Abstract
The influenza C virus CM2 protein is a small glycosylated integral membrane protein (115 residues) that spans the membrane once and contains a cleavable signal sequence at its N terminus. The coding region for CM2 (CM2 ORF) is located at the C terminus of the 342-amino acid (aa) ORF of a colinear mRNA transcript derived from influenza C virus RNA segment 6. Splicing of the colinear transcript introduces a translational stop codon into the ORF and the spliced mRNA encodes the viral matrix protein (CM1) (242 aa). The mechanism of CM2 translation was investigated by using in vitro and in vivo translation of RNA transcripts. It was found that the colinear mRNA derived from influenza C virus RNA segment 6 serves as the mRNA for CM2. Furthermore, CM2 translation does not depend on any of the three in-frame methionine residues located at the beginning of CM2 ORF. Rather, CM2 is a proteolytic cleavage product of the p42 protein product encoded by the colinear mRNA: a cleavage event that involves the recognition and cleavage of an internal signal peptide presumably by signal peptidase resident in the endoplasmic reticulum. Alteration of the predicted signal peptidase cleavage site by mutagenesis blocked generation of CM2. The other polypeptide species resulting from the cleavage of p42, designated p31, contains the CM1 coding region and an additional C-terminal 17 aa (formerly the CM2 signal peptide). Protein p31, in comparison to CM1, displays characteristics of an integral membrane protein.
Influenza A, B, and C viruses comprise three of the four genera within the family Orthomyxoviridae. Influenza A and B viruses contain separate spike glycoproteins encoded by discrete RNA segments, one having hemagglutinating (receptor binding) and fusion activities and the other having neuraminidase activity. In contrast, influenza C virus contains a single spike glycoprotein having hemagglutinating, esterase, and fusion activities (reviewed in ref. 1). In addition to the major spike glycoproteins, influenza A, B, and C viruses each encode a small integral membrane protein with a single membrane spanning domain: M2 (97 residues), NB (100 residues), and CM2 (115 residues), respectively. Each protein has a relatively small N-terminal extracellular domain and a much longer cytoplasmic tail (2–6). NB contains two, and CM2 one, N-linked carbohydrate chain, and the high mannose carbohydrates are modified by the addition of polylactosaminoglycan (4, 7). All three proteins, M2, NB, and CM2, are incorporated into virions (2, 8–10). The M2 protein has an ion channel activity that is required to permit protons to enter virions during uncoating of virus particles in endosomes and the ion channel activity is blocked by the antiviral drug amantadine (ref. 11; reviewed in refs. 12 and 13). It has been speculated that NB and CM2 may be the functional counterparts of M2 and have ion channel activities (reviewed in refs. 2, 4, and 13). Some recent electrophysiological measurements indicate NB forms an ion channel (14, 15).
Despite the similarity of size and topology of M2 and NB their coding strategies are completely different. M2 protein is encoded by a spliced mRNA derived from RNA segment 7 of influenza A virus (16) whereas NB protein is encoded in an overlapping reading frame on a bicistronic mRNA derived from influenza B virus RNA segment 6, that also encodes NA (17). In comparison, the coding strategy of CM2 has not been fully investigated. Two mRNA species are derived from influenza C virus RNA segment 6, a colinear mRNA transcript that contains a single ORF [segment 6 ORF], which has the potential to encode 374 amino acids (aa) (Mr ≈ 42 kDa) and a spliced mRNA that is predicted to encode the 242-aa (Mr, ≈28 kDa) residue viral matrix protein (CM1) (19) (see Fig. 1A). Seven amino acid residues upstream of the 5′ splice site in segment 6 ORF, three in-phase AUG codons are found and the region of segment 6 ORF bounded by the AUG codon at nucleotide 731–733 through the translational stop codon at nucleotides 1,148–1,150 has been designated CM2 ORF (2, 4, 18). It has been shown that CM2 ORF encodes the CM2 protein (4) by using synthetic RNA transcripts derived from this region of segment 6 ORF. The strategy used to synthesize CM2 in influenza C virus-infected cells remained to be determined and is reported here as being recognition and cleavage of an internal signal peptide present in the 374-aa product of the segment 6 ORF.
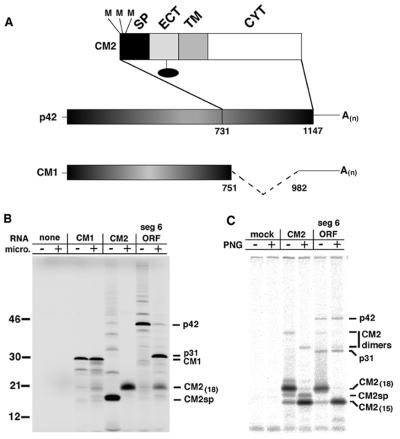
Figure 1.
