Abstract
PREFACE
The term 'prion' means an infectious protein that does not need an accompanying nucleic acid. There are six fungal prions, including four self-propagating amyloids and two enzymes that are necessary to activate their inactive precursors. Here we explore the scope of the prion phenomenon, the biological and evolutionary roles of prions, the structural basis of the amyloid prions, and the prominent role of chaperones (proteins that affect the folding of other proteins) and other cellular components in prion generation and propagation.
INTRODUCTION
The uniformly fatal mammalian transmissible spongiform encephalopathies (TSEs) were first widely recognized as scrapie of sheep in western Europe in the 1700s1, but may have existed much earlier2. The colourful history of the TSEs (Box 1) led to the widely3, but not universally4, accepted notion that these diseases are transmitted by a protein, without the need for an accompanying nucleic acid. Such an agent is called a prion, and the TSEs are believed to be caused by an amyloid (Glossary) form of the PrP protein. The known prions are altered forms of cellular proteins that are able to convert the unaltered form into the altered form. This positive-feedback feature is the basis for the self-propagation and infectivity of prions.
BOX 1: Early history of prions
~1000 B.C. Chinese character might suggest scrapie?

~1730 Earliest records of scrapie (die Reiberkrankheit, Ger.; la prurigo lombaire, Fr.; surlokor, Hung.) in Europe.
1920's Creutzfeldt, Jakob, … describe human spongiform encephalopathies
1936 Cuille and Chelle transmit scrapie from sheep to sheep by innoculation
1939 Propagation to goats: first trans-species transmission
1952 [Het-s] non-Mendelian gene of Podospora (Rizet)
1957 Zigas & Gajdusek describe Kuru among the cannibal Fore tribe of New Guinea
1959 Wm. Hadlow suggests Kuru similar to scrapie based on pathology
1960 Scrapie transmitted to mice (Chandler)
1965 [PSI+] non-Mendelian gene of yeast (Cox)
1966 Gibbs & Gajdusek show Kuru & CJD is infectious to monkeys: 'slow virus'
1966 Alper shows scrapie agent very UV - resistant. "Does the agent of scrapie replicate without nucleic acid?"
1967 Griffith suggests the prion mechanism essentially in its modern form!
1968 Dickinson describes Sinc (for scrapie incubation period) gene of mice. Later shown to be the PrP gene.
1971 [URE3] non-Mendelian gene of yeast (Lacroute)
1982 Prusiner purifies scrapie agent; names main protein PrP; coins term 'prion'
1985 Weissmann and Chesebro clone PrP gene of hamster and mouse
1986 Carlson & Prusiner: Sinc = PrP gene
1986 First case of BSE described (Wilesmith)
1989 Owen, Hsiao, Collinge & Prusiner: inherited CJD associated with mutant PrP gene
1993 Weissmann makes PrP knockout: cannot propagate scrapie, mice normal
1994 Cytoplasmic genes of yeast [URE3] and [PSI+] are prions (Wickner)
1996 First cases of nvCJD described (Collinge)
The properties expected for fungal prions were deduced from the known nature of infection by fungal viruses and from the concept of an infectious protein5. Fungal viruses are non-chromosomal genes or genetic elements transmitted from cell to cell by cytoplasmic mixing due to cell fusion in the process of sexual mating or asexual fusion of cellular processes6. Fungal prions are also expected to be non-chromosomal (cytoplasmic) genetic elements5. It was reasoned that fungal prions must have three genetic traits that are not found (and are not expected to be found) for nucleic acid replicons6. First, if a prion can be cured (Glossary), it can nonetheless arise again de novo in the cured strain because the protein is still present in the cell and can again (although it happens rarely) undergo the prion change. Second, transient overproduction of the protein should increase the frequency with which it undergoes the change to the prion form, simply because there is more of it to change, and once the change has occurred it propagates to the other molecules of the same protein. Third, for prion that are inactive forms of a normally active protein, the phenotype of mutants in the gene encoding the protein (which is necessary for the propagation of the prion) should be similar to that of the presence of the prion form, as in each case the normal form is deficient5. Two non-chromosomal genetic elements of Saccharomyces cerevisiae, [URE3] and [PSI+], whose molecular basis had long been mysterious, both satisfied all three of these genetic criteria for prions5, therefore initiating the fungal prion field. There are six known fungal prions, including four self-propagating amyloids and two enzymes; in this Review we will describe these prions, the identification of several with amyloids (therefore furthering their relationship to the mammalian prions), what is known about the amyloid structures, the roles of other cellular components and the biological roles of fungal prions.
SPECIFIC FUNGAL PRIONS
[URE3] and [PSI+], which are both non-chromosomal genes of Saccharomyces cerevisiae7,8, were shown to be prions of Ure2p and Sup35p (TABLE 1, FIG. 1), respectively, based on the three genetic criteria discussed above5. Ure2p is a regulator of nitrogen catabolism, repressing genes for the enzymes and transporters needed for using poor nitrogen sources, when a good source is available9. The [URE3] prion, similar to a ure2 mutation, results in inappropriate expression of (among many other genes) DAL5, which encodes the allantoate transporter, and this expression is usually used to indicate the presence of the [URE3] prion8,10–12.
Table 1.
