The crystal structure of the plasmid-mediated class C β-lactamase ACT-1 has been solved at 2.4 Å resolution.
Keywords: AmpC, ACT-1, β-lactamases
Abstract
The crystallographic structure of ACT-1, which is the first plasmid-mediated AmpC-type β-lactamase to have been completely analyzed in terms of nucleotide sequence and which has a high degree of sequence similarity to the chromosomal AmpC enzymes of Enterobacter cloacae and the plasmid-encoded MIR-1, has been solved at 2.4 Å resolution. The overall structure of ACT-1 is similar to those of other class C β-lactamases, such as the AmpC enzymes from E. cloacae P99 and Escherichia coli.
1. Introduction
The production of β-lactamases that hydrolyze and inactivate β-lactam antibiotics is the most widespread resistance mechanism to β-lactams such as penicillins, cephalosporins and related molecules (Wilke et al., 2005 ▶). β-Lactamases are classified into four classes, A, B, C and D, according to their primary structures (Ambler, 1980 ▶). In terms of substrate specificity, class C β-lactamases are classified into group 1 by the functional classification of Bush and coworkers and are described as cephalosporinases that are not inhibited by clavulanate (Bush et al., 1995 ▶). Class C enzymes were originally regarded as chromosomal enzymes produced by genera such as Enterobacter, Pseudomonas, Citrobacter and Serratia and were thus regarded as clinically less important enzymes. However, more than 20 plasmid-encoded class C enzymes have been identified in the last two decades (Philippon et al., 2002 ▶). Clinical isolates harbouring the plasmid-mediated class C enzymes have an increasing resistance toward β-lactam antibiotics such as cephamycin and monobactam. Many strains with plasmid-mediated AmpC enzymes also produce class A enzymes and plasmids encoding AmpC enzymes often carry multiple resistance, including resistance to aminoglycosides, chloramphenicol, sulfonamide, tetracycline, trimethoprim and mercuric ions (Philippon et al., 2002 ▶).
ACT-1 is the first plasmid-mediated AmpC-type β-lactamase derived from Enterobacter to have had its nucleotide sequence completely analyzed (Bradford et al., 1997 ▶). ACT-1 shares 86% identity to the ampC β-lactamase gene from E. cloacae P99, with 88% identity at the amino-acid level. It has also been revealed that ACT-1 has high similarity to plasmid-encoded MIR-1, with 91% amino-acid identity (Jacoby & Tran, 1999 ▶). Precise analysis of ACT-1 showed that the enzyme exhibited kinetic properties that were typical of class C β-lactamases as a whole, while its k cat/K m for cephaloridine was lower than that of AmpC from E. cloacae P99, the chromosomal counterpart of ACT-1 (Bauvois et al., 2005 ▶). The k cat/K m values of ACT-1 and of its chromosomal counterpart were 26 and 23 µM −1 s−1 for benzylpenicillin, 12 and 20 µM −1 s−1 for cephalothin and 18 and 31 µM −1 s−1 for nitrocefin, respectively, but the values for cephaloridine were 1.3 and 10 µM −1 s−1. Here, we describe the crystal structure of ACT-1 at 2.4 Å resolution and describe its structural features.
2. Material and methods
2.1. Bacterial strains and plasmids
Escherichia coli strains CJ236 [dut1 ung1 thi-1 relA1/pCJ105(F′ cam r)], MV1184 {ara, Δ(lac-proAB), rpsL, thi (ϕ80 lacZΔM15), Δ(srl-recA)306::Tn10(tet r)/F′[traD36, proAB+, lacIq, lacZΔM15]} and BL21(DE3)pLysS [F− dcm ompT hsdSB (rB −mB −) gal, λ(DE3) pLysS(Camr)] were used for site-directed mutagenesis, genetic construction and overproduction of proteins, respectively. The ACT-1 gene was cloned and the plasmid pBC1e was constructed for the expression of ACT-1 as described previously (Bauvois et al., 2005 ▶).