The CM2 glycoprotein is a translation product of the full length, colinear mRNA derived from influenza C virus RNA segment 6. (A) Schematic diagram depicting the previously established mRNAs derived from influenza C virus RNA segment 6. The colinear mRNA segment 6 ORF has the potential to encode a protein of Mr ≈ 42 kDa (p42). The CM2 ORF (nucleotides 731–1147) contains three in-frame methionine (M) residues (aa 236, 239, or 241 of the segment 6 ORF). The CM1 protein is translated from a spliced mRNA (19) and splicing introduces a translational stop codon after lysine residue 242 of the segment 6 ORF. The CM2 protein has been shown to contain a cleaved signal peptide (SP), an N-terminal extracellular domain (Ect) that is modified by addition of N-linked carbohydrate (solid oval), a hydrophobic transmembrane domain (TM), and an intracellular cytoplasmic tail (Cyt) (4). (B) RNA transcripts of cDNAs encoding CM1 ORF, CM2 ORF, or segment 6 ORF translated in vitro using rabbit reticulocyte lysate and incorporating [35S]methionine. Canine pancreatic microsomes were added to the translation reactions where indicated. (C) HeLa-T4 cells infected with Vac-T7 were transfected with plasmids (1 μg DNA per 3.5-cm-diameter plates) containing the CM2 ORF or segment 6 ORF, metabolically labeled with [35S]Pro-mix (50 μCi/plate), lysed, proteins immunoprecipitated with an α-CM2 serum, and an aliquot was treated with PNG.
MATERIALS AND METHODS
Plasmid Construction and Site-Directed Mutagenesis.
All plasmid constructs were based on the segment 6 ORF cDNA of influenza C/Ann Arbor/1/50 (C/AA) virus whose cloning and sequencing has been previously described (4). The cDNA for the influenza C virus matrix protein, CM1, was cloned from the entire segment 6 coding region by the PCR, using Vent DNA polymerase (New England Biolabs) and primers designed to create an ORF identical to the CM1 ORF encoded by the spliced mRNA derived from RNA segment 6 (19). All cDNAs were subcloned into pGEM 3 under the control of the bacteriophage T7 promoter (4). The eukaryotic expression vector pCAGGS (20) was used for transient expression of cDNAs.
Site-specific mutagenesis of segment 6 ORF cDNA was performed by using the unique site elimination mutagenesis kit (Pharmacia Biotech). To facilitate the identification of altered plasmids, restriction enzyme sites were introduced into all mutagenesis primers by utilizing translationally silent nucleotide substitutions. The plasmid DNA inserts were sequenced using an ABI Prism 310 genetic analyzer (Applied Biosystems).
CM1 Antiserum.
The CM1 coding region was fused in-frame to the C-terminus of the glutathione S-transferase (GST) coding region in plasmid pGEX4T-CM1 (Pharmacia Biotech). The glutathione S-transferase-CM1 fusion protein was expressed and purified as described (21), and estimated to be 95% pure as judged by SDS/PAGE and Coomassie brilliant blue staining. A female New Zealand White rabbit was immunized with the CM1-glutathione S-transferase fusion protein, using TiterMax adjuvant (Vaxcel, Norcross, GA).
In Vitro Transcription and Translation.
Bacteriophage T7 RNA polymerase transcripts were synthesized from HindIII linearized plasmids, by using the T7 mMessage mMachine kit (Ambion). RNA was translated in the presence of [35S]methionine (30 μCi per 100 μl reaction; 1 Ci = 37 GBq) (Amersham Life Science) by using rabbit reticulocyte lysates and canine pancreatic microsomes (3 μl per reaction) as indicated (4). Alkylation of the microsomes was accomplished by treatment with 5 mM N-ethyl-maleimide for 30 min at room temperature (22). The reactions were stopped by the addition of 0.5 mM DTT.
Infection, Transfection, and Metabolic Labeling of Cells.
HeLa-T4 cells were cultured as described previously (4). For expression of plasmid constructs, the vac-T7 expression system was used (23), essentially as described (4). Transient expression from pCAGGS plasmids was performed by transfecting plasmid DNA into HeLa-T4 cells in 3.5-cm-diameter plates using lipofectamine (GIBCO/BRL Life Sciences) according to the manufacturer’s protocol. Transfected cells were incubated for 18 h at 37°C before metabolic labeling. For influenza C virus infections, Madin-Darby canine kidney cells were infected with influenza C/AA virus at multiplicity of infection of 5–10 for 2 h at 37°C as described (4).
Pulse-chase labeling was performed essentially as described (4) by using 50 μCi [35S]Pro-mix per 3.5 cm-diameter tissue culture dish. Cell monolayers were lysed in RIPA buffer (10 mM Tris, pH 7.4/1% deoxycholate/1% Triton X-100/0.1% SDS) containing 0.15 M NaCl, 50 mM iodoacetamide, and protease inhibitors (24). The lysates were clarified by centrifugation for 10 min at 55,000 rpm in a Beckman TLA100.2 rotor and stored at −70°C.
