Alzheimer’s disease is characterized by proteolytic processing of the amyloid precursor protein (APP), which releases the aggregation-prone amyloid-β (Aβ) peptide and liberates the intracellular domain (AICD) that interacts with various adaptor proteins. The crystallized AICD–Fe65-PTB2 complex is of central importance for APP translocation, nuclear signalling, processing and Aβ generation.
Keywords: Alzheimer’s disease, amyloid precursor protein, Fe65, phosphotyrosine-binding domain
Abstract
Alzheimer’s disease is associated with typical brain deposits (senile plaques) that mainly contain the neurotoxic amyloid β peptide. This peptide results from proteolytic processing of the type I transmembrane protein amyloid precursor protein (APP). During this proteolytic pathway the APP intracellular domain (AICD) is released into the cytosol, where it associates with various adaptor proteins. The interaction of the AICD with the C-terminal phosphotyrosine-binding domain of Fe65 (Fe65-PTB2) regulates APP translocation, signalling and processing. Human AICD and Fe65-PTB2 have been cloned, overproduced and purified in large amounts in Escherichia coli. A complex of Fe65-PTB2 with the C-terminal 32 amino acids of the AICD gave well diffracting hexagonal crystals and data have been collected to 2.1 Å resolution. Initial phases obtained by the molecular-replacement method are of good quality and revealed well defined electron density for the substrate peptide.
1. Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder and is the most commonly occurring form of dementia worldwide. The diagnostic hallmarks are depositions of cerebral plaques that mainly contain neurotoxic amyloid β-peptides (Aβ), which are believed to play a pivotal role in AD (for a review, see Mattson, 2004 ▶). These peptides result from the proteolytic cleavage of the amyloid precursor protein (APP) via sequential cleavages by the aspartyl protease β-secretase and the large transmembrane γ-secretase complex (for reviews, see Selkoe, 2001 ▶; Haass, 2004 ▶).
APP is a type I transmembrane protein consisting of a large extracellular domain, a transmembrane domain and a short intracellular domain (AICD). The AICD contains the highly conserved 682-YENPTY-687 (neuronal APP695 numbering) motif to which a number of adaptor proteins including Fe65 bind via their phosphotyrosine-binding (PTB) domains (King & Turner, 2004 ▶). However, the interaction is independent of Tyr687 phosphorylation (Borg et al., 1996 ▶) and there is no evidence to date that Tyr687 is phosphorylated in vivo.
Fe65 is a neuronal adaptor protein involved in brain development and APP signalling (Guenette et al., 2006 ▶). It contains three protein–protein interaction domains: one WW domain and two PTB domains. The WW domain recognizes polyproline sequences in several proteins, including c-Abl tyrosine kinase (Zambrano et al., 2001 ▶) and the mammalian homologue of the Drosophila protein Enabled (Mena; Ermekova et al., 1997 ▶; Meiyappan et al., 2007 ▶). The PTB1 domain has been shown to bind to the histone acetyltransferase Tip60 (Cao & Sudhof, 2001 ▶), the transcription factor CP2/LSF/LBP1 (Zambrano et al., 1998 ▶), the cytoplasmic domains of the low-density lipoprotein receptor-related protein (LRP-1; Kounnas et al., 1995 ▶; Kinoshita et al., 2003 ▶; Pietrzik et al., 2004 ▶) and the ApoEr2 receptor (Hoe et al., 2006 ▶). The C-terminal PTB domain (Fe65-PTB2) is responsible for the binding of Fe65 to the AICD (Bressler et al., 1996 ▶; Borg et al., 1996 ▶). During the proteolytic processing of APP, the AICD together with Fe65 are released from the membrane and translocate into the nucleus, where the complex is involved in the modulation of transcription and gene expression (Cao & Sudhof, 2001 ▶).
Here, we report the cloning, overproduction and purification of the Fe65-PTB2 domain and of the complete AICD-C50 as well as of an AICD fragment consisting of the 32 C-terminal residues (AICD-C32). The complex of Fe65-PTB2 and the AICD-C32 fragment was crystallized and we present the preliminary X-ray analysis.
