Abstract
This study investigated regional variations in the contribution made by different human papilloma (HPV) types to invasive cervical cancer (ICC). A total of 85 studies using polymerase chain reaction to estimate HPV prevalence in ICC were identified. Data on HPV prevalence were extracted separately for squamous cell carcinoma (SCC) and for adeno- and adenosquamous-carcinoma (ADC). A total of 10 058 cases (8550 SCC, 1508 ADC) were included in pooled analyses. The most common HPV types in ICC were, in order of decreasing prevalence, HPV16, 18, 45, 31, 33, 58, 52, 35, 59, 56, 6, 51, 68, 39, 82, 73, 66 and 70. In SCC, HPV16 was the predominant type (46–63%) followed by HPV18 (10–14%), 45 (2–8%), 31 (2–7%) and 33 (3–5%) in all regions except Asia, where HPV types 58 (6%) and 52 (4%) were more frequently identified. In ADC, HPV prevalence was significantly lower (76.4%) than in SCC (87.3%), and HPV18 was the predominant type in every region (37–41%), followed by 16 (26–36%) and 45 (5–7%). The overall detection of HPV DNA was similar in different regions (83–89%). A majority of ICC was associated with HPV16 or 18 in all regions, but approximately a quarter of all ICC cases were associated with one of 16 other HPV types, their distribution varying by region.
Keywords: human papillomavirus, cervical carcinoma, squamous cell carcinoma, adenocarcinoma, epidemiology, literature review
Epidemiological studies have clearly established human papilloma-virus (HPV) infection as the central cause of invasive cervical cancer (ICC). This is the second most common cancer among women worldwide and the most common female cancer in large areas of the developing world where an estimated 80% of new cases arise (Parkin et al, 1999). Studies in 22 countries, coordinated by the International Agency for Research on Cancer (IARC), identified HPV DNA in almost all (99.7%) (of about 1000) cases of cervical cancer (Walboomers et al, 1999).
Approximately 40 distinct HPV types are known to infect the genital tract and epidemiological studies to date suggest that at least 14 of these, called oncogenic or high-risk (HR) types, are significantly associated with progression to ICC (Bosch et al, 1995). Most of these HR types are phylogenetically related to either HPV16 (31, 33, 35, 52 and 58) or HPV18 (39, 45, 59 and 68) (Chan et al, 1995). Limited evidence suggests that their distribution may vary by region (Bosch et al, 1995).
HPV vaccines hold great promise to reduce the global burden of ICC any potential vaccine be multivalent since prior infection with one type does not appear to decrease the risk of infection by another HPV type (Koutsky et al, 2002; Combita et al, 2002; Liaw et al, 2001). In this however, to collate all relevant published data to identify the most prevalent HPV types associated with ICC worldwide and within five geographic regions.
Materials and methods
Study selection
Source material was selected from citations listed in Medline and ISI Current Contents databases and from references cited in the selected papers. Key search terms included: cervical cancer, HPV, human, female, and polymerase chain reaction (PCR). The review was limited to studies that included a minimum of 20 ICC cases; carcinomas in situ were excluded. Studies had to provide a clear description of the use of PCR-based assays to identify HPV DNA. Studies using nonamplified hybridisation methods only were excluded based on the reduced sensitivity of such methods in comparison to PCR (Gravitt et al, 1991; Schiffman et al, 1991; Guerrero et al, 1992). Furthermore, articles were only included if type-specific prevalence of at least one HPV type other than HPV6, 11, 16 or 18 was reported. For articles where study methods suggested that additional type-specific data were available, these data were requested from the authors (Yang et al, 1997; Eluf-Neto et al, 1994; Chaouki et al, 1998; Meyer et al, 1998; Chen et al, 1999; Lin et al, 2001). If data or data subsets had been published in more than one article, only the publication with the largest sample size was included.
Data abstraction
For each study, the following key information was extracted: country of sample; sample size; mean age; study year; distribution of cases by histological type; type of cervical specimen (e.g., fresh/fixed biopsies or exfoliated cells) and PCR primers used to detect HPV positive samples; type-specific and overall prevalence of HPV infection. Where available, data on HPV-specific prevalence were extracted independently for squamous cell carcinoma (SCC) and for adeno- and adenosquamous carcinoma (henceforth collectively termed ADC). Where histology-specific HPV prevalence was not reported, cases were classified as being of ‘unspecified’ histology. Each study was classified into one of five geographical regions: Africa, Asia, Europe, North America and Australia, or South and Central America. For studies comparing HPV prevalence across regions (Munoz et al, 1992; Bosch et al, 1995; Sebbelov et al, 2000), data were separated into their regional components.
Studies included
Of studies published up to February 2002 on Medline identified by our search criteria, 82 qualified for inclusion (no additional studies were included from ISI Current Contents). Three studies were conference abstracts containing the detailed information required for inclusion (Illades-Aguiar et al, 2000; Nindl et al, 2001; Rabelo-Santos et al, 2001). In the course of contacting authors, additional data became available for two studies expanded since the original publication (Burger et al, 1996; Andersson et al, 2001). Detailed information on the design of each of the 85 included studies is listed in the appendix.
