The crystal structure of an acetyltransferase encoded by the gene PA1377 from Pseudomonas aeruginosa has been determined at 2.25 Å resolution. Comparison with a related acetyltransferase revealed a structural difference in the active site that was taken to reflect a difference in substrate binding and/or specificity between the two enzymes.
Keywords: PA1377, Pseudomonas aeruginosa, pitax, acetyltransferase
Abstract
Gene PA1377 from Pseudomonas aeruginosa encodes a 177-amino-acid conserved hypothetical protein of unknown function. The structure of this protein (termed pitax) has been solved in space group I222 to 2.25 Å resolution. Pitax belongs to the GCN5-related N-acetyltransferase family and contains all four sequence motifs conserved among family members. The β-strand structure in one of these motifs (motif A) is disrupted, which is believed to affect binding of the substrate that accepts the acetyl group from acetyl-CoA.
1. Introduction
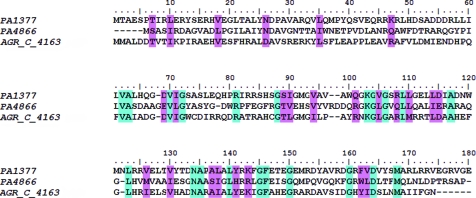
When the genome of Pseudomonas aeruginosa PAO1 was first published in 2000, approximately 45% of the ORFs were classed as representing genes for which no function could be determined (Stover et al., 2000 ▶; http://www.pseudomonas.com). Recently, we and others reported the crystal structure of a conserved hypothetical protein from P. aeruginosa (locus ID PA4866, termed pita; Davies et al., 2005 ▶, 2007 ▶; PDB code 1yvo; B. Nocek, X. Xu, A. Savchenko, A. Edwards & A. Joachimiak, unpublished work). Pita belongs to the GNAT (GCN5-related N-acetyltransferase) family and efforts have been made to identify its function (Davies et al., 2007 ▶). When the pita sequence was used to conduct a BLAST search of the Pseudomonas genome, a 177-amino-acid protein displaying 28% sequence identity to pita was returned (Fig. 1 ▶). This protein (locus ID PA1377, termed pitax) is also a conserved hypothetical protein. The function of pitax is unknown; however, preliminary studies (F. Chauviac, R. Tata & P. Brown, unpublished data) have revealed that it is an acetyltransferase, catalysing the transfer of an acetyl group from acetyl-CoA to an acceptor substrate. In this communication, we report the crystal structure of pitax to 2.25 Å resolution and compare it with that of pita.
Figure 1.
Multiple sequence alignment. PA1377, P. aeruginosa conserved hypothetical protein (pitax); accession No. Q9I3W7. PA4866, P. aeruginosa conserved hypothetical protein (pita); accession No. Q9HUU7. AGR_C_4163, A. tumefaciens probable acetyltransferase; accession No. Q8UD38. Residues coloured light blue are identical, while those coloured light pink display 70% conservation. The multiple sequence alignment was performed using ClustalW (Thompson et al., 1994 ▶) and the figure was generated with BioEdit (Hall, 1999 ▶).
2. Materials and methods
2.1. Cloning, protein expression, purification and crystallization
PA1377 (pitax) was amplified by PCR using primers 1377F (5′-GGATGACCCATATGACCGCCGAATCGCCGAC-3′) and 1377R (5′-AAAAGGATCCGTCGCGCCCGCCCCTCACT-3′) with chromosomal DNA from P. aeruginosa PAC1 as template. Amplifications were performed in 50 µl diluted RedTaq Readimix (Sigma Ltd) with 0.2 mM primers and 100 ng chromosomal DNA. The program was as follows: 60 s at 367 K, followed by 30 cycles of 30 s at 367 K, 30 s at 331 K, 60 s at 345 K and then 5 min at 345 K. The PCR product was cut with BamHI and NdeI and ligated with BamHI/NdeI-cut pET24a (Novagen Ltd). The ligation mixture was used to transform Escherichia coli JM109 and transformants containing recombinant plasmids were selected on LBkan medium and identified by colony PCR. After determination of the DNA sequence of the insert, the recombinant plasmid was used to transform E. coli BL21 (DE3).