2.2. Construction of a vector for overexpression of mature ACT-1
A new plasmid for the overexpression of mature ACT-1 was constructed by site-directed mutagenesis using the Kunkel method with the MUTA-GENE in vitro mutagenesis kit (BioRad, USA). An oligonucleotide primer, act-nde (5′-ACC TCC TGC TCG GTA CAT ATG ACC CCG ATG TCA GAA-3′), was used to insert an NdeI site and an initiating Met codon at the position corresponding to the cleavage site of the signal peptide. The nucleotide sequence of the mutated gene was confirmed by DNA sequencing with the Taq Dye Deoxy Terminator Cycle Sequencing Kit and a DNA sequencer (model 373A; Applied Biosystems, USA). The fragment was digested with the restriction enzymes NdeI and BamHI and inserted into the plasmid vector pET-28a digested with the same enzymes to yield pET28-ACT1Δss; this plasmid was used for the overexpression of mature ACT-1.
2.3. Expression and purification of the protein
The plasmid pET28-ACT1Δss was transformed into E. coli BL21(DE3)pLysS and protein production was performed in 100 ml 2-TY broth supplemented with 50 µg ml−1 kanamycin. The cells were grown at 303 K for 8 h and protein production was induced by addition of 0.1 mM IPTG when the absorbance of the cell culture at 600 nm was around 0.6. The cells were harvested by centrifugation, dissolved in 10 ml 15 mM Tris–HCl buffer pH 8.0 with 1 mM EDTA and then disrupted by sonication. After centrifugation, the enzyme in the supernatant was purified by ion-exchange chromatography on a SP-Toyopearl column (Tosoh, Japan) in 15 mM Tris–HCl buffer pH 8.0 and eluted with a 0–0.1 M linear NaCl gradient. The purity of the enzyme was assessed to be more than 95% by Coomassie blue staining after SDS–PAGE.
2.4. Crystal preparation and data collection
The purified protein was dialyzed against 5 mM sodium phosphate buffer pH 7.0 or 5 mM Tris–HCl buffer pH 7.0 and concentrated to 10 mg ml−1 for crystallization. Crystallization conditions were screened with Crystal Screens I and II (Hampton Research, USA) at 285 and 293 K and further refinement of the crystallization conditions was performed. The crystals were prepared by the hanging-drop or sitting-drop vapour-diffusion method with reservoir solutions consisting of 30% PEG 4000 in 100 mM Tris–HCl buffer pH 8.5 and 0.2 M sodium acetate at 285 K. Small plate-shaped crystals suitable for X-ray analysis were obtained within eight weeks. X-ray diffraction data were collected at Station NW12A of Photon Factory-AR, High Energy Accelerator Research Organization (KEK, Tsukuba, Japan) with λ = 1.0 Å at 100 K using an ADSC Quantum 210 charge-coupled device (CCD) detector with 1.0° rotation per image. The crystals were cryoprotected in a solution consisting of 20% ethylene glycol and 30% PEG 4000 in 100 mM Tris–HCl buffer pH 8.5 and 0.2 M sodium acetate and then flash-frozen in liquid nitrogen. The reflections were indexed, integrated and scaled using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶).
2.5. Structure determination
The initial model for refinement was the structure of E. cloacae P99 β-lactamase (PDB code 1blt, which has been replaced by 1xx2; Lobkovsky et al., 1993 ▶). The model was subjected to molecular replacement using the MOLREP program from CCP4 (Collaborative Computational Project, Number 4, 1994 ▶). Subsequently, rigid-body refinement, a simulated-annealing protocol with an initial temperature of 2000 K, positional minimization and individual B-factor refinement were performed using the CNS software package (Brünger et al., 1998 ▶). Manual model building was performed with O (Jones & Kjeldgaard, 1995 ▶). The stereochemical quality of the model was monitored periodically using the programs PROCHECK (Laskowski et al., 1993 ▶) and WHATIF (Vriend, 1990 ▶). After modelling of the protein structure, water molecules were automatically picked out using the CCP4 program.