Immunoprecipitation and Glycosidase Treatment.
Immunoprecipitations were performed essentially as described (24) using α-CM2 cytoplasmic tail rabbit serum (final dilution of 1:100) or α-CM1 rabbit serum (final dilution 1:50). To remove N-linked carbohydrate modifications, lysates were immunoprecipitated and digested for 16 h at 37°C with peptide-N- glycosidase F (PNG) (Boehringer Mannheim). Samples were then boiled in SDS/PAGE sample buffer (24).
Cellular Membrane Preparation and Protein Fractionation.
To distinguish between peripheral and integral membrane association of proteins, in vitro translation reactions performed in the presence of canine pancreatic microsomes were diluted to 0.4 ml with NTE buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.4 and 1 mM EDTA), divided into four 0.1 ml aliquots and diluted 1:1 with NTE buffer, 50 mM Tris⋅HCl (pH 7.4) containing 4 M KCl, 50 mM Tris⋅HCl (pH 7.4) containing 8 M urea, or 100 mM carbonate buffer (pH 11.5). The samples were incubated at 4°C for 60 min and microsomes were pelleted by centrifugation at 55,000 rpm in a TLA100.2 ultracentrifuge for 30 min at 4°C. The supernatant was removed, and the pelleted microsomes were resuspended in SDS/PAGE sample buffer. Proteins in the supernatant were precipitated with 10% trichloroacetic acid (24).
SDS/PAGE.
Polypeptides were analyzed on 17.5% acrylamide containing 4 M urea gels, as described (24) except where indicated. [14C] molecular weight standards (Amersham Life Sciences) were loaded on gels to approximate molecular weights. The gels were fixed, dried, and radioactivity was analyzed by using a Fuji BioImager 1000 and macbas software (Fuji Medical System) or gels were exposed to X-Omat AR film (Eastman Kodak).
RESULTS
CM2 Is Synthesized from the Influenza C Virus RNA Segment 6 Colinear mRNA Transcript.
The influenza C virus CM2 glycoprotein was shown to be encoded within RNA segment 6 nucleotides 731–1,150 by expression of synthetic RNA transcripts (4) (Fig. 1A). However, the natural mechanism for expression of CM2 had not been determined, and a protein product derived from segment 6 ORF had not been identified. Among the most likely scenarios for the mechanism of expression of CM2 ORF, were some form of proteolytic cleavage of a segment 6 ORF-derived precursor product or an internal ribosome entry event with initiation of translation occurring at one of the three internal AUG codons located immediately before the CM2 signal peptide.
To investigate the mechanism of CM2 translation, RNA transcripts were translated in vitro using rabbit reticulocyte lysates in the presence of canine pancreatic microsomes. RNA encoding the CM1 ORF yielded a predominant Mr ≈ 28 kDa polypeptide species that did not change mobility in the presence of microsomes (Fig. 1B). Further evidence that this protein is the authentic CM1 protein is provided below. RNA encoding the CM2 ORF in the absence of microsomes yielded the unglycosylated CM2 protein containing an uncleaved signal peptide [CM2sp] (4) (Fig. 1B), whereas translation of CM2 ORF in the presence of microsomes yielded glycosylated CM2 that lacks a signal peptide [CM2(18)] (4). Translation of RNA encoding the segment 6 ORF yielded a Mr ≈ 42 kDa protein (p42) but translation in the presence of microsomes yielded small amounts of p42, a predominant Mr ≈ 31 kDa species (p31) and glycosylated CM2(18): it was noted that p31 migrated slightly slower than CM1 (Fig. 1B).
CM2 ORF and segment 6 ORF were also expressed in HeLa-T4 cells using the vac-T7 expression system. Polypeptides were metabolically labeled with [35S]Pro-mix, immunoprecipitated using a sera specific for CM2 and carbohydrate chains digested with PNG as indicated (Fig. 1C). CM2 could be readily detected in cells transfected with DNA encoding CM2 ORF or segment 6 ORF. Interestingly, in cells expressing CM2 ORF, a species corresponding to unglycosylated CM2 containing a signal peptide (CM2sp) could be observed, in addition to the glycosylated and unglycosylated, signal peptide minus forms of CM2 [CM2(18) and CM2(15)]. CM2sp was not detected in cells expressing segment 6 ORF. However, small amounts of p42, whose protein sequence contains the peptide epitope used to generate the CM2 sera, were detected in cells expressing segment 6 ORF. Furthermore, small amounts of p31, which as described below does not contain the epitope specific for the CM2 sera, were detected. We do not know the reason for the precipitation of p31 but it is a fairly insoluble protein (see below) and we consider its presence to be due to nonspecific trapping in the immunoprecipitation procedure.
Translation of CM2 Does Not Depend on the Three AUG Codons Located at the N Terminus of the CM2 ORF.