E. coli BL21 (DE3) (pETpitax) was grown overnight in 1 l LBkan medum containing 1 mM IPTG. Cells were harvested by centrifugation and resuspended in 20 ml 50 mM Tris pH 8.5 and 1 mM EDTA (TE) at 277 K. All subsequent procedures were carried out at 273–277 K. Cells were broken by sonication using an MSE ultrasonic oscillator. 0.25 g streptomycin sulfate was added and the solution was centrifuged for 15 min at 10 000g to remove debris. The supernatant was loaded onto a Q-Sepharose column (2 × 15 cm) equilibrated with TE. Protein was eluted with a 500 ml linear 0–0.5 M gradient of NaCl in TE. Fractions containing pitax were bulked and concentrated by ultrafiltration (Amicon Ultra-15 device, Millipore) to 1 ml. The solution was loaded onto an FPLC MonoBead MonoQ HR 10/10 column equilibrated with TE and eluted with an NaCl gradient (87.5–175 mM) at a flow rate of 2.0 ml min−1. Fractions containing pitax were bulked and concentrated by ultrafiltration to 900 µl. Purification was monitored by SDS–PAGE and pitax was identified by its molecular weight (20 067 Da).
Crystals were grown using the hanging-drop vapour-diffusion method. The reservoir solution contained 500 µl 0.1 M Tris–HCl pH 6.5, 1.6 M ammonium sulfate and 0.1%(w/v) sodium azide. The drops contained 1 µl protein solution at 13 mg ml−1, to which an equal volume of reservoir solution was added. The drops were kept at 291 ± 0.5 K and crystals up to 370 µm in length appeared after 1–2 d. Crystals were flash-cooled in liquid nitrogen using a cryoprotectant comprising 0.1 M Tris–HCl pH 6.5, 1.6 M ammonium sulfate, 30%(v/v) glycerol and 0.1%(w/v) sodium azide.
2.2. Data collection, structure determination and refinement
Data were collected at Station 10.1, Synchrotron Radiation Source, Daresbury, UK (Cianci et al., 2005 ▶), integrated with MOSFLM (Leslie, 1992 ▶) and scaled with SCALA (Collaborative Computational Project, Number 4, 1994 ▶). Data-collection and processing statistics are presented in Table 1 ▶.
Table 1. Data-processing statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.488 |
| Space group | I222 |
| Unit-cell parameters (Å, °) | a = 79.71, b = 110.93, c = 134.48, α = β = γ = 90 |
| Resolution range (Å) | 64.3–2.25 (2.31–2.25) |
| Unique reflections | 28683 (2089) |
| Completeness (%) | 99.9 (99.9) |
| Redundancy | 6.7 (6.6) |
| I/σ(I) | 21.0 (2.8) |
| Rmerge (%) | 9.2 (61.3) |
| Solvent content (%) | 50.0 |
| Molecules per ASU | 3 |
| Wilson B factor (Å2) | 39.5 |
The pitax sequence was used to perform a BLAST search against the Protein Data Bank (Berman et al., 2000 ▶) in order to find a suitable search model for molecular replacement, as early attempts to solve the structure using the coordinates of pita were not successful. A probable acetyltransferase from Agrobacterium tumefaciens (gene AGR_C_4163; PDB code 2ge3; C. Chang, X. Xu, J. Gu, A. Savchenko, A. Edwards & A. Joachimiak, unpublished work) displaying 43% sequence identity to pitax was chosen (Fig. 1 ▶). Protein atoms from one subunit of the 2ge3 structure were used as a search model and the structure was solved with MOLREP (Vagin & Teplyakov, 1997 ▶). CNS v. 1.1 (Brünger et al., 1998 ▶) was used for refinement and manual model building was performed with QUANTA (Molecular Simulations Inc., 2000 ▶) and Coot (Emsley & Cowtan, 2004 ▶) using σA-weighted 2F obs − F calc and F obs − F calc electron-density maps. Protein atoms were modelled before water molecules, glycerol molecules, azide and sufate ions were incorporated into the structure. Topology and parameter files for glycerol and azide were obtained from the HIC-Up database (Kleywegt & Jones, 1998 ▶). The three molecules of the asymmetric unit were refined independently. The refined model was analysed using PROCHECK (Laskowski et al., 1993 ▶), SFCHECK (Vaguine et al., 1999 ▶), CNS (Brünger et al., 1998 ▶), Coot (Emsley & Cowtan, 2004 ▶) and MolProbity (Lovell et al., 2003 ▶). The refined model comprised protein residues 7–52 and 54–170 for chain A, 6–52 and 54–169 for chain B, and 5–52 and 54–170 for chain C. The model also contained 265 water molecules, five glycerol molecules, six sulfate ions and four azide ions. Refinement statistics are presented in Table 2 ▶.
