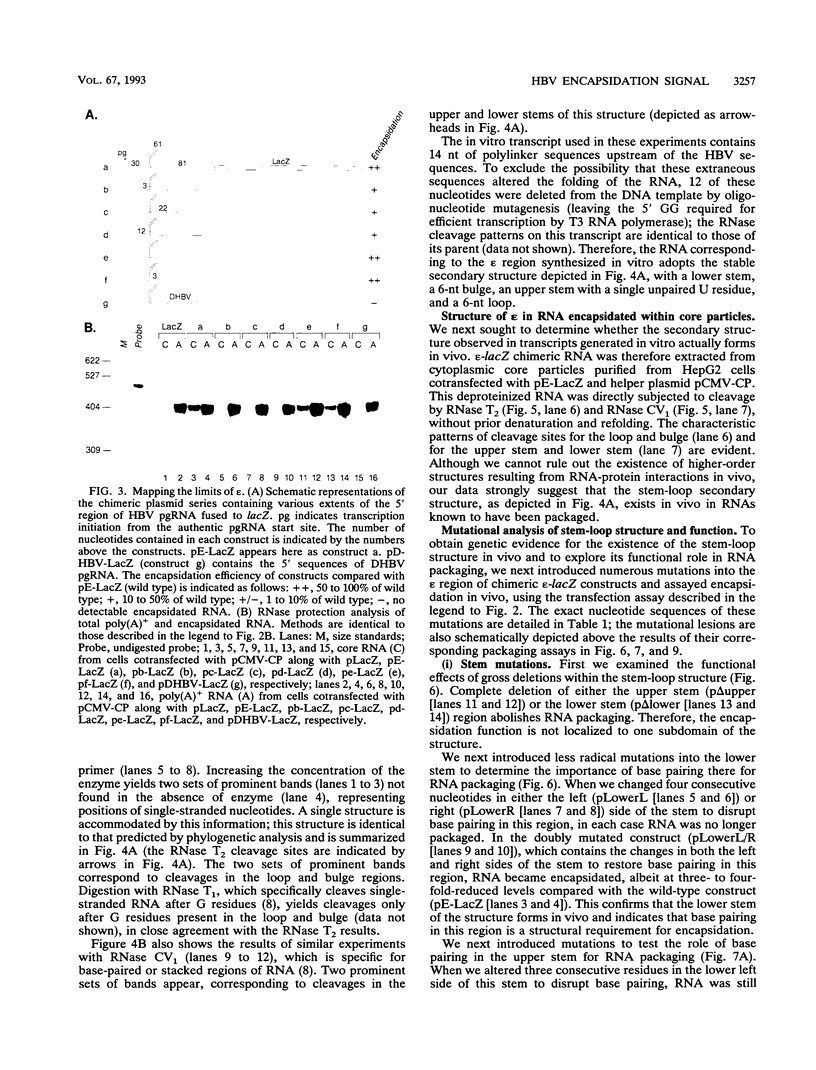
Abstract
Selective encapsidation of hepatitis B virus (HBV) genomic RNA within cytoplasmic core particles requires recognition of the cis-encapsidation signal, (termed epsilon) located at the 5' end of genomic RNA. By transfecting plasmids expressing chimeric RNAs bearing HBV sequences fused to lacZ, we have mapped the minimal region of epsilon to the 5' 94 nucleotides (nt) of genomic RNA. Enzymatic probing of the RNA secondary structure in this region (by using either in vitro transcripts or RNA extracted from HBV core particles) reveals a stem-loop structure containing a lower stem, a 6-nt bulge, an upper stem with a single unpaired U residue, and a 6-nt loop. The functional role of this structure in encapsidation was explored by examining the effects of mutations in epsilon on encapsidation of RNA in vivo. These studies reveal that (i) in the lower stem, base pairing but not specific primary sequence is required for function; (ii) there is no requirement for base pairing in the lower portion of the upper stem, but base pairing elsewhere in this stem contributes to packaging efficiency; (iii) the presence of the 6-nt bulge, but not its primary sequence, is important for function; and (iv) specific nucleotide sequences in the loop and in regions of the upper stem are critical for RNA encapsidation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990 Nov;64(11):5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992 Sep;11(9):3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum F., Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990 Jul;64(7):3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Pinto A. L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989 Aug;9(8):3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss V., Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang P. W., Jeng K. S., Hu C. P., Chang C. M. Characterization of a cis element required for packaging and replication of the human hepatitis B virus. Virology. 1992 Feb;186(2):701–711. doi: 10.1016/0042-6822(92)90037-p. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders G. H., Ganem D., Varmus H. E. 5'-terminal sequences influence the segregation of ground squirrel hepatitis virus RNAs into polyribosomes and viral core particles. J Virol. 1987 Jan;61(1):35–41. doi: 10.1128/jvi.61.1.35-41.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders G. H., Ganem D., Varmus H. Mapping the major transcripts of ground squirrel hepatitis virus: the presumptive template for reverse transcriptase is terminally redundant. Cell. 1985 Aug;42(1):297–308. doi: 10.1016/s0092-8674(85)80125-2. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Geballe A. P., Spaete R. R., Mocarski E. S. A cis-acting element within the 5' leader of a cytomegalovirus beta transcript determines kinetic class. Cell. 1986 Sep 12;46(6):865–872. doi: 10.1016/0092-8674(86)90068-1. [DOI] [PubMed] [Google Scholar]
- Hirsch R. C., Lavine J. E., Chang L. J., Varmus H. E., Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990 Apr 5;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- Hirsch R. C., Loeb D. D., Pollack J. R., Ganem D. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J Virol. 1991 Jun;65(6):3309–3316. doi: 10.1128/jvi.65.6.3309-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R., Colgrove R., Ganem D. Replication of duck hepatitis B virus in two differentiated human hepatoma cell lines after transfection with cloned viral DNA. Virology. 1988 Nov;167(1):136–142. doi: 10.1016/0042-6822(88)90062-1. [DOI] [PubMed] [Google Scholar]
- Junker-Niepmann M., Bartenschlager R., Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990 Oct;9(10):3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Köchel H. G., Kann M., Thomssen R. Identification of a binding site in the hepatitis B virus RNA pregenome for the viral Pol gene product. Virology. 1991 May;182(1):94–101. doi: 10.1016/0042-6822(91)90652-r. [DOI] [PubMed] [Google Scholar]
- Lavine J., Hirsch R., Ganem D. A system for studying the selective encapsidation of hepadnavirus RNA. J Virol. 1989 Oct;63(10):4257–4263. doi: 10.1128/jvi.63.10.4257-4263.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M. An RNA secondary structure workbench. Nucleic Acids Res. 1988 Mar 11;16(5):1789–1798. doi: 10.1093/nar/16.5.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M., Junker-Niepmann M., Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990 Dec 21;63(6):1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992 Jul;66(7):4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury S., Faruqi A. F., Shih C. Pregenomic RNA encapsidation analysis of eleven missense and nonsense polymerase mutants of human hepatitis B virus. J Virol. 1991 Jul;65(7):3617–3624. doi: 10.1128/jvi.65.7.3617-3624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. A., Racaniello V. R., Dunn G., Cooper J., Minor P. D., Almond J. W. New model for the secondary structure of the 5' non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989 May 20;207(2):379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- Sprengel R., Kuhn C., Will H., Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985 Apr;15(4):323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Will H., Reiser W., Weimer T., Pfaff E., Büscher M., Sprengel R., Cattaneo R., Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987 Mar;61(3):904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]