2. Material and methods
2.1. Cloning and protein overproduction
A gene fragment encoding the human Fe65-PTB2 domain comprising residues 534–666 was amplified by the polymerase chain reaction (PCR) with the primers Fe65PTB2_F and Fe65PTB2_R (Table 1 ▶). The PCR product was cloned into the NcoI and XhoI sites of the pET21d vector (Novagen). The construct contains a C-terminal hexahistidine tag introduced by the reverse primer.
Table 1. Primer sequences used for cloning.
| Oligonucleotide | Sequence, 5′–3′ |
|---|---|
| Fe65PTB2_F | GTACCCATGGGCGCGCCTAAGAATGAGTTGG |
| Fe65PTB2_R | AGCTAAGCTTTCAATGGTGATGGTGATGGTGCTGGGAACGGGCATCC |
| APP_C50_F | GTACCATATGGTGATGCTGAAGAAG |
| APP_C32_F | GTACCCATGGACGCCGCTGTCACCCCAGAG |
| APP_C_R | GTACAAGCTTCTAGTTCTGCATCTGCTCAAAG |
The gene coding for the human AICD (residues 646–695 with respect to neuronal APP695 numbering) was amplified using the primers APP_C50_F and APP_C50_R (Table 1 ▶) and cloned into the NdeI and HindIII sites of the pET25b vector (Novagen). The construct comprising the final 32 C-terminal residues (664–695) of APP was amplified using the primers APP_C32_F and APP_C_R and cloned into the NcoI and HindIII cleavage sites of the pETtrx_1b vector (the vector was a kind gift from Gunter Stier, EMBL Heidelberg, Germany). While the AICD was untagged, the untagged AICD-C32 construct could not be produced in significant amounts and was therefore fused to thioredoxin with a hexahistidine tag containing a TEV cleavage site in between. All constructs were verified by DNA sequencing.
All recombinant proteins were overproduced in Escherichia coli BL21 (DE3) RIL cells. Cells overproducing Fe65-PTB2 or the AICD were grown in Terrific broth (TB) medium containing ampicillin (100 mg l−1), while cells overproducing the AICD-C32 construct were grown in Lysogeny broth (LB) medium containing kanamycin (30 mg l−1). All cells were incubated at 310 K until the optical density reached ∼0.8 and overproduction was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After induction, cells overproducing the AICD or the AICD-C32 fragment were incubated at 310 K for 4 h, while the overproduction of Fe65-PTB2 was performed at 293 K for 16 h. Cells were harvested by centrifugation for 20 min at 5000g and 277 K and frozen at 193 K.
2.2. Purification
Cell pellets were resuspended in 10 ml lysis buffer per gram of cells and the protein was extracted by a combination of sonification and passage through an M1-10L Microfluidizer (Microfluidics). For Fe65-PTB2 and AICD-C32 the lysis buffer contained 300 mM NaCl, 50 mM Tris pH 8.0, 10 mM imidazole and 0.02%(v/v) 1-thioglycerol. The cell lysate was centrifuged at 125 000g at 277 K and the supernatant was applied onto a 1 ml His-Trap HP column (GE Healthcare). The column was washed with ten column volumes of lysis buffer for AICD-C32 and with lysis buffer containing 30 mM imidazole for Fe65-PTB2. Both proteins were eluted from the column in lysis buffer containing 300 mM imidazole.
To remove the thioredoxin and the hexahistidine tag from AICD-C32, the eluted protein was concentrated to a final volume of 2.5 ml using an Amicon Ultracel-5K (Millipore) and applied onto a PD10 column (GE Healthcare) equilibrated in a buffer containing 200 mM NaCl, 20 mM Tris pH 8.0 and 1 mM DTT. The fusion protein was cleaved overnight at 277 K by adding 200 µg recombinant TEV protease. After digestion the sample was again applied onto a 1 ml His-Trap HP column and the flowthrough containing the AICD-C32 peptide was collected. Both Fe65-PTB2 and AICD-C32 were further purified on an S30 16/60 size-exclusion column equilibrated in 150 mM NaCl, 10 mM Tris pH 8.0.
