time and Tom Caradoc-Davies for assistance. This work formed part of the Grain Protection Genes initiative of CSIRO and the Australian Grains Research and Development Corporation.
References
- Amadasi, A., Bertoldi, M., Contestabile, R., Bettati, S., Cellini, B., di Salvo, M. L., Borri-Voltattorni, C., Bossa, F. & Mozzarelli, A. (2007). Curr. Med. Chem.14, 1291–1324. [DOI] [PubMed]
- Biswal, B. K., Cherney, M. M., Wang, M., Garen, C. & James, M. N. G. (2005). Acta Cryst. D61, 1492–1499. [DOI] [PubMed]
- Canaan, S., Sulzenbacher, G., Roig-Zamboni, V., Scappuccini-Calvo, L., Frassinetti, F., Maurin, D., Cambillau, C. & Bourne, Y. (2005). FEBS Lett.579, 215–221. [DOI] [PubMed]
- Christen, P. & Mehta, P. K. (2001). Chem. Rec.1, 436–447. [DOI] [PubMed]
- di Salvo, M. L., Safo, M. K., Musayev, F. N., Bossa, F. & Schirch, V. (2003). Biochim. Biophys. Acta, 1647, 76–82. [DOI] [PubMed]
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.33, 22–24.
- Griese, J. J., Jakob, R., Schwarzinger, S. & Dobbek, H. (2006). J. Mol. Biol.361, 140–152. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Safo, M. K., Mathews, I., Musayev, F. N., di Salvo, M. L., Thiel, D. J., Abraham, D. J. & Schirch, V. (2000). Structure, 8, 751–762. [DOI] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol.229, 105–124. [DOI] [PubMed]