Estimation of type-specific prevalence
HPV prevalence data were expressed as percentages of all cases tested for HPV. Multiple infections (3.7% of all ICC cases) were separated into constituent types, thus type-specific prevalence represents that in either single or multiple infections. Cases with specimens considered to be inadequate for PCR testing were excluded. Type-specific prevalence is presented for the 18 most common HPV types as identified by this review (HPV types 6, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 70, 73 and 82 also known as MM4, W13B or IS39) in order of descending prevalence for each subgroup analysis. Consensus PCR primers My09/11 (Bernard et al, 1994), GP5+/6+ (Chaouki et al, 1998) and SPF10 (Kleter et al, 1999) were considered to amplify all 18 HPV types, L1C1/L1C2 (Nakagawa et al, 1996) to amplify all types, but HPV73 and 82, GP5/6 to amplify types 6, 11, 16, 18, 31, 33, 35 and 45 only (Roda Husman et al, 1995), and pU1M/2R (Harima et al, 2002) to amplify types 6, 16, 18, 31, 33, 35, 52, 56, 58 and 59 only. For other consensus and type-specific PCR primers, only those HPV types specified in the individual reports were considered amplifiable. For HPV-specific prevalence, only studies testing for a particular HPV type contribute to the analysis for that type, and therefore sample size varies between the type-specific analyses.
Statistical analyses
Sources of variation in overall HPV prevalence were investigated by unconditional multiple logistic regression analysis (Breslow and Day, 1980). The final model included the following sources of variation: geographical region, histological type of ICC, type of specimen for HPV DNA testing, and type of PCR primers used. Mean age and study year were found not to be significantly related to overall HPV prevalence. Adjustment of overall HPV prevalence for these variables was done using the adjust command in Stata version 7.0, based on probability estimates from the logistic regression model. Confidence intervals for overall HPV prevalence were calculated assuming the nonindependence of cases within the same study using the cluster option in Stata (White, 1980). P-values comparing the prevalence of particular HPV types in subsets of ICC cases refer to χ2 tests.
Results
Meta-analysis of overall HPV prevalence
A total of 10 058 ICC cases from the 85 identified studies were included in this meta-analysis of HPV prevalence (Table 1). A majority of cases came from studies performed in Asia (31%) and Europe (33%), with African studies representing the smallest proportion of cases (6%). HPV prevalence was reported stratified by histological type for 73% of the cases: 5825 SCC cases and 1508 ADC cases. In total, 12 studies included only SCC and seven studies included only ADC.
Table 1. Region- and histology-specific distribution of included studies and ICC cases.
|
Number (%) of cases with histology-specific HPV data |
||||||
|---|---|---|---|---|---|---|
| Region | No. of studies | Countries represented | No. of cases | SCC | ADC | Unspecified |
| Africa | 6 | Algeria, Benin, Guinea, Mali, Morocco, Senegal, South Africa, Tanzania, Uganda | 609 | 204 (33.5) | 21 (3.4) | 384 (63.1) |
| Asia | 28 | Mainland China, India, Indonesia, Japan, Korea, Malaysia, Philippines, Taiwan, Thailand | 3091 | 2273 (73.5) | 381 (12.3) | 437 (14.1) |
| Europe | 32 | Austria, Czech Republic, Denmark, Finland, France, Germany, Greece, Greenland, Holland, Hungary, Ireland, Italy, Norway, Poland, Russia, Sweden, UK | 3336 | 2010 (60.3) | 603 (18.1) | 723 (21.7) |
| North America and Australia | 13 | Australia, Canada, USA | 1562 | 914 (58.5) | 450 (28.8) | 198 (12.7) |
| South and Central America | 12 | Argentina, Bolivia, Brazil, Chile, Colombia, Costa Rica, Cuba, Honduras, Mexico, Panama, Paraguay, Peru | 1460 | 424 (29.0) | 53 (3.6) | 983 (67.3) |
| Total | 85 | 10 058 | 5825 (57.9) | 1508 (15.0) | 2725 (27.1) | |
HPV=human papillomavirus; ICC=invasive cervical cancer; SCC=squamous cell carcinoma; ADC=adeno/adenosquamous carcinoma.
Adjusted overall HPV prevalence ranged from 79.3% in Asia to 88.1% in North America and Australia, but did not differ significantly between regions (Table 2). HPV DNA was significantly less likely to be detected in ADC (76.5%) than in SCC (87.3%) (P<0.001). DNA detection in ICC of unspecified histology (89.2%) was similar to that in SCC.
Table 2. Prevalence of HPV by region, histological type, HPV DNA specimen and PCR primers used.
| Variable | No. of studies | No. of cases | Crude HPV prevalence (%) | Adjusteda HPV prevalence (%) | 95% confidence intervals |
|---|---|---|---|---|---|
|
Region | |||||
| Africa | 6 | 609 | 88.8 | 86.5 | (76.4–92.7) |
| Asia | 28 | 3091 | 83.1 | 79.3 | (73.7–84.0) |
| Europe | 32 | 3336 | 85.9 | 86.7 | (82.5–90.0) |
| North America and Australia | 13 | 1562 | 87.5 | 88.1 | (83.6–91.5) |
| South and Central America | 12 | 1460 | 89.3 | 87.7 | (83.1–91.2) |
|
Histological type | |||||
| Squamous cell carcinoma | 47 | 5825 | 86.9 | 87.3 | (84.8–89.5) |
| Adeno(squamous) carcinoma | 45 | 1508 | 76.7 | 76.5 | (72.3–80.3) |
| Unspecified | 48 | 2725 | 89.0 | 89.2 | (85.1–92.3) |
|
HPV DNA specimen | |||||
| Fixed biopsies | 34 | 3324 | 84.3 | 83.3 | (78.9–86.9) |
| Fresh biopsies | 27 | 3992 | 88.4 | 87.8 | (84.9–90.2) |
| Unspecified biopsies | 8 | 496 | 87.1 | 87.2 | (82.3–90.9) |
| Cervical exfoliated cells | 13 | 1444 | 80.1 | 78.9 | (68.5–86.5) |
| Cells and biopsies | 3 | 802 | 93.5 | 92.5 | (87.3–95.7) |
|
Primers | |||||
| MY09/11 | 31 | 4355 | 85.9 | 83.3 | (80.1–86.0) |
| GP5/6 | 6 | 506 | 80.8 | 77.8 | (64.9–87.0) |
| GP5+/6+ | 14 | 1681 | 92.2 | 90.1 | (85.0–93.6) |
| SPF10 | 3 | 275 | 96.7 | 97.2 | (87.9–99.4) |
| PU1M/2R | 6 | 376 | 80.9 | 79.4 | (68.8–87.0) |
| L1C1/C2 | 5 | 655 | 91.2 | 88.0 | (77.2–94.1) |
| Combination | 9 | 1351 | 88.4 | 86.4 | (77.9–92.0) |
| Other | 4 | 166 | 84.3 | 89.3 | (75.2–95.9) |
| TS-PCR only | 7 | 693 | 73.6 | 74.7 | (63.8–83.2) |
Adjusted for histological type, region, HPV DNA specimen and PCR primers.