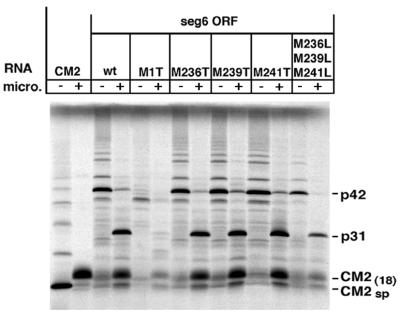
The finding that in vitro translation of segment 6 ORF mRNA in the presence of microsomal membranes yielded p31 and CM2, suggested a proteolytic cleavage event generated CM2. However, to rule out translational initiation at one of the three AUG codons located adjacent to the CM2 signal peptide, site-directed mutagenesis was performed on these three AUG codons as well as on the AUG codon that defines the beginning of segment 6 ORF and is the presumed initiation codon for p42. In the single mutants, AUG codons were changed to ACG encoding threonine and in a triple mutant, to UUG encoding leucine. As shown in Fig. 2, in vitro translation of synthetic RNA transcripts containing the single codon changes M236T, M239T, or M241T and the triple mutant M236L, M239L, and M241L had no effect on the nature of the protein products synthesized in the presence or absence of microsomal membranes. In each case p42 was observed in the absence of membranes and in the presence of membranes the predominant species were p31 and CM2(18). Translation of RNA transcripts containing the change M1T abolished production of p42 but greatly reduced amounts of a Mr ≈ 37 kDa species (p37) were observed in the absence of membranes, a species compatible with initiation at the second AUG (aa residue 51) from the 5′ end of the segment 6 ORF. In the presence of membranes, translation of M1T yielded very low amounts of CM2(18) consistent with proteolytic processing of p37.
Figure 2.
Synthesis of CM2 is independent of the three AUG codons located at the 5′ end of the CM2 ORF. Site directed mutagenesis was performed on segment 6 ORF cDNA, substituting Thr for Met residues at segment 6 ORF residues 1, 236, 239, or 241. A triple mutant was also constructed substituting Leu for Met at residues 236, 239, and 241. RNA transcripts synthesized from the indicated cDNAs were translated in vitro, in the presence or absence of canine pancreatic microsomes.
Mutagenesis of the CM2 Signal Sequence Cleavage Site Blocks Production of p31 and CM2 from p42.
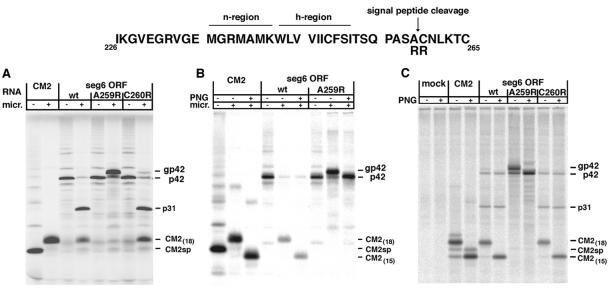
As p31 and CM2 appeared to be derived from p42 by a proteolytic cleavage event it seemed possible that p42 would be inserted into the membrane of the endoplasmic reticulum (ER) and p42 would be cleaved by signal peptidase to yield p31 and CM2. From the algorithm of von Heijne (25) it can be deduced that cleavage of the CM2 signal sequence would most likely occur after alanine residue 259. To test this hypothesis, an arginine residue was substituted for alanine (A259R) and as a control the adjacent cysteine residue was changed to arginine (C260R). Arginine does not occur on the N-terminal side of signal peptidase cleavage sites (25, 26) and it has been shown experimentally that introduction of arginine at the N-terminal side of a cleavage site blocks cleavage (27). Translation in vitro of a synthetic RNA transcribed from A259R segment 6 ORF cDNA in the presence of microsomes did not yield p31 and CM2 (Fig. 3A). Instead, a major new Mr ≈ 44 kDa species, designated gp42, and some p42 were observed. The mutation C260R had little effect on the production of p31 and CM2. Immunoprecipitation of the in vitro translation products, using a sera raised to a CM2 peptide, confirmed that p42 and gp42 contained the CM2 epitope (Fig. 3B). The shift in mobility of gp42 to that of p42 after PNG treatment, indicated that gp42 is a glycosylated from of p42 (Fig. 3B), and these data are consistent with glycosylation occurring on residue 270 of segment 6 ORF (Fig. 1A). A parallel experiment was performed by using the vac-T7 expression system in HeLa-T4 cells transfected with cDNAs encoding wt, A259R or C260R segment 6 ORF, and comparable data to that obtained for translation in vitro was obtained (Fig. 3C). The inclusion of 0.5 mM N-ethyl maleimide in the in vitro translation reaction containing microsomal membranes blocked the processing of wt p42 to p31 and CM2 (data not shown), a finding consistent with requiring translocation across the ER membrane for cleavage to occur (22).
Figure 3.