Prions and Their Effects
| Prion | protein affected | phenotype | mechanism of phenotype |
|---|---|---|---|
| TSEs | PrP | neuronal vacuolation, astrocytosis, neuronal loss, ataxia, dementia, death | possibly internalized oligomeric PrP interference with intracellular endosomes and secretory system |
| [URE3] | Ure2p | derepression of genes for utilizing poor nitrogen sources, slowed growth | failure to retain Gln3p in cytoplasm |
| [PSI+] | Sup35p | read-thru of translation termination codons | inactive translation termination factor |
| [PIN+] | Rnq1p | ability to induce [PSI+] appearance by Sup35p overproduction | cross-seeding by Rnq1p amyloid of Sup35p amyloid |
| [Het-s] | HETs | heterokaryon incompatibility | ? |
| [β] | vacuolar protease B | death in stationary phase, failure in meiosis | failure to degrade cellular proteins under N starvation |
| [C] | MAP kinases | slow growth, increased pigment | ? |
Figure 1. Yeast and fungal amyloid prions.
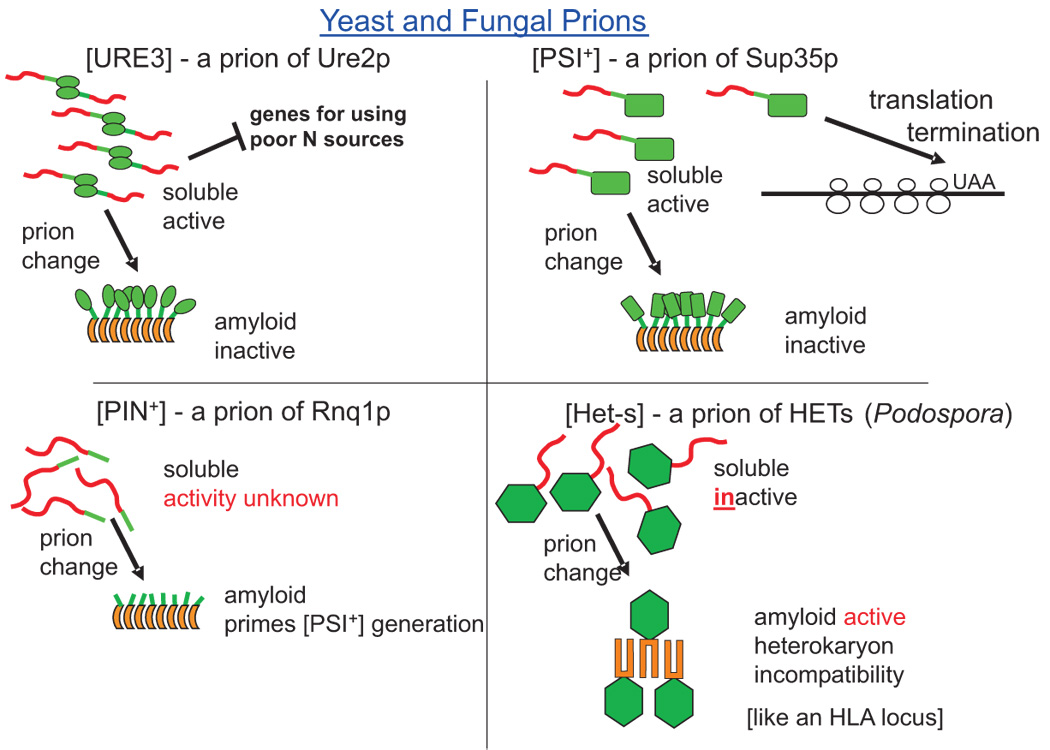
The soluble forms of Ure2p and Sup35p function in nitrogen regulation and transcription termination, respectively. Their amyloid forms are non-functional. Soluble Rnq1p has no known cellular function and the amyloid form can sporadically prime polymerization of Sup35p or Ure2p resulting in generation of the [PSI+] and [URE3] prions. The soluble form of the HETs protein has no known function, but its amyloid form is necessary for heterokaryon incompatibility, a limitation on fusion of neighboring colonies. Red domains are apparently unstructured in the native form and become amyloid in the prion form. Green shapes are natively structured domains.
Sup35p is a translation-termination factor of S. cerevisiae, and, similar to sup35 mutations, the [PSI+] prion results in increased read-through of termination codons7. The ade2-1 premature termination mutation with the weak serine-inserting SUQ5 suppressor tRNA or the ade1-14 nonsense mutant are adenine auxotrophs, but also accumulate a red pigment due to oxidation of an accumulated precursor. This red color is useful in genetic tests for [PSI+]7 and has been adapted to the [URE3] system as well in the form of DAL5-ADE2 fusion genes11,12.
The [PIN+] non-chromosomal gene was identified by its requirement for induction of [PSI+] by overproducing Sup35p13, and is a self-propagating amyloid of Rnq1p14 (FIG. 1). Rnq1 means rich in N and Q residues, and this protein carries out a self-propagating aggregation in vivo, before its relationship to the [PIN+] gene was known15. Deletion of RNQ1 does not produce any known phenotype15.