Adjusted HPV prevalence was significantly higher from studies testing both cells and biopsies for HPV DNA (92.5%) than from studies testing either cervical exfoliated cells (78.9%) or fixed biopsies (83.3%) only. For PCR primers, highest HPV prevalence was obtained in studies using SPF10 primers (97.2%) and the lowest in studies using type-specific PCR (TS-PCR) only (74.7%). Adjusted overall HPV prevalence varied between 77.8 and 90.1% for other primer sets, but these differences were not statistically significant.
Meta-analysis of HPV type-specific prevalence
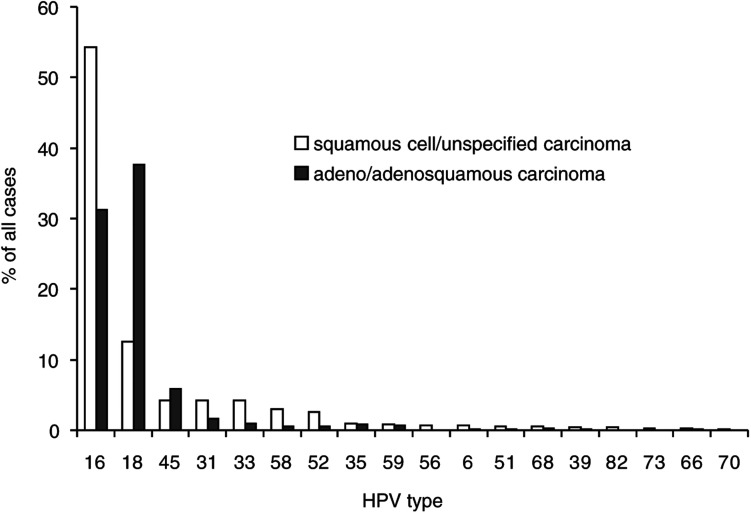
Owing to their similar overall and type-specific HPV prevalence, ICC of unspecified histology were combined with SCC for comparison of HPV type-specific prevalence by histological type (Figure 1). The most common HPV types identified were, in order of decreasing prevalence, HPV16, 18, 45, 31, 33, 58, 52, 35, 59, 56, 6, 51, 68, 39, 82, 73, 66 and 70. Other HPV types were detected in no more than 0.2% of ICC cases. There was considerable variation in HPV-specific prevalence between SCC and ADC. HPV16 was identified more often in SCC (55.2%) than in ADC (31.3%) (P<0.001). The same was found for the HPV16 phylogenetically related types 31, 33, 52 and 58 (P<0.001), but not 35. Conversely, HPV18 was more prevalent in ADC (37.7%) than in SCC (12.3%) (P<0.001). The HPV18 phylogenetically related type 45 was also more prevalent in ADC (5.8%) than in SCC (3.4%) (P=0.04).
Figure 1.
Type-specific prevalence of HPV in 10 058 worldwide cases of invasive cervical cancer by histological type.
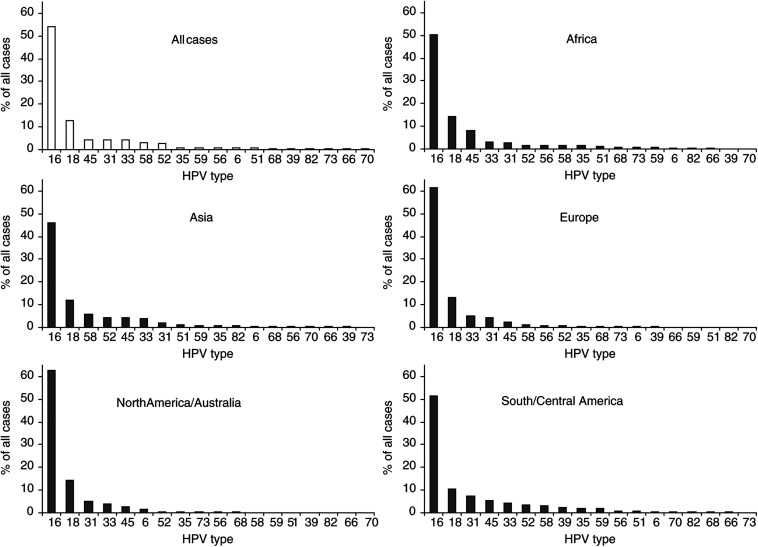
Comparison of HPV-specific prevalence in SCC by region is shown in Figure 2. In SCC, HPV16 was the predominant type in all regions studied, varying from 45.9% in Asia to 62.6% in North America and Australia. HPV18 was found consistently in 10–14% of SCC cases. In most regions, HPV45 (2–8%), 31 (2–7%) and 33 (3–5%) were the most prevalent types in SCC after types 16 and 18. In cases from Africa, the prevalence of HPV45 (8.0%) was more than twice that of either 31 (2.7%) or 33 (3.2%). In cases from Asia, HPV58 (5.8%) and 52 (4.4%) were found more commonly than HPV45, 31 and 33. Other HPV types varied considerably in their prevalence from region to region, but accounted for no more than 2% of ICC cases from any region.