Cleavage of gp42 in the presence of microsomal membranes to yield p31 and CM2 is blocked by altering the signal peptidase cleavage site. The aa sequence of segment 6 ORF around the signal peptide of CM2 is shown, indicating the charged N terminus (n-) and hydrophobic core (h-) regions of the signal peptide (43), together with the predicted signal peptide cleavage site (arrow). Two mutants, A259R and C260R were generated in segment 6 ORF. (A) T7 RNA transcripts expressing CM2 ORF, segment 6 ORF (wt), A259R segment 6 ORF or C260R ORF were translated in vitro using rabbit reticulocyte lysate in the presence and absence of canine pancreatic microsomes. (B) Polypeptides from the indicated in vitro translation reactions from (A) were immunoprecipitated with α-CM2 serum and an aliquot was treated with PNG at 37°C for 18 h. (C) Plasmids encoding CM2 ORF, segment 6 ORF (wt), A259R segment 6 ORF or C260R ORF were expressed by using the vac-T7 expression system in HeLa-T4 cells and polypeptides were immunoprecipitated using α-CM2 serum. An aliquot was treated with PNG.
Expression of Influenza C Virus RNA Segment 6 Encoded Polypeptides in Virus-Infected and Transiently Transfected Cells.
We were interested in examining whether the p31 could be detected in influenza C virus-infected cells. A sera specific for CM1 was generated (Materials and Methods). This sera immunoprecipitated CM1 from influenza C virus-infected Madin-Darby canine kidney cells and, using the vac-T7 expression system, CM1 expressed from CM1 ORF, and p31 expressed from segment 6 ORF (Fig. 4A). The sera also immunoprecipitated gp42-expressed from A259R segment 6 ORF (data not shown). However, we could not identify the p31 species in influenza C virus-infected cells.
Figure 4.
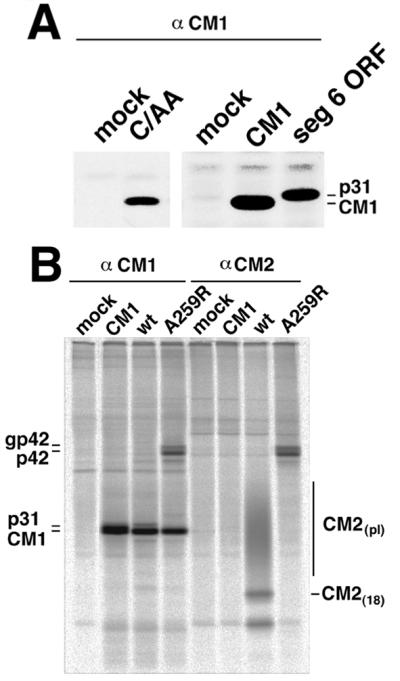
Expression of influenza C virus RNA segment 6-encoded polypeptides in virus-infected and transiently-transfected cells. (A) Immunoprecipitation of CM1 from C/AA and mock-infected Madin-Darby canine kidney cells and, using the vac-T7 expression system, CM1 expressed from CM1 ORF, and p31 expressed from segment 6 ORF. Madin-Darby canine kidney cells were mock or influenza C/AA virus-infected, at 24 h postinfection cells metabolically labeled with [35S]Pro-mix and lysates immunoprecipitated using a sera (α-CM1) raised in rabbits to a glutathione S-transferase-CM1 fusion protein. Polypeptides were separated by SDS/PAGE on a 10% acrylamide gel. (B) Expression of polypeptides encoded by segment 6 ORF mRNAs after transient expression of segment 6 ORF. HeLa-T4 cells were transfected with a nuclear expression plasmid, pCAGGS, containing wt segment 6 ORF, A259R segment 6 ORF and as a control, CM1 ORF. At 18 h posttransfection, the cells were metabolically labeled with [35S]Pro-mix, lysed, and proteins immunoprecipitated with the indicated antiserum.
Influenza C virus transcription and splicing occurs in the nucleus of infected cells (reviewed in ref. 1). Thus, to increase the expression levels over those found in influenza C virus-infected cells we turned to a surrogate system that permits the nuclear expression and splicing of influenza C virus RNA species, an approach similar to that used previously to express spliced influenza A virus RNA transcripts (28). Wild-type (wt) segment 6 ORF, A259R segment 6 ORF, and—as a control—CM1 ORF, were transiently expressed in the nucleus of HeLa-T4 cells using the vector pCAGGS (20). At 18 h posttransfection, cells were metabolically labeled with [35S]Pro-mix, lysed, and immunoprecipitated with antisera specific for CM1 or CM2 proteins. As shown in Fig. 4B, wt segment 6 ORF-expressing cells accumulated large amounts of CM1 protein and accumulated smaller amounts of p31. In these cells CM2(15), CM2(18), and CM2 modified by addition of polylactosaminoglycan (CM2pl) could also be detected. In A259R segment 6 ORF-expressing cells, large amounts of CM1 protein accumulated and smaller amounts of gp42 and p42 could be detected but not p31 and CM2 proteins, as expected if cleavage of the precursor were blocked. Thus, the protein products predicted to be translated from spliced and unspliced segment 6 derived RNAs were found in transfected cells.