[Het-s] was first described as a non-chromosomal gene necessary for the heterokaryon incompatibility in the filamentous fungus Podospora anserina16. In this process, two converging fungal colonies carry out trial fusion of cellular processes and test the identity of alleles at a dozen polymorphic loci (called het loci) to limit fusion to genetically identical individuals17,18. The het-s locus has alleles s and S, approximately equally represented in the population, and fusion of het-s and het-S hyphae leads to incompatibility, but only if the [Het-s] non-chromosomal gene is present. The [Het-s] non-chromosomal gene has the genetic properties expected of a prion of the HETs protein encoded by the het-s allele19.
The prion concept is not limited to amyloids. An enzyme for which the active form is necessary for activation of its own inactive precursor can also be a prion20. The [β] prion in S. cerevisiae is the self-activating vacuolar protease B20. Protease B is made as an inactive precursor for which cleavage (by mature protease B) activates it21,22. The [β] prion is necessary for meiosis in yeast and for optimal survival in stationary phase20. In P. anserina, a non-chromosomal gene called [C], for 'crippled growth' is apparently based on a self-activating MAP kinase cascade23.
THE AMYLOID OF PRION PROTEINS
Infection with amyloid of recombinant proteins
Amyloid is a filamentous and typically protease-resistant protein structure with a ‘cross-β-sheet’ architecture, meaning that the β-strands of the β-sheet run perpendicular to the long axis of the filaments (FIG. 2). A great deal of evidence indicated that [PSI+], [URE3], [Het-s] and [PIN+] are amyloid forms of Sup35p, Ure2p, HET-s and Rnq1p, respectively (reviewed in24). Each of these prions has now been shown to be transmissible to uninfected cells by the introduction of amyloid formed in vitro from the corresponding recombinant protein. For [Het-s], nearly 100% of colonies subjected to "gene gun" [G] introduction of amyloid HETs protein became infected, but only background rates are obtained with soluble protein or heat-denatured or acid-denatured aggregates25. Distinct prion variants of [PSI+] (see below) were faithfully transmitted by infection with amyloid formed from recombinant Sup35p primed with amyloid seeds from extracts of distinct strains26. Different in vitro conditions of amyloid formation can also lead to distinct amyloid variants, which are transmitted to cells by infection as prion variants27. [URE3] can likewise be transmitted to cells by infection with amyloid of Ure2p, but only rarely by soluble Ure2p12. Amyloid of recombinant Ure2p was nearly as infectious as extracts of [URE3] strains, and no infectivity was present in particles smaller than about 40-mers12. Recently transmission of [PIN+] to yeast by amyloid of Rnq1p has similarly been documented28.
Figure 2. Sup35NM structure model.
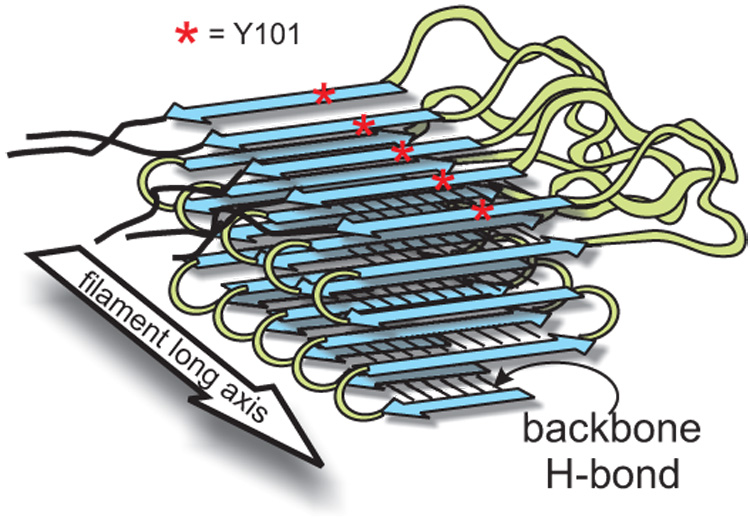
Parallel in-register β-sheet structure of the prion domain of Sup35p43. β-strands (blue arrows) run perpendicular to the long axis of the filaments and are connected by loops (green). A given residue (such as Tyr101) is aligned with the same residue in the adjacent strand (red). This structure can explain the transmission of prion variant information, as the entirety of each prion domain contacts those of the next and previous molecules in the filament.
Shuffled prion domains can still be prions
The prion domain of Ure2p is the Gln(Q)/Asn(N)-rich N-terminal 65 to 89 residues that is unstructured in its native (soluble) form 29,30. Sup35p is comprised of a Q/N-rich N-terminal 123 residue prion domain (N), a 130 residue highly charged domain (M) and a C-terminal domain sufficient for translation termination (C)31. Point mutations in N32,33, like single amino acid polymorphisms of PrP34 can block the propagation of prions, even though both protein sequences can form prions themselves35. This sequence-specificity for prion transmission, long known in studies of the species barrier in mammals36,37, suggests a relationship between apposed residues in the amyloid β-sheets that constitute the infectious material38.
Surprisingly, random shuffling of the prion domains of Ure2p or Sup35p did not prevent the formation of prions by the shuffled proteins39,40. These results showed that for Ure2p and Sup35p it is the amino acid composition of the prion domain, not its sequence, that determines its ability to form a prion. Any complementarity or similarity between paired residues in an antiparallel β-sheet or a β-helix would certainly be destroyed by random shuffling. However, the pairing of identical residues in a parallel in-register β-sheet (see Glossary) would remain possible in the shuffled sequence38,41. Therefore, shuffleability of a prion domain suggests it has a parallel in-register β-sheet structure.