Figure 2.
Type-specific prevalence of HPV in 8550 cases of squamous cell and unspecified cervical carcinoma by region.
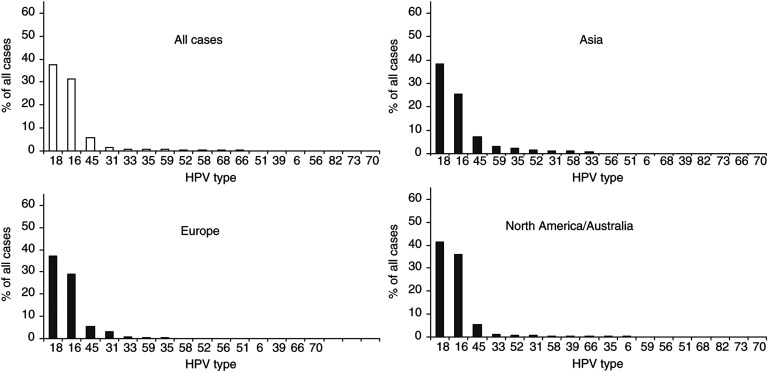
Sufficient ADC-specific data existed for the comparison of HPV-specific prevalence across Asia, Europe and North America and Australia (Figure 3). HPV18 was the predominant type (37.7%), found consistently in 37–41% of ADC cases in these regions, with HPV16 accounting for a smaller proportion (26–36%). HPV45 was the third most prevalent in each region, present in 5–7% of ADC cases vs only in 2–4% of SCC cases from these regions. The HPV16 phylogenetically related types 31, 33, 52 and 58 (but not 35) were all less prevalent in ADC cases than in SCC cases from each region.
Figure 3.
Type-specific prevalence of HPV in 1508 cases of adeno- and adenosquamous cervical carcinoma by region.
Discussion
Two-thirds of ICC cases included in this meta-analysis were associated with HPV16 (51.0%) or 18 (16.2%) infection. However, more than 16 other HPV types were also associated with ICC, of which the most prevalent were types 45, 31, 33, 58 and 52 (collectively accounting for 18.3% of cases). The HPV16 family of viruses were more commonly found in SCC than ADC, whereas the HPV18 family were more common in ADC.
Geographical region
Overall detected HPV prevalence varied little between geographical regions (83–89%), but was low compared to the almost 100% HPV prevalence identified in studies using the most sensitive HPV detection methods (Walboomers et al, 1999). This reflects the fact that many studies used HPV DNA detection strategies of suboptimal sensitivity. When comparing prevalence by region and histology, we attempted to take account of alternative HPV DNA sources and PCR primers by adjustment. However, it is not known to what extent other unknown sources of variation such as sample storage conditions, specific PCR conditions and quality of histopathology may affect these comparisons. Residual differences in prevalence between regions could also be because of the yet unknown HPV types not amplified by the existing PCR primers.
There were many similarities in HPV type-specific distribution across the regions studied. In SCC, HPV16 was clearly the predominant type varying from 45.9% in Asia to 62.6% in Europe, with HPV18 being found consistently in 10–14% of the cases. Other rarer types appeared to vary in their distribution. In most regions, HPV45, 31 and 33 were the third, fourth and fifth most common genotypes, although not necessarily in that order. Asia appeared to be different with a larger proportion of cancers associated with HPV58 and 52, as highlighted by a recent study in China of 786 cases in which HPV58 and 52 were the third (10%) and fourth (9%) most common genotypes in ICC (Wong et al, 2000). Other types in SCC were too rare to make inferences on region-specific variations.
Histological type of ICC
This meta-analysis shows that overall HPV prevalence detected in ADC was significantly lower than that detected in SCC. This intriguing finding does not appear to be because of differences with respect to region or HPV detection methods as it persisted even after adjusting for these factors. ADC arises from tissue deeper in the interior of the cervix uteri than SCC, and it has been reported to be more difficult to appropriately sample exfoliated cells of ADC than SCC (Sasieni and Adams, 2001). However, most HPV detected in the present review was based on biopsy specimens (77%). A proportion of cervical ADC could be misclassified ADC arising from the endometrium or other rare histological variants of ADC, for example, clear cell and mesonephric, which have been suggested to be HPV-independent (O'Leary et al, 1998; Pirog et al, 2000).
Whereas HPV16 was the most common type in SCC followed by HPV18, the situation was reversed in ADC where HPV18 was the most common type, followed closely by HPV16. This difference has been described independently by many of the studies in this analysis and by studies outside the scope of this review (IARC, 1995). Compared to HPV16, HPV18 has been shown to be associated with increasing oncogenic potential in cell culture (Barbosa and Schlegel, 1989), as well as a more rapid transition to malignancy (Burger et al, 1996) and a poorer prognosis of cancer patients (Nakagawa et al, 1996; Hildesheim et al, 1999; Schwartz et al, 2001). Given the fact that columnar tissue giving rise to ADC is less accessible, and possibly less susceptible to HPV infections, than the squamous tissue of SCC, the establishment of ADC may require a relatively more aggressive infection. In addition to HPV16 and 18, this large meta-analysis facilitated the identification of differences for some rarer phylogenetically related types: the HPV16-related types 31, 33, 35, 52, and 58 were more prevalent in SCC (15.0% collectively) than in ADC (4.4% collectively); and HPV18-related 45 was more prevalent in ADC (5.8%) than SCC (4.2%). All these differences were seen consistently in all regions where the comparison was possible.