For the complete AICD construct the lysis buffer contained 150 mM NaCl, 50 mM Tris pH 7.5 and 1 µg ml−1 lysozyme. Prior to protein extraction, the cells were incubated for 1 h at 277 K. The cell lysate was centrifuged at 20 000g and 277 K. The pellet fraction containing the protein in inclusion bodies (IBs) was washed five times in lysis buffer without lysozyme but with 1.0%(v/v) lauryldimethylamine N-oxide (LDAO) and then twice in lysis buffer without detergent. The purified IBs were solubilized in 6 M guanidinium hydrochloride (GdnHCl) and then diluted with water to a final concentration of 4 M GdnHCl. Refolding of the AICD was performed by the rapid dilution method with 150 mM NaCl, 100 mM Tris pH 7.5 and 0.8 M l-arginine as the refolding buffer. The refolded protein was finally purified on an S30 16/60 size-exclusion column as described above.
Complex formation between Fe65-PTB2 and AICD or AICD-C32 was performed by addition of the respective AICD construct in a 1.5:1 ratio and incubation of the binding partners for 1 h at 277 K. In both cases the resulting complex was applied onto an S30 16/60 size-exclusion column as described above. Prior to crystallization experiments, the protein was concentrated using an Amicon Ultracel-5K (Millipore).
2.3. Crystallization
Initial crystallization trials of the complex containing Fe65-PTB2 and AICD or AICD-C32 were performed using the Nextal screen formulations (Qiagen) and crystallization drops (100 nl protein solution and 100 nl reservoir) were set up using a Phoenix crystallization robot (Art Robbins). Crystallization optimization was performed by the hanging-drop vapour-diffusion method at 277, 284 or 293 K. The reservoir was mixed with protein solution in a 1:1 ratio (2 µl each). After rapid soaking in mother liquor supplemented with 20%(v/v) ethylene glycol, crystals were flash-cooled in liquid nitrogen prior to diffraction experiments.
2.4. Data collection and phasing
Diffraction data were collected from the Fe65-PTB2–AICD-C32 complex crystals at cryogenic temperature (100 K) using an ADSC Q4 CCD detector on beamline ID14-eh2 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. Data were processed with the program MOSFLM and scaled with SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). For the molecular-replacement method, we used the program Phaser (McCoy, 2007 ▶) with PDB entry 1oqn as a search model.
3. Results and discussion
The Fe65-PTB2 domain comprising residues Ala534–Gln666 has been cloned and its overproduction in BL21 (DE3) RIL cells resulted in a typical yield of 20 mg per litre of bacterial cell culture. Owing to the C-terminal hexahistidine tag, purification proved to be straightforward and after nickel-column chromatography the protein was already almost pure on Coomassie-stained SDS–PAGE. The remaining impurities could be removed by size-exclusion chromatography, resulting in a pure and homogeneous sample.
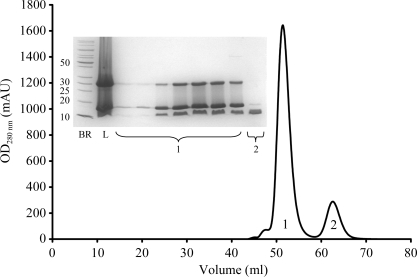
The AICD was cloned without a tag into the pET25b vector and overproduced in typical amounts of 3 mg per litre of cell culture in BL21 (DE3) RIL cells. The protein turned out to form inclusion bodies (IBs) located in the pellet fraction after cell rupture and subsequent centrifugation. The IBs were purified in lysis buffer containing 1%(v/v) LDAO, solubilized in 6 M GdnHCl and finally diluted to a final concentration of 4 M GdnHCl. Screens varying in pH, salt and l-arginine concentration were performed to reveal possible refolding conditions. The optimal refolding buffer contained 150 mM NaCl, 100 mM Tris pH 7.5 and 0.8 M l-arginine. Size-exclusion chromatography and static light-scattering (SLS) analysis showed the protein to be monomeric in solution. After size-exclusion chromatography the protein was homogeneous and pure as judged by SDS–PAGE. Purified Fe65-PTB2 and AICD formed a complex that could be purified by size-exclusion chromatography (Fig. 1 ▶). The monomeric state of the AICD sample and its ability to bind to Fe65-PTB2 with high affinity indicated that the refolded protein was biologically active and that a native Fe65-PTB2–AICD complex was formed.