Parallel in-register β-sheet structure of Sup35NM
Solid-state nuclear magnetic resonance (NMR) has been crucial in elucidating the structure of amyloids42. Tyrosine residues are scattered throughout the prion domain (N) of Sup35p, and none are in the adjacent highly charged M domain. Using 13C-1-tyrosine labeled amyloid of Sup35NM, solid-state NMR experiments showed that the distance from one labeled tyrosine to its next closest tyrosine neighbor was about 5 Å, approximately the 4.7 Å distance between β-strands43. This result strongly supports the parallel in-register β-sheet model for Sup35NM amyloid (Fig. 2). Although the N domain is sufficient to propagate [PSI+]31, labeling leucine residues, which are largely in the M domain, showed that they too were largely in parallel in-register β-sheet structure43.
X-ray diffraction analysis of an amyloid - like structure formed by a seven-residue peptide from Sup35N, GNNQQNY, showed a parallel in-register β-sheet structure, the first atomic-level structure of an amyloid44. However, small fragments of other amyloids may have architectures different from the full peptide. Using pyrene maleimide modification, another study proposed a β-helix structure for Sup35NM45, but the large probe size (~10 × 5 Å) may have affected the outcome. A β-helix involves β-bonds within each molecule, which is inconsistent with the solid-state NMR results43 and with mass per length measurements46.
The parallel in-register β-sheet structure implies that each residue of the prion domain is in intimate contact with the same residue of the adjacent molecules in the filament. This provides a simple templating mechanism for the transmission of prion variant information, which is presumed to be a difference in amyloid structure, during growth of the filament.
The prion domain of HETs is the C-terminal residues 218–28947. Solid-state NMR studies of HETs filaments show remarkably higher resolution than has been previously found for other amyloids, suggesting greater uniformity in structure48. The prion domain (residues 218–289) includes four β-strand segments, with homology between pairs of segments48.
PRION PROPAGATION AND CHAPERONES
Starting with the disaggregating chaperone Hsp10449,50, many chaperones and their co-factors are crucial to prion propagation, including Hsp70s, Hsp40s, and their co-chaperones 51–58(Fig. 3).
Figure 3. Chaperones and prions.
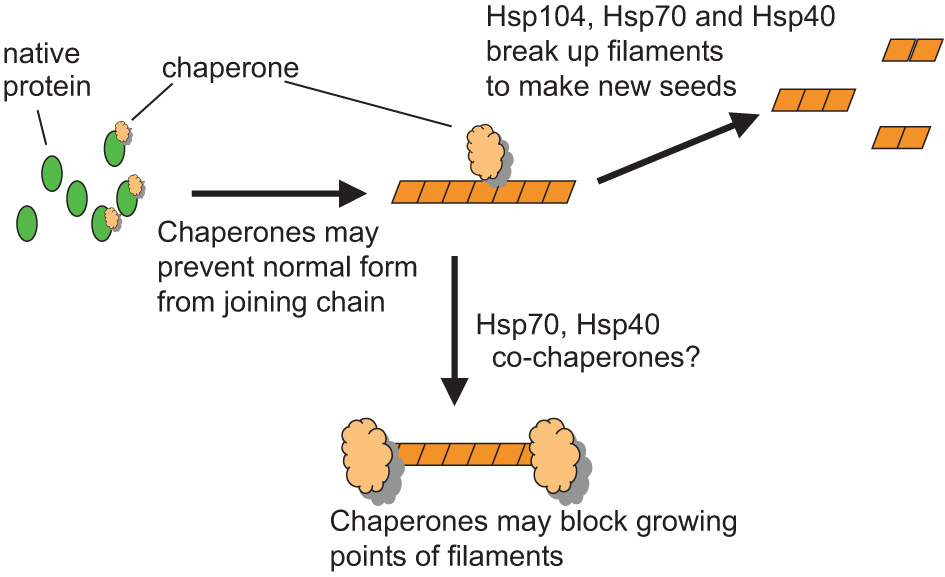
Chaperones (tan shapes) may help prion propagation by breaking long amyloid filaments into shorter ones thereby creating new growth points for amyloid formation (seeds). Chaperones may also hinder prion propagation by binding to the ends of filaments thereby blocking their growth or by binding to the soluble form of the protein thereby preventing the protein from joining the chain. Certainly Hsp104, and probably the cytoplasmic Ssa Hsp70s, have a role in filament breakage.
Hsp104 is required for each of the amyloid-based yeast prions13,50,54, and study of its role in prion propagation is facilitated by a surprisingly specific inhibitor, millimolar guanidine55,59–61. In cooperation with Hsp70s and Hsp40s, Hsp104 can disaggregate heat-denatured proteins62, and is believed to promote prion propagation by breaking up long amyloid filaments to create new seeds63–66. Overproduction of Hsp104 cures [PSI+], but not [URE3] or [PIN+].