For all regions where histological comparison was possible, the ratio of ADC to SCC was higher than that reported by cancer registries (Parkin et al, 1997). For example, ADC represent 23.1% of histologically verified cases from Europe in this study, but only 15.3% of ICC cases reported to European cancer registries (Parkin et al, 1997). Since all seven studies of only ADC cases were from Europe, Japan or USA, ADC is over-represented in this meta-analysis, particularly in developed countries. No material differences in results were observed when SCC was compared with cancers of unspecified histology.
Study limitations
The different PCR primers employed by the studies covered in this analysis varied in their overall detection of HPV DNA, with the highest prevalence being obtained with SPF10 and GP5+/6+ primers, supporting findings from previous studies (Davies et al, 2001). Such variation is partly because of known differences in the range of HPV DNA types amplifiable by each primer set, and this was taken into consideration when estimating type-specific prevalence. However, there is also evidence that not all primer sets amplify individual HPV types with the same sensitivity (Qu et al, 1997; Kleter et al, 1999), and such differences are a potential source of bias in this analysis.
The type-specific prevalence reported for each individual type includes that in multiple infections, which were reported in a total of 3.7% of our ICC cases. Since many of the included studies tested for only a subset of HPV types, many multiple infections will have been missed. Hence, this meta-analysis was unable to estimate how often individual HPV types were found in the presence of other types, which limits the conclusions that can be made about individual HPV-type oncogenicity. In particular, a large proportion of cases positive for HPV6, which is not thought to be oncogenic, may be coinfected with an undetected HR HPV type.
The cases included in this meta-analysis are not representative of the worldwide distribution of ICC. The proportion of cases contributed by Africa (6.1%) and Asia (30.7%) in this study under-represent their proportional burdens of worldwide cervical cancer, which are 14.1 and 49.4%, respectively (Parkin et al, 1999). In contrast, the proportion of cases contributed by Europe (33.2%) and North America and Australia (15.5%) over-represent their proportional burdens, which are 15.7 and 4.4%, respectively. Adjustment of type-specific prevalence in all ICC cases (Figure 1) by weighting each region according to their cancer burden, however, did not materially effect the results, highlighting the general similarity of HPV-type distribution across regions (data not shown).
Furthermore, the cases in this meta-analysis were not drawn uniformly from across each region. Large areas have not been included (e.g., the Middle East and Indian subcontinent in Asia), while other specific populations such as Japan in Asia are over-represented. There is also evidence of inter-regional variation in HPV-type distribution; the high prevalence of HPV52 and 58 in Asia is more apparent in cases from China/Korea/Japan than in those from South East Asia. Hence, for the comparison of alternative regional groupings, HPV-specific prevalence is presented by study in the Appendix (Table A1).
Table AI. Study methods and prevalence of human papillomavirus by study and by region.
|
HPV prevalence (% of all cases tested) |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First author | Reference | Country | HPV DNA source | PCR primers used to identify all HPV +ve | No. cases | SCC (incl. unspec)/ADC | Any | 16 | 18 | 45 | 31 | 33 | 58 | 52 | 35 | 59 | 56 | 51 | 6 | 68 | 39 | 82 | 73 | 66 | 70 |
|
Africa | |||||||||||||||||||||||||
| Bosch FX | JNCI (1995) | Algeria, Benin, Guinea, Mali,Uganda,Tanzania | Fresh biopsies | My09/11 | 186 | 186/0 | 89.8 | 42.5 | 17.7 | 12.4 | 2.7 | 2.7 | 2.7 | 2.2 | 2.2 | 0.0 | 3.2 | 1.1 | 0.0 | 2.2 | 0.5 | 0.0 | |||