CM1 and p31 Differ in Their Association with Membranes.
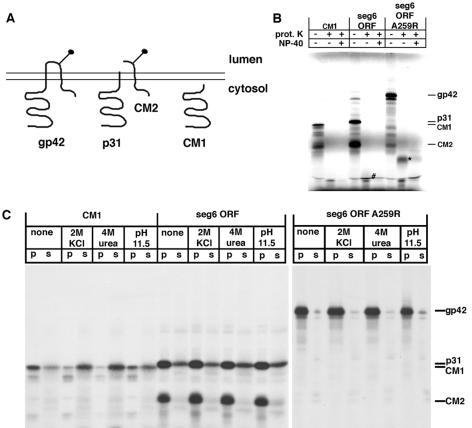
To determine whether p31 was translocated into the ER or remained cytosolic (Fig. 5A), in vitro protein translocation across the membrane was examined by determining proteinase K sensitivity of the proteins. It was found that CM1 and p31 were sensitive to proteinase K digestion indicating the bulk of both proteins were located in the cytoplasm (Fig. 5B). The predicted CM2-derived protease protected fragment contains only a single methionine residue rendering it difficult to visualize and it migrated very close to the gel dye front. However, the predicted gp42 protease protected fragment (observed on expression of A259R segment 6 ORF) contains at least three more methionine residues and a fragment of Mr ≈ 10 kDa was detected (Fig. 5B). Thus these data support the membrane orientations for p31 and gp42 shown in Fig. 5A.
Figure 5.
(A) Schematic diagram of the predicted membrane topology of gp42, p31 and CM2. Stick and ball indicates N-linked glycosylation. (B) RNA transcripts encoding CM1, segment 6 ORF, and A259R segment 6 ORF were translated in vitro in the presence of canine pancreatic microsomes and treated as indicated with 50 μg/ml proteinase K and 1% Nonidet P-40. #, protease protected fragment of CM2; ∗, the protease protected fragment of gp42. (C) RNA transcripts encoding CM1, segment 6 ORF, and A259R segment 6 ORF were translated in vitro in the presence of canine pancreatic microsomes and treated with 2 M KCl, 4 M urea, or pH 11.5 buffer as described in Materials and Methods. lanes s; supernatants; lanes p; pellets.
Although the protease protection data suggests that p31 is located in the cytoplasm after cleavage from p42, the presence of the additional C-terminal 17 residue hydrophobic CM2 signal peptide would be expected to alter its biochemical properties as compared with that of CM1. To assess the ability of CM1, p31, and gp42 to associate with membranes, the proteins were translated in vitro in the presence of microsomes and the membranes treated with strong protein denaturants. Treatment of membranes with urea or alkaline pH has been shown to extract peripheral membrane proteins whereas integral membrane proteins remain associated with the lipid bilayer (29). Whereas the CM1 protein had properties of a peripheral membrane protein in that it was found to be soluble after 2 M KCl, 4 M urea, or pH 11.5 treatments (Fig. 5C), gp42, p31, and CM2 had properties of integral membrane proteins and could not be solubilized with these treatments (Fig. 5C). In comparison to CM2 and gp42 that under all conditions were found in the pellet fraction, some p31 was found in the supernatant, but the amount did not change under the conditions used. Taken together, the data indicates that the additional 17 C-terminal hydrophobic residues from the CM2 signal sequence are sufficient to cause the association of p31 with membranes.
DISCUSSION
The data indicate that the influenza C virus RNA segment 6 derived colinear mRNA, which contains the RNA segment 6 ORF of 374 aa residues, is translated to yield a precursor glycoprotein, gp42. Targeting of gp42 to the ER membrane is thought to occur by an internal signal sequence (residues 236–259, approximately) and transfer across the membrane is presumably halted by the second hydrophobic transmembrane domain (residues 283–305). gp42 is cleaved to yield p31 and the glycosylated integral membrane protein, CM2. Cleavage of gp42 depends on the presence of pancreatic microsomes in the translation reaction and N-ethyl maleimide-treated microsomes lost their ability to promote CM2 expression, indicating an activity of the microsomes is responsible for CM2 production. Mutagenesis of the predicted site (A259R) for cleavage of the precursor by signal peptidase blocked cleavage, strongly suggesting the cleavage event is mediated by signal peptidase. p31 that contains the entire sequence of the influenza C virus matrix protein, CM1, plus 17 C-terminal hydrophobic residues, has properties of an integral membrane protein. It was observed that the signal peptide of CM2 was less readily cleaved when CM2 was expressed from the CM2 ORF as compared with expression of CM2 from gp42. Analysis of internal deletion mutants of gp42 suggest that cleavage is more efficient if 30 residues of gp42 precede the signal sequence (data not shown), a finding that may reflect the efficiency of formation of the presumptive signal peptide loop for insertion into the ER membrane translocon (30).