Cytoplasmic Hsp70s, (Ssa1 to Ssa4, Ssb1, 2) bind exposed hydrophobic protein segments and help refold the protein in an ATP-regulated process. The Hsp70•ADP form binds tightly, whereas the Hsp70•ATP form rapidly binds and releases the peptide substrate. Mutants of Ssa1 lose [PSI+]52 and mutants of Ssa2 lose [URE3]20. Overproduction of Ssa1 inhibits curing of [PSI+] by Hsp104 overproduction51, but Ssa1 itself cures [URE3]67. Detailed genetic analysis shows that the Ssa1•ADP form inhibits [PSI+] propagation, whereas the Ssa1•ATP form promotes it58. Therefore, overproduction of the co-chaperone Sti1p or depletion of the nucleotide exchange factor Fes1p, both of which favor the Ssa•ADP form, impair [PSI+] propagation, whereas depletion of Sti1p or overproduction of Fes1p have the opposite effects58. It is possible that the tightly binding Ssa•ADP form binds to the growing ends of filaments or to unstructured Sup35p prion domains and prevents filament growth.
As breakage of filaments to form new seeds is believed to be a prime role of Hsp104 in prion propagation63,65,66, filament breakage by shearing also plays a prominent role in amyloid propagation in vitro68,69. Direct observations of fibre elongation show that it occurs by monomer addition69, and the less than expected dependence on monomer concentration of the time lag in amyloid formation is explained best by fibre fragmentation69, not by addition of oligomers70.
PRION VARIANTS AND THE SPECIES BARRIER
Mammalian prion 'strains' were identified by differences in incubation period, disease symptoms and signs and distribution of brain lesions despite having the identical PrP sequence71. Likewise, variants of yeast prions have been identified based on differing stability and intensity of phenotype11,12,72,73. Different variants of [PSI+] are based on different amyloid structures26,27,74, but the precise differences in structure are not yet known. Different prion variants can also have distinct chaperone requirements for propagation53.
In mammals, the 'species barrier' is the elongated incubation period or inefficient transmission of TSEs from one species to another75, due to differences in the sequence of PrP36. Bovine spongiform encephalitis (BSE) is a distinct variant of TSE that has a reduced species barrier compared with sheep scrapie strains (reviewed by76). Collinge has proposed that the PrP of each species is capable of a different range of prion conformations, and that a given prion variant (conformation) can infect those species whose PrP can assume that conformation76. Therefore, species barrier is a variant-specific phenomenon (Fig. 4). A similar species barrier with variant-dependence has been shown between, for example, [PSI+] based on S. cerevisiae, and Pichia methanolica77–81.
Figure 4. Prion variants & species barrier.
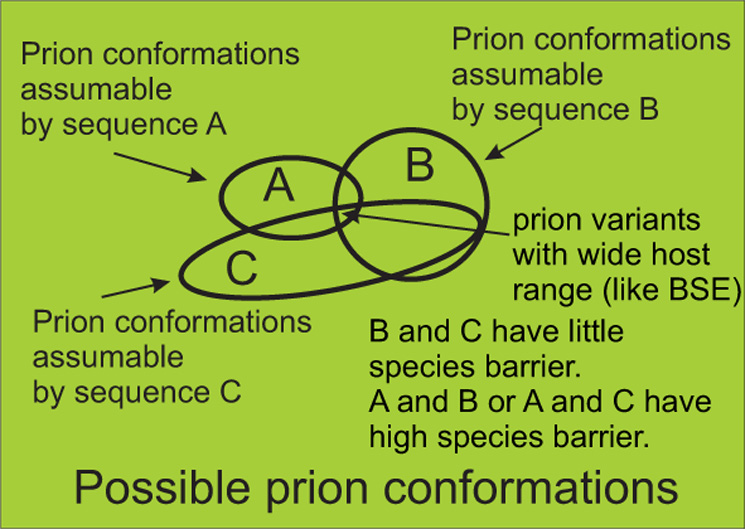
Sheep scrapie shows limited infectivity for goats, a phenomenon called the species barrier. The overlap of conformations that donor and recipient proteins can assume determine the strength of the species barrier76. As species B and C have prion proteins able to assume many similar amyloid conformations, there will be little species barrier between them. Prion protein of species A and C have few common conformations and so will have a high species barrier. A prion variant (such as bovine spongiform encephalopathy) due to an amyloid conformation that can be assumed by the protein sequence of many species will have a broad host range.
PRION GENERATION
High frequency induction of [PSI+] by overproduction of Sup35p reqiures [PIN+]13 or [URE3] 73 or excess of one of many Q/N-rich proteins, even without forming prions14,82. This suggests cross-seeding as the likely mechanism83. Each yeast amyloid-based prion can also partially interfere with the propagation of others in some cases67,84.
Depletion of Ssb1 and Ssb2, two similar Hsp70s associated with ribosomes or of Ubc4, one of the major ubiquitin-conjugating enzymes, can also increase the frequency with which [PSI+] arises de novo85,86. As Ssb1 and Ssb2 are believed to promote proper folding of proteins as they are synthesized, their absence might result in misfolded forms of Sup35p that are more prone to become prions. The Ubc4 defect would be assumed to result in failure to destroy misfolded Sup35p molecules but ubiquitin-conjugated Sup35p was not detectable85,86.