| Bayo S | Int J Epidemiol (2002) | Mali | Fresh biopsies | GP5+/6+ | 65 | 65/0 | 96.9 | 47.7 | 12.3 | 10.8 | 0.0 | 1.5 | 3.1 | 0.0 | 1.5 | 0.0 | 0.0 | 6.2 | 0.0 | 0.0 | 0.0 | 3.1 | 0.0 | ||
| Chaouki N | Int J Cancer (1998) | Morocco | Exfol. cells | GP5+/6+ | 186 | 173/13 | 94.6 | 67.7 | 12.4 | 4.8 | 3.8 | 2.7 | 0.0 | 1.1 | 0.5 | 1.6 | 0.5 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 |
| Lin P | Cancer Epid Biomark Prev (2001) | Senegal | Exfol. cells | MY09/11 +HMB01 | 51 | 51/0 | 64.7 | 37.3 | 7.8 | 9.8 | 0.0 | 5.9 | 2.0 | 3.9 | 2.0 | 0.0 | 2.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| Williamson AL | J Med Virol (1994) | South Africa | Fresh biopsies | MY09/11 | 68 | 60/8 | 80.9 | 45.6 | 1.5 | 1.5 | 5.9 | 5.9 | 0.0 | ||||||||||||
| ter Meulen J | Int J Cancer (1992) | Tanzania | Exfol. cells | GP 5/6 | 53 | 53/0 | 88.7 | 37.7 | 32.1 | 5.7 | 0.0 | 1.9 | |||||||||||||
| Region subtotal | 609 | 588/21 | 88.8 | 50.2 | 14.1 | 7.9 | 2.6 | 3.1 | 1.6 | 1.6 | 1.4 | 0.6 | 1.6 | 1.2 | 0.4 | 0.8 | 0.0 | 0.3 | 0.7 | 0.2 | 0.0 | ||||
|
Asia | |||||||||||||||||||||||||
| Huang S | Int J Cancer (1997) | China | Fresh biopsies | MY09/11 | 40 | 35/5 | 87.5 | 27.5 | 30.0 | 0.0 | 0.0 | 0.0 | 27.5 | 27.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Lin QQ | Int J Cancer (1998) | China | Fresh/fixed biopsies | MY09/11, GP5+/6+ | 77 | 77/0 | 93.5 | 48.1 | 5.2 | 1.3 | 2.6 | 3.9 | 18.2 | 5.2 | 0.0 | 2.6 | 0.0 | 0.0 | 10.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 |
| Lo KWK | Gynecol Obstet Invest (2001) | China | Fresh biopsies | MY09/11 | 121 | 107/14 | 78.5 | 48.8 | 11.6 | 0.0 | 0.8 | 5.0 | 6.6 | 0.8 | 0.0 | 0.0 | 0.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Peng H | Int J Cancer (1991) | China | Exfol. cells | TS-PCR only | 101 | 92/9 | 34.7 | 31.7 | 3.0 | ||||||||||||||||
| Stephen AL | Int J Cancer (2000) | China | Fixed biopsies | GP5+/6+ of TS-PCR neg samples only | 34 | 24/10 | 88.2 | 61.8 | 8.8 | 2.9 | 0.0 | 2.9 | 2.9 | 0.0 | 0.0 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| Munirajan AK | Gynecol Oncol (1998) | India | Fresh biopsies | pU-1M/pU-2R | 43 | 43/0 | 69.8 | 53.5 | 9.3 | 0.0 | 2.3 | 2.3 | 0.0 | 2.3 | 0.0 | 0.0 | 0.0 | ||||||||
| Bosch FX | JNCI (1995) | Indonesia, Philippines, Thailand | Fresh biopsies | My09/11 | 98 | 98/0 | 96.9 | 42.9 | 31.6 | 8.2 | 1.0 | 2.0 | 2.0 | 2.0 | 1.0 | 1.0 | 3.1 | 0.0 | 0.0 | 1.0 | 4.1 | 0.0 | |||
| Fujinaga Y | J Gen Virol (1991) | Japan | Fresh biopsies | pU-1M/pU-2R | 39 | 39/0 | 84.6 | 48.7 | 12.8 | 5.1 | 5.1 | 7.7 | 2.6 | 0.0 | |||||||||||
| Harima Y | Int J Radiat Oncol Biol Phys (2002) | Japan | Fresh biopsies | pU-1M/pU-2R | 84 | 79/5 | 76.2 | 26.2 | 4.8 | 2.4 | 2.4 | 7.1 | 2.4 | 0.0 | 0.0 | 0.0 | 1.2 | ||||||||
| Kashiwabara K | Acta Pathol Japan (1992) | Japan | Fixed biopsies | L1C1/C2 | 93 | 68/25 | 58.1 | 48.4 | 6.5 | 0.0 | 1.1 | 0.0 | 3.2 | 0.0 | |||||||||||
| Maki H | Jpn J Cancer Res (1991) | Japan | Biopsies | L1 PCR | 29 | 29/0 | 82.8 | 44.8 | 20.7 | 6.9 | 6.9 | 6.9 | 0.0 | ||||||||||||
| Nagai Y | Am J Clin Oncol (2001) | Japan | Exfol. cells | L1C1/C2 | 293 | 239/54 | 85.3 | 25.3 | 3.1 | 3.8 | 4.1 | 3.1 | 1.7 | ||||||||||||
| Nakagawa S | Cancer (1996) | Japan | Fresh biopsies | L1C1/C2 +C2M | 146 | 116/30 | 88.4 | 37.7 | 18.5 | 0.7 | 2.1 | 6.2 | 8.2 | 10.3 | 2.1 | 0.0 | 0.0 | 0.7 | 0.0 | 2.1 | 0.0 | 0.0 | 0.7 | ||
| Nawa A | Cancer (1995) | Japan | Fresh/fixed biopsies | E6C1/C2 | 23 | 23/0 | 87.0 | 73.9 | 13.0 | 0.0 | |||||||||||||||
| Saito J | Gynecol Obstet Invest (2000) | Japan | Fixed biopsies | pU-1M/pU-2R | 66 | 66/0 | 75.8 | 34.8 | 12.1 | 10.6 | 1.5 | 6.1 | |||||||||||||
| Sasagawa T | Cancer EpidBiomark Prev (2001) | Japan | Exfol. cells | LCR-E7 | 84 | 72/12 | 89.3 | 42.9 | 14.3 | 1.2 | 6.0 | 1.2 | 3.6 | 10.7 | 1.2 | 0.0 | 0.0 | 4.8 | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Yamakawa Y | Gynecol Oncol (1994) | Japan | Fixed biopsies | TS-PCR only | 64 | 0/64 | 67.2 | 32.8 | 39.1 | 1.6 | 0.0 | 7.8 | 0.0 | ||||||||||||