Table 2. Refinement statistics.
| Resolution range (Å) | 64.3–2.25 |
| No. of reflections | 28683 |
| No. of protein atoms per ASU | 3843 |
| No. of water molecules per ASU | 265 |
| Average B factor (Å2) | |
| Protein atoms | 34.8 |
| Water molecules | 38.1 |
| Glycerol molecules | 52.6 |
| Azide ions | 50.3 |
| Sulfate ions | 50.3 |
| Rcryst (%) (all reflections) | 19.1 |
| Rfree (%) (5% of reflections) | 22.6 |
| R.m.s.d. bond lengths (Å) | 0.014 |
| R.m.s.d. bond angles (°) | 1.37 |
| σA coordinate error (Å) | 0.29 |
| Ramachandran plot analysis (structure analysed with MolProbity) | |
| No. of residues in favoured regions (%) | 97.9 |
| No. of residues in allowed regions (%) | 99.8 |
| Outliers (%) | 0.2 |
2.3. DALI server search
The pitax dimer was used to perform a structural alignment search with the DALI server (Holm & Sander, 1994 ▶) and hits from the search were returned in order of Z score. The Z score represents the statistical significance of the structural alignment and a score of less than 2 indicates structural dissimilarity (Holm & Sander, 1994 ▶). 128 alignments with Z scores ranging from 2 (the lowest) to 20.1 (the highest) were returned from the search; of these, structures with alignments with Z scores above 5 (55 structures in total) were used for more detailed comparison with pitax.
3. Results and discussion
3.1. Pitax is a GNAT-family member
Searching the Pfam database (Finn et al., 2006 ▶) revealed that pitax is a member of the GCN5-related N-acetyltransferase (GNAT) superfamily (PF00583). Four conserved sequence motifs (motifs A–D) have been identified in GNAT-family members (Neuwald & Landsman, 1997 ▶) and examination of the sequence of pitax confirmed the presence of all four of these motifs.
3.2. Dimerization
Many GNAT-family members exist in a dimeric form [e.g. pita from P. aeruginosa (Davies et al., 2007 ▶), glucosamine-6-phosphate N-acetyltransferase GNA1 from Saccharomyces cerevisiae (Peneff et al., 2001 ▶), aminoglycoside acetyltransferase AAC9(6′)Ii from Enterococcus faecium (Burk et al., 2003 ▶) and RimL N α-acetyltransferase from Salmonella typhimurium (Vetting et al., 2005 ▶)]. The crystal structure of pitax suggests that it may also exist as a dimer. In pitax, two of the three molecules (A and B) of the asymmetric unit form a dimer and are related to one another by an approximate twofold rotation axis of 179.2° (Fig. 2 ▶), burying an area of approximately 2600 Å2 at their interface, as calculated by CNS (Brünger et al., 1998 ▶). The third molecule (C) of the asymmetric unit forms a structurally similar dimer with another molecule related by twofold crystallographic symmetry.
Figure 2.
The pitax dimer. Molecules A and B of the asymmetric unit are shown. The GNAT motifs defined by Neuwald & Landsman (1997 ▶) are coloured, from the N-terminus to the C-terminus, as follows: purple (motif C), green (motif D), blue (motif A) and red (motif B). All other atoms are coloured grey. This figure was generated with PyMOL (DeLano, 2002 ▶).
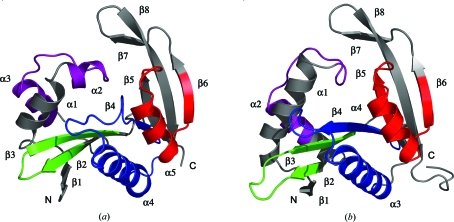
3.3. Overall fold
Pitax is an α/β-fold protein and its overall structure is best described as two regions of antiparallel β-sheet, each flanked by α-helices (Fig. 3 ▶ a). The first antiparallel β-sheet (strands β1–4) wraps around helix α4 on one side and is flanked on its opposite face by helices α1–3. The second β-sheet (strands β5–8) is flanked by helix α5. The four GNAT-superfamily motifs are highlighted in Figs. 2 ▶ and 3 ▶(a).