[PSI+] prion generation is also affected by components of the actin cytoskeleton87. Interactions of Sla1p, Sla2p, End3p, Arp2p and Arp3p with Sup35NM are detected by two hybrid methods, while sla1 or end3 mutants show decreased generation of [PSI+] on overproduction of Sup35p87. The same mutants show decreased Sup35p aggregates, which may account for the effect on prion generation. The authors suggest that this cytoskeleton - assembly apparatus may be acting like the mammalian aggresome, a perinuclear structure where aggregates are accumulated.
ARE FUNGAL PRIONS A HELP OR A HINDRANCE?
Although most amyloids are associated with pathogenic processes, several are known to be functional for the host. The 'curli' amyloids on the surface of certain bacteria promote adhesion that is important in colonization88. Amyloids provide a stable outer coat to certain fish eggs89 and amyloid 'hydrophobins' coat fungal cells90. Amyloid may also play a role in melanin biosynthesis91. Are any yeast or fungal prions similarly advantageous?
The [Het-s] prion carries out heterokaryon incompatibility, a process used by most (or all) filamentous fungi apparently to prevent infection with debilitating fungal viruses. Therefore, the demonstration that [Het-s] was a prion19 suggested that this was the first prion to have a role for the cell92. The [β] prion is necessary for meiosis and for survival in stationary phase20, indicating that this prion is quite beneficial. Based on subtle differences in growth rates under various laboratory conditions, it was suggested that [PSI+] helps yeast to evolve93. However, this approach would require determining to what extent these growth conditions are represented in the yeast ecological niche, and whether in such conditions [PSI+] yeast is more likely to be found94. Moreover, survival under non-growth conditions may be as important as rates of growth.
An alternate approach was to survey for yeast prions in wild strains95. Infectious agents can be widespread in nature in spite of often being a severe detriment to their hosts. Prions are no exception to this rule, as scrapie of sheep and chronic wasting disease of deer and elk can be frequent enough to seriously impact herds in captivity or in the wild. Certainly, an infectious entity which is also an advantage to its host would become widespread in natural populations, particularly one which, like the yeast prions [URE3] and [PSI+], arises de novo at rates as high as 10−6, precluding absence because of failure of exposure of the population. Therefore, it was reasoned, a prion that is not found in wild yeast must be detrimental to its host95. As controls, the mildly detrimental nucleic acid replicons 20S RNA, 23S RNA, L-A dsRNA virus, L-BC dsRNA virus, and 2 µm DNA plasmid were readily found in wild isolates. However, neither [URE3] nor [PSI+] was identified in any of 70 wild isolates. A few wild strains examined by others also failed to turn up [PSI+]78,96. This indicates that these prions cause diseases in yeast95. However, [PIN+] is occasionally found in wild strains, similar in frequency to the mildly growth-slowing nucleic acid replicons95,96. Since [PIN+] arises de novo at rates many orders of magnitude higher than do the DNA and RNA parasites, but is limited in its occurence, it is probably mildly detrimental.
In contrast to the yeast [PSI+] and [URE3] prions which are at least rare in wild strains (if not absent entirely), [Het-s] is found in 80% of wild isolates with the het-s allele97. This is consistent with the idea that [Het-s] is benefiting its host, but another possibility has emerged from genetic analysis of [Het-s]. In crosses of female het-s [Hets] cells with male het-S strains, there is selective lethality of het-S segregants in a reaction like the incompatibility reaction of vegetative cells97. This apparently constitutes a meiotic drive system in which het-s promotes its inheritance not by benefiting the organism but by killing off individuals that inherit the alternate allele.
Another argument advanced for a functional role of yeast prions is that 'prion domains', N-terminal regions of Sup35p and Ure2p homologues not essential for function of the protein, have been maintained in evolution, and some have been shown to be capable of prion conversion in S. cerevisiae77,78,98, and so prion formation must be important for the cell. However, Aigle's group99 showed that although the Saccharomyces paradoxus Ure2p has an N-terminal Q/N-rich region only slightly differing from that of S. cerevisiae, it does not undergo a prion change at detectable frequency in S. paradoxus. In addition, the C-terminal domain complementation of ure2Δ is incomplete without overexpression, showing that the prion domain functions in nitrogen regulation, like the rest of the molecule100. Moreover, studies in S. cerevisiae and P. anserina indicate functions for the prion domain of Sup35p independent of prion formation101,102. Therefore, these N-terminal extensions do not necessarily enable prion formation and are involved in the function of the protein without forming prions. Prion formation may be viewed as a rare unfavorable consequence of these important domains, much as the occurrence of Creutzfeldt-Jakob disease, Alzheimer's disease, Parkinson's disease and amyotrophic lateral sclerosis do not explain the conservation in evolution of PrP, Aβ precursor protein, α-synuclein and superoxide dismutase. In summary, the [URE3], [PSI+] and probably [PIN+] prions are a hindrance, the evidence for [Het-s] favors it being a help, and [β] is clearly helpful.
Future prospects
What is the scope of the prion phenomenon? The presence of four prions in S. cerevisiae and two in Podospora anserina argues that there are more to be found. There are many self-modifying enzymes; might some of these, as in the case of [β] and [C], under some circumstances become prions? Are there more useful amyloids like [Het-s] or more debilitating ones? Yeast prions are already being used to screen for anti-prion drugs that are active against mammalian prions103. What is the structural basis of the amyloids that are central to the prion phenomena? The parallel in-register β sheet structure of [PSI+] still leaves open the issues of the details of this structure, the structural basis of prion variants, whether other amyloid prions will have similar structures, and the difference(s) between infectious and non-infectious amyloids. How does the bewildering array of chaperone effects on prions translate into mechanisms of promoting propagation or curing? This area will likely clarify the nature of prions, chaperones and the wider problem of amyloid diseases.