| Hwang T | J Korean Med Sci (1999) | Korea | Exfol. cells | pU-1M/pU-2R | 41 | 39/2 | 92.7 | 36.6 | 9.8 | 7.3 | 9.8 | 17.1 | 7.3 | 2.4 | |||||||||||
| Kim KH | Yonsei Med J (1995) | Korea | Fixed biopsies | WD72/76 + WD66/67/154 | 30 | 30/0 | 70.0 | 53.3 | 16.7 | 0.0 | 0.0 | ||||||||||||||
| Yadav M | Med J Malaysia (1995) | Malaysia | Fresh biopsies | MY09/11 | 23 | 23/0 | 95.7 | 73.9 | 65.2 | 13.0 | 4.3 | ||||||||||||||
| Ngelangel C | JNCI (1998) | Philippines | Fresh biopsies+exfol. cells | GP5+/6+ | 356 | 323/33 | 93.5 | 38.8 | 25.6 | 12.9 | 0.6 | 0.0 | 2.5 | 2.8 | 0.0 | 2.0 | 0.6 | 2.5 | 0.0 | 0.6 | 0.3 | 0.3 | 0.3 | 1.1 | 0.0 |
| Chen SL | Cancer (1993) | Taiwan | Fresh biopsies | L1C1/C2, E6C1/C2/C3 | 43 | 40/3 | 72.1 | 46.5 | 4.7 | 0.0 | 2.3 | 2.3 | 2.3 | 0.0 | |||||||||||
| Chen TM | Int J Cancer (1994) | Taiwan | Fresh biopsies | MY09/11, pU-1M/pU-2R | 433 | 382/51 | 79.0 | 46.2 | 12.2 | 6.5 | 0.0 | ||||||||||||||
| Lai HC | Int J Cancer (1999) | Taiwan | Fresh biopsies | MY09/11 | 94 | 87/7 | 86.2 | 47.9 | 8.5 | 0.0 | 2.1 | 4.3 | 18.1 | 2.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Yang YC | Gynecol Oncol (1997) | Taiwan | Fixed biopsies | MY09/11 of TS-PCR neg samples only | 136 | 120/16 | 83.1 | 63.2 | 6.6 | 0.7 | 5.1 | ||||||||||||||
| Bhattarakosol P | J Med Assoc Thai (1996) | Thailand | Fixed biopsies | MY09/11 | 100 | 100/0 | 82.0 | 35.0 | 17.0 | 3.0 | 0.0 | ||||||||||||||
| Chichareon S | JNCI (1998) | Thailand | Fresh biopsies+exfol. cells | GP5+/6+ | 377 | 338/39 | 94.7 | 54.4 | 20.7 | 1.6 | 1.9 | 1.3 | 2.7 | 2.4 | 0.5 | 1.6 | 0.0 | 0.0 | 0.5 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.5 |
| Siritantikorn S | Southeast Asian J Trop Med Public Health (1997) | Thailand | Lavage | My09/11 | 23 | 21/2 | 60.9 | 56.5 | 4.3 | 4.3 | 4.3 | 0.0 | 0.0 | ||||||||||||
| Region subtotal | 3091 | 2710/381 | 83.1 | 43.4 | 15.3 | 4.5 | 2.0 | 3.4 | 5.4 | 4.2 | 1.0 | 1.1 | 0.4 | 1.0 | 0.5 | 0.5 | 0.2 | 0.4 | 0.1 | 0.3 | 0.3 | ||||
|
Europe | |||||||||||||||||||||||||
| Birner P | Mod Pathol (2001) | Austria | Fixed biopsies | GP 5+/6+ | 86 | 86/0 | 88.4 | 68.6 | 8.1 | 7.0 | 8.1 | 41.9 | 2.3 | 0.0 | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 | 0.0 | 0.0 |
| Baay MFD | J Clin Microbiol (2001) | Belgium | Fixed biopsies | GP5+/6+ of TS-PCR neg samples only | 115 | 95/20 | 87.8 | 67.0 | 13.0 | 1.7 | 2.6 | 1.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 |
| Tachezy R | J Med Virol (1999) | Czech Republic | Fresh biopsies | MY09/11 | 49 | 49/0 | 73.5 | 59.2 | 10.2 | 2.0 | 4.1 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.1 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Hording U | APMIS (1997) | Denmark | Fixed biopsies | TS-PCR only | 50 | 0/50 | 70.0 | 18.0 | 52.0 | 0.0 | |||||||||||||||
| Sebbelov AM | Microbes Infect (2000) | Denmark | Fixed biopsies | GP5/6 of TS-PCR neg samples only | 34 | 34/0 | 85.3 | 70.6 | 0.0 | 0.0 | 0.0 | 5.9 | 0.0 | ||||||||||||
| Iwasawa A | Cancer (1996) | Finland | Fixed biopsies | MY09/11 | 460 | 352/108 | 88.0 | 63.5 | 25.0 | 2.6 | 0.0 | ||||||||||||||
| Lombard I | J Clin Oncol (1998) | France | Fresh biopsies | TS-PCR of SBH neg samples only | 297 | 269/28 | 82.8 | 50.5 | 10.1 | 0.3 | 1.0 | 2.0 | 1.0 | 0.3 | 0.3 | 0.0 | 0.0 | ||||||||
| Riou G | Lancet (1990) | France | Biopsies | TS-PCR of SBH neg samples only | 106 | 89/17 | 84.0 | 54.7 | 16.0 | 5.7 | 0.0 | ||||||||||||||
| Milde-Langosch K | Int J Cancer (1995) | Germany | Biopsies | MY09/11 | 51 | 25/26 | 80.4 | 51.0 | 25.5 | 3.9 | 0.0 | 0.0 | 0.0 | ||||||||||||
| Nindl I | International Papillomavirus Conference Proceedings (2001) | Germany | Fixed biopsies | My09/11 | 77 | 77/0 | 89.6 | 42.0 | 16.0 | 18.0 | 6.0 | 0.0 | 1.0 | 1.0 | 0.0 | 8.0 | 1.0 | 1.0 | |||||||