Except in proteins that span membranes more than once, the presence of an internal signal peptide is not commonly found in cellular proteins. Ovalbumin contains an internal signal peptide required for translocation of the nascent polypeptide into the lumen of the ER and this domain is not cleaved from the mature protein (31, 32). Also with some artificially constructed proteins such as a fusion protein consisting of the normally cytosolic α-globin fused via its C terminus to the N terminus of normally secreted bovine preprolactin it was found that efficient cleavage of the precursor protein by signal peptidase occurred and that both proteins were translocated into the ER (33, 34). However in experiments with other artificially constructed proteins it has been found that adding aa residues N-terminal to a signal sequence signal does not result in translocation of the N-terminal cleaved fragment into the ER lumen (27). The data obtained with p31, the N-terminal cleavage product of gp42, indicates it has properties of an integral membrane protein and it may be that the charged residues N-terminal to the signal sequence act to block membrane translocation.
Although internal signal peptides as the first hydrophobic membrane spanning domain are not commonly found in cellular proteins there are some parallels in other viral systems. Cleavage of the capsid (C) protein from the precursor polypeptides of rubella virus (35–38) and hepatitis C virus (39, 40) occur via cleavage of an internal signal peptide, located between the capsid protein and the integral membrane protein coding region. In the case of rubella virus (reviewed in ref. 41), the mature C protein maintains the signal peptide at its C terminus, and this signal peptide was shown to increase the association of C with intracellular membranes (36).
The p31 protein could not be identified in influenza C virus-infected cells but small amounts of p31 were detected when RNA segment 6 ORF was expressed using the surrogate nuclear expression system. It is possible that p31 undergoes degradation in influenza C virus-infected cells and this remains to be determined. If p31 were not degraded in influenza C virus infected cells, and assuming p31 folds in a manner similar to CM1, then there would be two forms of the matrix protein, one peripherally associated with membranes and one firmly attached to membranes.
The coding strategies for M2, NB, and CM2, proteins that are speculated to be functional counterparts of influenza A, B, and C viruses respectively, are widely divergent and may represent a case of parallel or convergent evolution (Fig. 6). For influenza A virus, the M2 protein is translated from a low abundance spliced mRNA derived from RNA segment 7 and the high abundance RNA segment 7 derived colinear transcript encodes the viral matrix protein, M1 (16). For influenza C virus, the CM1 matrix protein is translated from a highly abundant spliced mRNA (19) whereas the colinear RNA transcript that encodes CM2 is in low abundance (18, 19). The NB protein is translated from a bicistronic mRNA derived from influenza B virus RNA segment 6 using a reading frame that overlaps that of the viral neuraminidase protein (5, 17). However, each of the different coding strategies for the three small integral membrane proteins has a common denominator, the limitation of the synthesis of the small integral membrane protein with respect to the other major protein encoded by the same RNA segment. It is known that over expression of M2 in eukaryotic cells leads, through its ion channel activity, to a delay in intracellular transport and morphological changes to the Golgi apparatus (42). Thus, controlling the level of M2, NB, and CM2 expression may be important in preventing deleterious effects on the virus-infected host cell.
Figure 6.
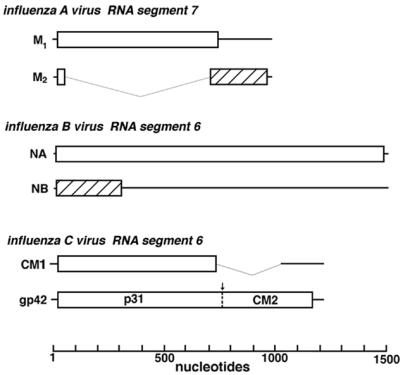
Schematic diagram of the coding strategies used for expression of the influenza A virus M2 protein, the influenza B virus NB protein, and the influenza C virus CM2 protein. Open boxes indicate the ORF for the matrix protein (influenza A and C viruses) or the neuraminidase protein (influenza B virus), whereas hatched boxes indicate overlapping coding sequences. The CM1 ORF is identical to the gp42 ORF until amino acid 242, after which a stop codon introduced by the splice junction terminates the ORF.