Acknowledgement
This research was supported by the Intramural Research Program of the NIH, NIDDK.
Glossary
- Prion
Infectious protein (with no needed nucleic acid for infectivity).
- Prion seeds
Amyloid fragments that can grow, be again fragmented and thus propagate the prion. Similarly, active enzyme molecules of the [β] and [C] prions act as seeds.
- Non-Mendelian (or non-chromosomal or cytoplasmic) genetic element
A gene or replicon that in inherited or transmitted independent of the chromosomes, such as the mitochondrial genome, the 2 micron plasmid, a yeast virus or, as discussed here, a prion.
- Amyloid
A filamentous form of protein with a cross β-sheet structure. That is, the β-strands are perpendicular to the long axis of the filaments
- Parallel β-sheet
Adjacent β-strands are oriented in the same N- to C- terminal direction.
- In-register parallel β-sheet
Each residue is aligned with the same residue of the adjacent strand:
- Nuclear magnetic resonance
(NMR). Using solid-state NMR distances between labeled nuclei can be measured by the rate of decay of signal due to dipole-dipole coupling.
- Gene gun
A device using a pneumatic gun to propel gold particles coated with DNA or protein into cells to genetically transform them.
References
- 1.M'Gowan JP. Investigation into the disease of sheep called "scrapie". Edinburgh: Blackwood; 1914. [Google Scholar]
- 2.Wickner RB. Scrapie in ancient China? Science. 2005;309:874. doi: 10.1126/science.309.5736.874b. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB, editor. Prion Biology and Diseases. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 4.Chesebro B. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull. 2003;66:1–20. doi: 10.1093/bmb/66.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170.Original description of yeast prions, including genetic criteria that distinguish prions from nucleic acid replicons.
- 6.Wickner RB. In: Fields Virology. Fifth Edition. Knipe DM, Howley PM, editors. Lippincott: Williams & Wilkins; 2006. [Google Scholar]
- 7.Cox BS. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 8.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: connecting the dots. FEMS Microbiol. Revs. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turoscy V, Cooper TG. Ureidosuccinate is transported by the allantoate transport system in Saccharomyces cerevisiae. J. Bacteriol. 1987;169:2598–2600. doi: 10.1128/jb.169.6.2598-2600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. Embo J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5.Q/N-rich protein aggregates can prime [PSI+] prion generation.
- 15.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Molec. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 16.Rizet G. Les phenomenes de barrage chez Podospora anserina: analyse genetique des barrages entre les souches s et S. Rev. Cytol. Biol. Veg. 1952;13:51–92. [Google Scholar]
- 17.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Revs. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benkemoun L, Saupe SJ. Prion proteins as genetic material in fungi. Fungal Genet. Biol. 2006;43:789–803. doi: 10.1016/j.fgb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773.Original identification of [Het-s] as a prion.
- 20.Roberts BT, Wickner RB. A class of prions that propagate via covalent auto-activation. Genes Dev. 2003;17:2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones EW. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- 22.Zubenko GS, Park FJ, Jones EW. Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics. 1982;102:679–690. doi: 10.1093/genetics/102.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kicka S, Bonnet C, Sobering AK, Ganesan LP, Silar P. A mitotically inheritable unit containing a MAP kinase module. Proc. Natl. Acad. Sci. USA. 2006;103:13445–13450. doi: 10.1073/pnas.0603693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickner RB, et al. Prions: proteins as genes and infectious entities. Genes Dev. 2004;18:470–485. doi: 10.1101/gad.1177104. [DOI] [PubMed] [Google Scholar]
- 25.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci U S A. 2002;99:7402–7407. doi: 10.1073/pnas.072199199.First demonstrated transmission of prion by amyloid of recombinant protein.
- 26.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391.Amyloid structure determines prion variant
- 27.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392.Amyloid structure determines prion variant
- 28.Patel BK, Liebman SW. "Prion proof" for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.10.069. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93.First biochemical evidence for yeast prions; prion domains.
- 30.Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured? Biochemistry. 2005;44:321–328. doi: 10.1021/bi047964d. [DOI] [PubMed] [Google Scholar]
- 31.TerAvanesyan A, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2- mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 34.Mead S, et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science. 2003;300:640–643. doi: 10.1126/science.1083320. [DOI] [PubMed] [Google Scholar]
- 35.Kochneva-Pervukhova NV, et al. Mechanism of inhibition of Ψ+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. Embo J. 1998;17:5805–5810. doi: 10.1093/emboj/17.19.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prusiner SB, et al. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 37.Priola SA, Caughey B, Race RE, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross ED, Minton AP, Wickner RB. Prion domains: sequences, structures and interactions. Nat. Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 39.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan JCC, Oyler NA, Yau W-M, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10--39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Quart. Revs. Biophys. 2006;1:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 43.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103.First evidence - based prion amyloid structure.