3. Results and discussion
3.1. Overall structure
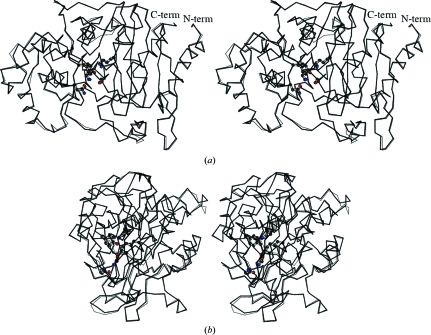
The ACT-1 crystal structure was solved by molecular replacement and refined at 2.4 Å resolution. Four protein molecules were present in the asymmetric unit. The statistics are summarized in Table 1 ▶. Four polypeptide chains A–D form crystallographically independent monomers in the asymmetric unit. The quality of the model was analyzed with the programs PROCHECK and WHATIF. 89.6% of the nonproline and nonglycine residues were in the most favoured regions, 10.1% were in additionally allowed regions and 0.3% were in generously allowed regions of the Ramachandran plot. The model is comprised of 359 residues (Pro2–Leu360) in all four chains. In all the polypeptide chains models were not built for two residues, namely the initiating Met residue and Thr1 in the N-terminal region, as the electron densities for this region were not clear. No disorder was observed in the C-terminal region. The structures of the four independent molecules in the asymmetric unit are almost identical (Fig. 1 ▶). The root-mean-square deviation (r.m.s.d.) for Cα atoms between any two molecules from the four polypeptide chains varies in the range 0.36–0.41 Å. The distance between two corresponding Cα atoms is often larger than 0.8 Å for residues 92–100, 123–128, 135–139, 203–204, 239–243 and 300–301. All of these regions are coincident with the regions with high temperature-factor values. This suggests that these regions, which are conformationally diverse in the four independent ACT-1 molecules, are more flexible than other regions.
Table 1. Crystallographic data and refinement statistics.
Values in parentheses are for the last resolution shell.
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 77.05, b = 80.52, c = 86.88, α = 76.2, β = 68.9, γ = 63.5 |
| Resolution limits (Å) | 20.00–2.40 (2.42–2.40) |
| Unique reflections | 67758 (1658) |
| Completeness | 98.5 (97.9) |
| Redundancy | 2.0 (2.0) |
| Rmerge (%) | 5.4 (17.3) |
| I/σ(I) | 14.8 (3.90) |
| Refinement | |
| No. of protein atoms | 11088 |
| No. of water molecules | 356 |
| Resolution limits (Å) | 20.0–2.4 (2.42–2.40) |
| Total No. of reflections used | 66758 (1361) |
| R factor | 0.239 (0.281) |
| Rfree | 0.247 (0.282) |
| Mean temperature factor (Å2) | |
| Protein atoms | 28.06 |
| Water atoms | 33.20 |
| R.m.s. deviations in bond length (Å) | 0.019 |
| R.m.s. deviations in bond angles (°) | 2.32 |
Figure 1.
(a) Overlay of the two independent ACT-1 molecules A and B of the four molecules A–D in the asymmetric unit. Molecule A is represented by a bold solid line and molecule B by a thin solid line. The side chains of Ser64, Lys67, Gln120, Tyr150, Asn152 and Lys315 are shown in ball-and-stick representations. (b) The side view in an orientation 90° from that in (a).
3.2. Comparison with E. cloacae P99 β-lactamase
As expected from the high degree of amino-acid sequence homology between ACT-1 and E. cloacae P99 AmpC, the crystal structure of ACT-1 shows remarkable similarity to that of the E. cloacae P99 β-lactamase, which is a chromosomal counterpart of ACT-1. The r.m.s.d. values for Cα atoms between each of the four ACT-1 molecules and the E. cloacae P99 enzyme range from 0.58 to 0.63 Å.