Figure 3.
Structure of the pitax and pita monomers. (a) Structure of the pitax monomer. α-Helices and β-strands are labelled α1–5 and β1–8. Molecule B of the structure is shown. (b) Structure of the pita monomer. α-Helices and β-strands are labelled α1–4 and β1–8. Coordinates from molecule B of the pita native structure are shown (PDB code 2j8m; Davies et al., 2007 ▶). In both (a) and (b) the GNAT motifs defined by Neuwald & Landsman (997) are coloured, from the N-terminus to the C-terminus, as follows: purple (motif C), green (motif D), blue (motif A) and red (motif B). All other atoms are coloured grey. This figure was generated with PyMOL (DeLano, 2002 ▶).
Comparison of the structures of pitax and pita showed that they are similar (r.m.s.d. of 2.1 Å over 163 Cα atoms; Fig. 3 ▶). However, structural differences were observed in regions contained within motifs C and A. In pitax motif C comprises two helices (α2 and α3) separated by a loop region (Fig. 3 ▶ a), whereas in pita this motif comprises a loop region followed by a helix (α2) (Fig. 3 ▶ b). Structural variation in motif C is not unprecedented as this motif is not as well conserved as the other motifs among GNAT-family members, at least in terms of sequence similarity (Neuwald & Landsman, 1997 ▶). Motif A, on the other hand, is highly conserved (Neuwald & Landsman, 1997 ▶). This motif comprises a β-strand followed by an α-helix and some of its residues provide important contacts for binding of acetyl-CoA (Modis & Wierenga, 1998 ▶; Angus-Hill et al., 1999 ▶; Wybenga-Groot et al., 1999 ▶). The β-strand and α-helix comprising motif A (β4 and α3) are clearly visible in pita (Fig. 3 ▶ b), whereas in pitax, while helix α4 is present, strand β4 is considerably shortened (Fig. 3 ▶ a). Since the catalytic site of pita lies in this region, this difference may well be significant in determining the location or specificity of the acetyl acceptor-binding site.
3.4. The conformation of strand β4 in pitax may affect substrate binding
Preliminary kinetic studies have demonstrated that pitax is an acetyltransferase and weakly catalyses the acetylation of l-methionine and l-methionine sulfoxide (F. Chauviac, R. Tata & P. Brown, unpublished data). The substrates of pitax bear some structural similarity to the substrates of pita: l-methionine sulfoximine and l-methionine sulfone. Because of the overall structural similarity between these proteins and their substrates, the structure of pitax was superposed onto the structure of pita in complex with l-methionine sulfoximine (Davies et al., 2007 ▶) in order to try to locate the substrate-binding site of pitax (Fig. 4 ▶).
Figure 4.
The location of the pitax active site may differ from that in pita. The structure of pitax (yellow) was superposed globally onto the structure of pita in complex with one of its substrates, l-methionine sulfoximine (blue; PDB code 2j8r; Davies et al., 2007 ▶). Residues are numbered according to their respective protein sequences. Met91 in pitax clearly occludes the equivalent region occupied by l-methionine sulfoximine (labelled MSX) in pita. This figure was generated with PyMOL (DeLano, 2002 ▶).
In pitax, strand β4, which constitutes part of motif A, is disrupted. The protein main chain deviates from the plane of the β-strand (in pita) and causes a methionine residue (Met91) to occlude the region that in pita is the acceptor substrate-binding site (Fig. 4 ▶). Thus, either the location of the substrate-binding site in pitax must be different from that of pita or a conformational change is required to allow substrate to bind.
A surface representation of pitax generated in PyMOL (DeLano, 2002 ▶) using protein atoms only revealed the presence of a pocket. When the structure of a probable acetyltransferase from A. tumefaciens in complex with acetyl-CoA (PDB code 2ge3; Chang et al., unpublished work) was superposed onto pitax, acetyl-CoA was found to occupy this pocket. However, this region in pitax putatively occupied by acetyl-CoA did not provide any clues as to where the acceptor substrate might bind, as potential access to substrate by the acetyl group was blocked by the presence of Met91. However, Met91 is flanked by two glycine residues, which may facilitate conformational change, allowing the acetyl group access to a bound substrate molecule.