- 44.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnan R, Lindquist S. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diaz-Avalos R, King CY, Wall JS, Simon M, Caspar DLD. Strain-specific morphologies of yeast prion amyloids. Proc Natl Acad Sci U S A. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balguerie A, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. Embo J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chernoff YO, Ono B-I. In: Protein synthesis and targeting in yeast. Brown AJP, Tuite MF, McCarthy JEG, editors. Berlin: Springer-Verlag; 1992. pp. 101–107. [Google Scholar]
- 50.Chernoff YO, Lindquist SL, Ono B-I, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373.First demonstration of chaperone involvement in prion propagation.
- 51.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 54.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 56.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 Mutations Affect [PSI(+)] Prion Propagation and Cell Growth Differently and Implicate Hsp40 and Tetratricopeptide Repeat Cochaperones in Impairment of [PSI(+)] Genetics. 2003;163:495–506. doi: 10.1093/genetics/163.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones G, Song Y, Chung S, Masison DC. Propagation of yeast [PSI+] prion impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 61.Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299.Guanidine cures prions by inhibiting Hsp104
- 62.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4.Chaperones interact in disaggregating proteins.
- 63.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 64.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion "seeds" but not prion protein aggregation in yeast. Mol. Cell. Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cox BS, Ness F, Tuite MF. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuite MF, Koloteva-Levin N. Propagating prions in fungi and mammals. Mol. Cell. 2004;14:541–552. doi: 10.1016/j.molcel.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 67.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hall D, Edskes HK. Silent prions lying in wait: a two-hit model of prion/amyloid formation and infection. J. Mol. Biol. 2004;336:775–786. doi: 10.1016/j.jmb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 69.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. Plos Biol. 2004;2:1582–1590. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 71.Bruce ME, McConnell I, Fraser H, Dickinson AG. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 72.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375.Original description of [PIN+] prion.
- 73.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion "strains" in yeast. Proc Natl Acad Sci U S A. 2002;99 Suppl. 4:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- 75.Pattison IH. In: Slow, Latent and Temperate Virus Infection. Gajdusek DC, C. J. Gibbs J, Alpers M, editors. Washington, DC: US Government Printing Office; 1965. pp. 249–257. [Google Scholar]
- 76.Collinge J. Variant Creutzfeldt-Jakob disease. Lancet. 1999;354:317–323. doi: 10.1016/S0140-6736(99)05128-4. [DOI] [PubMed] [Google Scholar]
- 77.Kushnirov VV, Kochneva-Pervukhova NV, Cechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chernoff YO, et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Molec. Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 79.Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 80.Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Yeast [PSI+] "prions" that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell. 2001;7:1121–1130. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 81.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 82.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 83.Vitrenko YA, Gracheva EO, Richmond JE, Leibman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J. Biol. Chem. 2007;282:1779–1787. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 84.Bradley ME, Liebman SW. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI+] prion. Mol. Cell. Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 2006 doi: 10.1074/jbc.M609597200. in press. [DOI] [PubMed] [Google Scholar]
- 87.Ganusova EE, et al. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chapman MR, et al. Role of Escherichia coli Curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Podrabsky JE, Carpenter JF, Hand SC. Survival of water stress in annual fish embryos: dehydration avoidance and egg amyloid fibers. Am. J. Physiol. Regulatory Integrative Comp. Physiol. 2001;280:R123–R131. doi: 10.1152/ajpregu.2001.280.1.R123. [DOI] [PubMed] [Google Scholar]
- 90.Wosten HA, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 91.Berson JF, et al. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 2003;161:521–533. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wickner RB. A new prion controls fungal cell fusion incompatibility. Proc. Natl. Acad. Sci. USA. 1997;94:10012–10014. doi: 10.1073/pnas.94.19.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 94.Partridge L, Barton NH. Evolving evolvability. Nature. 2000;407:457–458. doi: 10.1038/35035173. [DOI] [PubMed] [Google Scholar]
- 95.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102.[URE3] and [PSI+] are disease agents: selfish proteins.
- 96.Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol. Microbiol. 2003;49:1005–1017. doi: 10.1046/j.1365-2958.2003.03608.x. [DOI] [PubMed] [Google Scholar]
- 97.Dalstra HJP, Swart K, Debets AJM, Saupe SJ, Hoekstra RF. Sexual transmission of the [Het-s] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A. 2003;100:6616–6621. doi: 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Edskes HK, Wickner RB. Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full - length protein. Proc. Natl. Acad. Sci. USA. 2002;99 Suppl. 4:16384–16391. doi: 10.1073/pnas.162349599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Talarek N, Maillet L, Cullin C, Aigle M. The [URE3] prion is not conserved among Saccharomyces species. Genetics. 2005;171:23–54. doi: 10.1534/genetics.105.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetcs. 2007 doi: 10.1534/genetics.107.074153. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gagny B, Silar P. Identification of the genes encoding the cytosolic translation release factors from Podospora anserina and analysis of their role during the life cycle. Genetics. 1988;149:1763–1775. doi: 10.1093/genetics/149.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Urakov VN, et al. N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol. Biol. 2006;7:34–46. doi: 10.1186/1471-2199-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bach S, et al. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat. Biotechnol. 2003;21:1075–1081. doi: 10.1038/nbt855.Using yeast to find drugs against prion diseases of mammals.