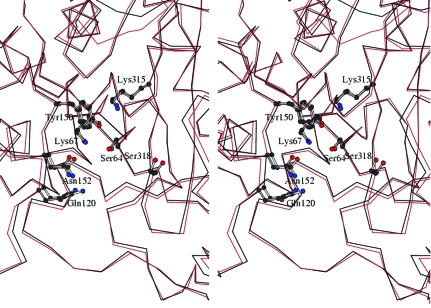
The active site of ACT-1 is positioned in the cleft between two domains, as is observed in other class C β-lactamses. Superposition of the active-site structure of ACT-1 molecule A on that of E. cloacae P99 β-lactamase also indicates high similarity in the conformations of the residues that are important in catalysis and substrate binding (Fig. 2 ▶). The Cα positions and side-chain conformation of the catalytic residues, such as Ser64, Lys67 and Lys315, are well conserved between the two structures. This result is consistent with the fact that the kinetic parameters of ACT-1 do not show major differences from those of E. cloacae P99 AmpC (Bauvois et al., 2005 ▶). However, we have noticed that the width of the active-site cleft is slightly smaller in ACT-1 than in E. cloacae P99 AmpC (Fig. 2 ▶). The distance between Asn152 Cα and Ser318 Cα is in the range 11.6–12.0 Å in the four independent molecules of ACT-1, with a mean value of 11.9 Å. In the two independent molecules of the E. cloacae P99 enzyme the distance is between 12.4 and 12.5 Å and in the GC1 enzyme the distance is 13.2 Å. Similarly, the distance between Gln120 Cα and Ser318 Cα is in the range 12.6–13.0 Å in the ACT-1 molecules, while it is between 13.5 and 13.3 Å in the E. cloacae P99 enzyme and 14.8 Å in the GC1 enzyme. Such differences in the active-site cleft seem to be caused not by the specific amino-acid residues and/or secondary-structural elements that compose the cleft, but by the relative movement of the two domains to slightly narrow the cleft that is positioned between them. The structural features mentioned above may explain the kinetic results, which show that the value of k cat/K m for cephaloridine for the ACT-1 enzyme is somewhat smaller than that of its chromosomal counterpart (Bauvois et al., 2005 ▶).
Figure 2.
Stereoview of the active-site structure of ACT-1 in comparison with that of E. cloacae P99 β-lactamase. The Cα trace of the ACT-1 structure (molecule A) is represented by a black line and that of E. cloacae P99 enzyme (molecule A) by a red line.
Supplementary Material
PDB reference: Class C β-lactamase ACT-1, 2zc7, r12zc7sf
Acknowledgments
Thanks are extended to Drs Noriyuki Igarashi and Takashi Tonozuka for kind help with data collection and data processing.
References
- Ambler, R. P. (1980). Philos. Trans. R. Soc. Lond. B Biol. Sci.289, 321–331. [DOI] [PubMed]
- Bauvois, C., Ibuka, A. S., Celso, A., Alba, J., Ishii, Y., Frere, J. M. & Galleni, M. (2005). Antimicrob. Agents Chemother.49, 4240–4246. [DOI] [PMC free article] [PubMed]
- Bradford, P. A., Urban, C., Mariano, N., Projan, S. J., Rahal, J. J. & Bush, K. (1997). Antimicrob. Agents Chemother.41, 563–569. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Bush, K., Jacoby, G. A. & Medeiros, A. A. (1995). Antimicrob. Agents Chemother.39, 1211–1233. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Jacoby, G. A. & Tran, J. (1999). Antimicrob. Agents Chemother.43, 1759–1760. [DOI] [PMC free article] [PubMed]
- Jones, T. A. & Kjeldgaard, M. (1995). O – The Manual, v.5.10.3. Uppsala University, Sweden.
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lobkovsky, E., Moews, P. C., Liu, H., Zhao, H., Frere, J. M. & Knox, J. R. (1993). Proc. Natl Acad. Sci. USA, 90, 11257–11261. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Philippon, A., Arlet, G. & Jacoby, G. A. (2002). Antimicrob. Agents Chemother.46, 1–11. [DOI] [PMC free article] [PubMed]
- Vriend, G. (1990). J. Mol. Graph.8, 52–56. [DOI] [PubMed]
- Wilke, M. S., Lovering, A. L. & Strynadka, N. C. (2005). Curr. Opin. Microbiol.8, 525–533. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Class C β-lactamase ACT-1, 2zc7, r12zc7sf