Acknowledgments
This research was supported in part by National Institute of Allergy and Infectious Diseases Research Grant R37 AI-20201. A.P. is an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- ER
endoplasmic reticulum
- PNG
peptide-N-glucosidase
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Lamb R A, Krug R M. In: Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, editors. New York: Lippincott-Raven; 1996. pp. 1353–1394. [Google Scholar]
- 2.Hongo S, Sugawara K, Muraki Y, Kitame F, Nakamura K. J Virol. 1997;71:2786–2792. doi: 10.1128/jvi.71.4.2786-2792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb R A, Zebedee S L, Richardson C D. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Pekosz A, Lamb R A. Virology. 1997;237:439–451. doi: 10.1006/viro.1997.8788. [DOI] [PubMed] [Google Scholar]
- 5.Williams M A, Lamb R A. Mol Cell Biol. 1986;6:4317–4328. doi: 10.1128/mcb.6.12.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zebedee S L, Richardson C D, Lamb R A. J Virol. 1985;56:502–511. doi: 10.1128/jvi.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams M A, Lamb R A. Mol Cell Biol. 1988;8:1186–1196. doi: 10.1128/mcb.8.3.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brassard D L, Leser G P, Lamb R A. Virology. 1996;220:350–360. doi: 10.1006/viro.1996.0323. [DOI] [PubMed] [Google Scholar]
- 9.Betakova T, Nermut M V, Hay A J. J Gen Virol. 1996;77:2689–2694. doi: 10.1099/0022-1317-77-11-2689. [DOI] [PubMed] [Google Scholar]
- 10.Zebedee S L, Lamb R A. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto L H, Holsinger L J, Lamb R A. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 12.Hay A J. Semin Virol. 1992;3:21–30. [Google Scholar]
- 13.Lamb R A, Pinto L H. Virology. 1997;229:1–11. doi: 10.1006/viro.1997.8451. [DOI] [PubMed] [Google Scholar]
- 14.Sunstrom N A, Premkumar L S, Premkumar A, Ewart G, Cox G B, Gage P W. J Membr Biol. 1996;150:127–132. doi: 10.1007/s002329900037. [DOI] [PubMed] [Google Scholar]
- 15.Chizhmakov I, Ogden D, Betakov T, Phillips A, Hay A. Biophys J. 1998;74:A-Pos372. [Google Scholar]
- 16.Lamb R A, Lai C-J, Choppin P W. Proc Natl Acad Sci USA. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw M W, Choppin P W, Lamb R A. Proc Natl Acad Sci USA. 1983;80:4879–4883. doi: 10.1073/pnas.80.16.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hongo S, Sugawara K, Nishimura H, Muraki Y, Kitame F, Nakamura K. J Gen Virol. 1994;75:3503–3510. doi: 10.1099/0022-1317-75-12-3503. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita M, Krystal M, Palese P. J Virol. 1988;62:3348–3355. doi: 10.1128/jvi.62.9.3348-3355.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 21.Horvath C M, Williams M A, Lamb R A. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter P, Ibrahimi I, Blobel G. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuerst T R, Niles E G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterson R G, Lamb R A. In: Molecular Virology: A Practical Approach. Davidson A, Elliott R M, editors. Oxford: IRL–Oxford Univ. Press; 1993. pp. 35–73. [Google Scholar]
- 25.von Heijne G, Gavel Y. Eur J Biochem. 1988;174:6711–6718. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Shaw A S, Rottier P J, Rose J K. Proc Natl Acad Sci USA. 1988;85:7592–7596. doi: 10.1073/pnas.85.20.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb R A, Lai C-J. Virology. 1982;123:237–256. doi: 10.1016/0042-6822(82)90258-6. [DOI] [PubMed] [Google Scholar]
- 29.Gilmore R, Blobel G. Cell. 1983;35:677–685. doi: 10.1016/0092-8674(83)90100-9. [DOI] [PubMed] [Google Scholar]
- 30.Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 31.Palmiter R D, Gagnon J, Walsh K A. Proc Natl Acad Sci USA. 1978;75:94–98. doi: 10.1073/pnas.75.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lingappa V R, Lingappa J R, Blobel G. Nature (London) 1979;281:117–121. doi: 10.1038/281117a0. [DOI] [PubMed] [Google Scholar]
- 33.Simon K, Perara E, Lingappa V R. J Cell Biol. 1987;104:1165–1172. doi: 10.1083/jcb.104.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perara E, Lingappa V R. J Cell Biol. 1985;101:2292–2301. doi: 10.1083/jcb.101.6.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suomalainen M, Garoff H, Baron M D. J Virol. 1990;64:5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hobman T C, Gillam S. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 37.Oker-Blom C, Jarvis D L, Summers M D. J Gen Virol. 1990;71:3047–3053. doi: 10.1099/0022-1317-71-12-3047. [DOI] [PubMed] [Google Scholar]
- 38.Marr L D, Sanchez A, Frey T K. Virology. 1991;180:400–405. doi: 10.1016/0042-6822(91)90046-e. [DOI] [PubMed] [Google Scholar]
- 39.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frey T K. In: Advances in Virus Research. Maramorosch K, Murphy F A, Shatkin A J, editors. Vol. 44. San Diego: Academic; 1994. pp. 69–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi T, Leser G P, Lamb R A. J Cell Biol. 1996;133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. In: Genetic Engineering. Setlow J K, editor. Vol. 14. New York: Plenum; 1992. pp. 1–11. [DOI] [PubMed] [Google Scholar]