Figure 1.
Size-exclusion chromatography profile and SDS–PAGE gel for the Fe65-PTB2–AICD complex. The fractions corresponding to the elution peaks are indicated. Abbreviations in the SDS–PAGE insert: BR, broad-range protein markers (kDa); L, load. Fe65-PTB2 appears in two bands corresponding to the monomer (16 kDa) and dimer (32 kDa). The appearance of the dimer is an SDS–PAGE artefact.
Using the purified Fe65-PTB2–AICD complex, about 5000 crystallization experiments were set up with protein concentrations varying between 10 and 100 mg ml−1 and at temperatures of 277, 284 and 293 K. A Phoenix nanodrop crystallization robot and common commercial crystallization screens were used in the experiments. However, no crystals could be obtained of this complex.
The AICD-C32 APP fragment was cloned into the pETtrx_1b vector and overproduced in typical amounts of 60 mg per litre of bacterial cell culture. After Ni-affinity chromatography, TEV digestion and a second Ni-affinity chromatography step to remove thioredoxin and TEV protease the protein was almost pure, with only a small amount of impurities resulting from the thioredoxin. These impurities could be removed by size-exclusion chromatography, resulting in about 5 mg of homogenous and highly pure AICD-C32 per litre of bacterial cell culture. Like the complete AICD, AICD-C32 forms a stable complex with Fe65-PTB2 which could be purified by size-exclusion chromatography and which was concentrated to 60 mg ml−1 prior to crystallization.
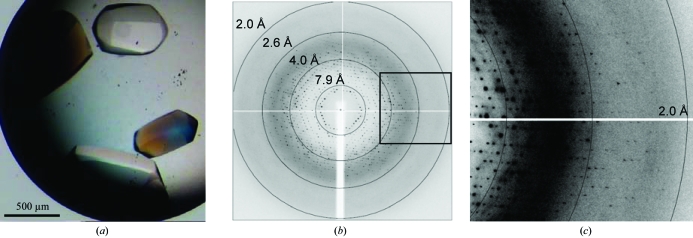
A crystallization hit obtained from Nextal cation screen formulation condition No. 51 gave rise to large (500 × 500 × 700 µm) and well diffracting crystals which grew within a few days in 3.2 M NaCl and 100 mM sodium acetate pH 4.6 at 293 K (Fig. 2 ▶ a). The crystals diffracted to 2.1 Å resolution using synchrotron radiation and a complete data set was collected on beamline ID14-eh2 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France with an oscillation increment of 0.45° (over a total of 180°; Figs. 2 ▶ b and 2 ▶ c). Data-collection statistics are shown in Table 2 ▶.
Figure 2.
(a) Hexagonal crystals of the Fe65-PTB2–AICD-C32 complex. (b) A 0.45° oscillation image collected on beamline ID14-eh2 at the ESRF. The resolution rings are indicated. (c) Magnification of the boxed area in (b) to show the resolution limit.
Table 2. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Beamline | ID14-eh2, ESRF |
| Wavelength (Å) | 1.00 |
| Space group | P61 |
| Unit-cell parameters (Å) | a = b = 114.3, c = 74.8 |
| Total No. of reflections | 363360 |
| No. of unique reflections | 32612 |
| Resolution (Å) | 33–2.10 (2.21–2.10) |
| Completeness (%) | 100 (100) |
| 〈I/σ(I)〉 | 28.8 (4.7) |
| Redundancy | 11.1 (11.2) |
| Molecules per ASU | 2 |
| Rmerge† (%) | 6.0 (46.5) |
R
merge = 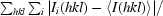
 × 100.
× 100.