The structures returned from the DALI server search were compared with pitax in order to investigate whether any other acetyltransferase possessed a similar conformation to that of the disrupted strand β4. However, it was found that while the majority contained small differences in strand β4, none of these caused the plane of the β-strand to deviate. The only protein with a similar conformation to pitax in this particular region was FemA from Staphylococcus aureus (Z score 10, r.m.s.d. 3.7 Å, 10% sequence identity over 138 residues; Benson et al., 2002 ▶). FemA belongs to the N-acetyltransferase clan (CL0257 in the Pfam database; a clan being a group of families with a single evolutionary origin; Finn et al., 2006 ▶) but does not bind acetyl-CoA.
4. Conclusions
We have solved the crystal structure of a conserved hypothetical protein (PA1377, termed pitax) from P. aeruginosa to 2.25 Å resolution. We are examining further the substrate specificity of pitax and will soak putative substrates into crystals in order to obtain enzyme–substrate complex structures. Residues important for substrate binding and catalysis will also be studied by site-directed mutagenesis. Parallel studies to determine the physiological role of pitax are being carried out by observing the properties of a mutant bacterium in which the pitax gene has been inactivated.
Supplementary Material
PDB reference: pitax, 2vi7, r2vi7sf
Acknowledgments
We thank Michele Cianci and Mark Ellis (SRS) and Mary Holdom (King’s College London) for assistance during data collection.
References
- Angus-Hill, M. L., Dutnall, R. N., Tafrov, S. T., Sternglanz, R. & Ramakrishnan, V. (1999). J. Mol. Biol.294, 1311–1325. [DOI] [PubMed]
- Benson, T. E., Prince, D. B., Mutchler, V. T., Curry, K. A., Ho, A. M., Sarver, R. W., Hagadorn, J. C., Choi, G. H. & Garlick, R. L. (2002). Structure, 10, 1107–1115. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Burk, D. L., Ghuman, N., Wybenga-Groot, L. E. & Berghuis, A. M. (2003). Protein Sci.12, 426–437. [DOI] [PMC free article] [PubMed]
- Cianci, M. et al. (2005). J. Synchrotron Rad.12, 455–466. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davies, A. M., Tata, R., Agha, R., Sutton, B. J. & Brown, P. R. (2005). Proteins, 61, 677–679. [DOI] [PubMed]
- Davies, A. M., Tata, R., Beavil, R. L., Sutton, B. J. & Brown, P. R. (2007). Biochemistry, 46, 1829–1839. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Finn, R. D., Mistry, J., Schuster-Böckler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., Moxon, S., Marshall, M., Khanna, A., Durbin, R., Eddy, S. R., Sonnhammer, E. L. L. & Bateman, A. (2006). Nucleic Acids Res.34, D247–D251. [DOI] [PMC free article] [PubMed]
- Hall, T. A. (1999). Nucleic Acids Symp. Ser.41, 95–98.
- Holm, L. & Sander, C. (1994). Nucleic Acids Res.22, 3600–3609. [PMC free article] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1998). Acta Cryst. D54, 1119–1131. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EAMCB Newsl. Protein Crystallogr.26
- Lovell, S. C., Davis, I. W., Arendall, W. B. III, de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S. & Richardson, D. C. (2003). Proteins, 50, 437–450. [DOI] [PubMed]
- Modis, Y. & Wierenga, R. (1998). Structure, 6, 1345–1350. [DOI] [PubMed]
- Molecular Simulations Inc. (2000). QUANTA2000. Molecular Simulations Inc., San Diego, USA.
- Neuwald, A. F. & Landsman, D. (1997). Trends Biochem. Sci.22, 154–155. [DOI] [PubMed]
- Peneff, C., Mengin-Lecreulx, D. & Bourne, Y. (2001). J. Biol. Chem.276, 16328–16334. [DOI] [PubMed]
- Stover, C. K. et al. (2000). Nature (London), 406, 959–964.
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Vetting, M. W., de Carvalho, L. P. S., Roderick, S. L. & Blanchard, J. S. (2005). J. Biol. Chem.280, 22108–22114. [DOI] [PubMed]
- Wybenga-Groot, L. E., Draker, K., Wright, G. D. & Berghuis, A. M. (1999). Structure, 7, 497–507. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: pitax, 2vi7, r2vi7sf