The crystals of the Fe65-PTB2–AICD-C32 complex belong to the hexagonal space group P61, with unit-cell parameters a = b = 114.3, c = 74.8 Å. Calculation of the Matthews coefficient (Matthews, 1968 ▶) suggested that either two (V M = 3.62 Å3 Da−1, 66% solvent content) or three (V M = 2.41 Å3 Da−1, 49% solvent content) protein monomers are present in the asymmetric unit. However, molecular replacement with the program Phaser was only successful with two monomers in the asymmetric unit, resulting in a reasonable crystal packing with neither steric clashes nor space for a third monomer. A polyalanine model of one monomer of the Disabled1 PTB domain structure (PDB code 1oqn, residues 1031–1175), which shares 17% sequence identity to Fe65-PTB2 (residues 534–667), was used as a search model. The initial electron-density map calculated from the starting model (R cryst = 49.2%, figure of merit of 0.37) already revealed clear density for the substrate peptide. We therefore expect the structure to be solvable by the molecular-replacement method. Model building and crystallographic refinement are currently in progress.
Revealing the three-dimensional structure of the Fe65-PTB2 domain in complex with the AICD-C32 APP fragment is an important step in understanding the protein network surrounding APP since APP processing and signalling as well as amyloid-β generation are dependent on the interaction with Fe65.
Acknowledgments
We thank A. Scholz for crystallization using the Phoenix robot at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany. We acknowledge access to beamline ID14-eh2 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France and the excellent support by the beamline scientists. This work was financially supported by Deutsche Forschungsgemeinschaft (DFG) grants WI2649/1-1 and WI2649/1-2 to KW.
References
- Borg, J. P., Ooi, J., Levy, E. & Margolis, B. (1996). Mol. Cell. Biol.16, 6229–6241. [DOI] [PMC free article] [PubMed]
- Bressler, S. L., Gray, M. D., Sopher, B. L., Hu, Q., Hearn, M. G., Pham, D. G., Dinulos, M. B., Fukuchi, K., Sisodia, S. S., Miller, M. A., Disteche, C. M. & Martin, G. M. (1996). Hum. Mol. Genet.5, 1589–1598. [DOI] [PubMed]
- Cao, X. & Sudhof, T. C. (2001). Science, 293, 115–120. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Ermekova, K. S., Zambrano, N., Linn, H., Minopoli, G., Gertler, F., Russo, T. & Sudol, M. (1997). J. Biol. Chem.272, 32869–32877. [DOI] [PubMed]
- Guenette, S., Chang, Y., Hiesberger, T., Richardson, J. A., Eckman, C. B., Eckman, E. A., Hammer, R. E. & Herz, J. (2006). EMBO J.25, 420–431. [DOI] [PMC free article] [PubMed]
- Haass, C. (2004). EMBO J.23, 483–488. [DOI] [PMC free article] [PubMed]
- Hoe, H. S., Magill, L. A., Guenette, S., Fu, Z., Vicini, S. & Rebeck, G. W. (2006). J. Biol. Chem.281, 24521–24530. [DOI] [PubMed]
- King, G. D. & Turner, R. S. (2004). Exp. Neurol.185, 208–219. [DOI] [PubMed]
- Kinoshita, A., Shah, T., Tangredi, M. M., Strickland, D. K. & Hyman, B. T. (2003). J. Biol. Chem.278, 41182–41188. [DOI] [PubMed]
- Kounnas, M. Z., Moir, R. D., Rebeck, G. W., Bush, A. I., Argraves, W. S., Tanzi, R. E., Hyman, B. T. & Strickland, D. K. (1995). Cell, 82, 331–340. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mattson, M. P. (2004). Nature (London), 430, 631–639. [DOI] [PMC free article] [PubMed]
- Meiyappan, M., Birrane, G. & Ladias, A. A. (2007). J. Mol. Biol.372, 970–980. [DOI] [PMC free article] [PubMed]
- Pietrzik, C. U., Yoon, I. S., Jaeger, S., Busse, T., Weggen, S. & Koo, E. H. (2004). J. Neurosci.24, 4259–4265. [DOI] [PMC free article] [PubMed]
- Selkoe, D. J. (2001). Physiol. Rev.81, 741–766. [DOI] [PubMed]
- Zambrano, N., Bruni, P., Minopoli, G., Mosca, R., Molino, D., Russo, C., Schettini, G., Sudol, M. & Russo, T. (2001). J. Biol. Chem.276, 19787–19792. [DOI] [PubMed]
- Zambrano, N., Minopoli, G., de Candia, P. & Russo, T. (1998). J. Biol. Chem.273, 20128–20133. [DOI] [PubMed]