| Bosch FX | JNCI (1995) | Germany, Poland, Spain | Fresh biopsies | MY09/11 | 86 | 86/0 | 95.3 | 65.1 | 8.1 | 2.3 | 5.8 | 1.2 | 1.2 | 3.5 | 1.2 | 0.0 | 2.3 | 0.0 | 0.0 | 3.5 | 0.0 | 0.0 | 1.0 | ||
| Dokianakis DN | Oncol Rep (1999) | Greece | Pap smears | GP 5/6 | 77 | 75/2 | 58.4 | 2.6 | 36.4 | 3.9 | |||||||||||||||
| Koffa M | Int J Oncol (1994) | Greece | Fixed biopsies | GP 5/6 | 39 | 32/7 | 76.9 | 35.9 | 38.5 | 10.3 | |||||||||||||||
| Labropoulou V | Sex Transmis Dis (1997) | Greece | Fresh biopsies | MY09/11 | 35 | 35/0 | 97.1 | 54.3 | 22.9 | 0.0 | 5.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| Sebbelov AM | Microbes Infect (2000) | Greenland | Fixed biopsies | GP5/6 of TS-PCR neg samples only | 32 | 32/0 | 84.4 | 81.3 | 0.0 | 0.0 | 3.1 | 3.1 | 0.0 | ||||||||||||
| Konya J | J Med Virol (1995) | Hungary | Fresh biopsies | L1C1/C2 +C2M | 47 | 41/6 | 97.9 | 55.3 | 40.4 | 2.1 | 0.0 | 0.0 | 2.1 | 0.0 | |||||||||||
| O'Leary JJ | J Clin Pathol (1998) | Ireland | Fixed biopsies | GP5/6, GP1/2 | 20 | 20/0 | 90.0 | 80.0 | 10.0 | 0.0 | 0.0 | ||||||||||||||
| Sjyldberg BM | Mod Pathol (1999) | Ireland, Sweden | Fixed biopsies | GP 5+/6+ | 38 | 0/38 | 60.5 | 23.7 | 26.3 | 0.0 | 0.0 | ||||||||||||||
| Garzetti GG | Cancer (1998) | Italy | Fresh biopsies | GP 5/6 | 32 | 32/0 | 68.8 | 50.0 | 15.6 | 6.3 | 0.0 | 0.0 | 0.0 | ||||||||||||
| Voglino G | Pathologica (2000) | Italy | Fixed biopsies | MY09/11, pU1M/2R | 145 | 120/25 | 98.6 | 71.0 | 6.9 | 16.6 | 2.1 | 0.6 | |||||||||||||
| Karlsen F | J Clin Microbiol (1996) | Norway | Fresh biopsies | My09/11, GP5+6+, Oli1b/2I, CpI/II | 361 | 361/0 | 98.3 | 68.4 | 14.1 | 1.1 | 8.3 | 0.6 | |||||||||||||
| Kleter B | J Clin Microbiol (1999) | Russia | Fixed biopsies | SPF10 | 180 | 129/51 | 100.0 | 64.4 | 9.4 | 7.8 | 3.9 | 1.1 | 1.7 | 1.1 | 1.7 | 0.0 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Munoz N | Int J Cancer (1992) | Spain | Exfol. cells | MY09/11 | 142 | 142/0 | 69.0 | 45.8 | 3.5 | 3.5 | 3.5 | 1.4 | 0.0 | 0.0 | 0.0 | ||||||||||
| Rodriguez JA | Diag Mol Pathol (1998) | Spain | Fixed biopsies | GP 5+/6+ | 54 | 54/0 | 85.2 | 61.1 | 7.4 | 5.6 | 5.6 | 1.9 | 1.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Andersson S | Eur J Cancer (2001) | Sweden | Fixed biopsies | My09/11 | 173 | 0/173 | 68.2 | 23.9 | 36.4 | 5.2 | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 | |||||
| Hagmar B | Med Oncol Tumor Pharmacother (1992) | Sweden | Fixed biopsies | MY09/11 | 71 | 71/0 | 74.6 | 38.0 | 9.9 | 16.9 | 8.5 | 0.0 | |||||||||||||
| Wallin KL | NEJM (1999) | Sweden | Fixed biopsies | MY09/11, GP 5+/6+ | 104 | 85/19 | 76.9 | 47.1 | 25.0 | 0.0 | 2.9 | 5.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| Zehbe I | J Pathol (1997) | Sweden | Fixed biopsies | GP 5+/6+ | 45 | 38/7 | 95.6 | 53.3 | 20.0 | 6.7 | 0.0 | 13.3 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |||||||
| Baay MFD | Eur J Gynaec Oncol (1996) | The Netherlands | Fixed biopsies | MY09/11, GP5/6, CpI/II | 162 | 162/0 | 87.7 | 61.7 | 14.2 | 0.6 | 1.9 | 4.3 | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| van den Brule AJC | Int J Cancer (1991) | The Netherlands | Fresh biopsies | GP5/6 + GP1/2 | 50 | 50/0 | 100.0 | 84.0 | 26.0 | 4.0 | 2.0 | 0.0 | |||||||||||||
| Arends MJ | Hum Pathol (1993) | UK | Fixed biopsies | TS-PCR only | 47 | 26/21 | 78.7 | 53.2 | 29.8 | 0.0 | 0.0 | ||||||||||||||
| Crook T | Lancet (1992) | UK | Fresh biopsies | TS-PCR of SBH neg samples only | 28 | 23/5 | 89.3 | 71.4 | 17.9 | 0.0 | 0.0 | 0.0 | |||||||||||||
| Giannudis A | Int J Cancer (1999) | UK | Fixed biopsies | GP 5+/6+ | 43 | 43/0 | 100.0 | 81.4 | 9.3 | 2.3 | 0.0 | 4.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.3 | 0.0 | |||
| Region subtotal | 3336 | 2733/603 | 85.9 | 56.0 | 17.5 | 2.9 | 4.2 | 4.4 | 0.8 | 0.5 | 0.5 | 0.1 | 0.6 | 0.0 | 0.3 | 0.4 | 0.3 | 0.0 | 0.3 | 0.1 | 0.0 | ||||
|
North America and Australia | |||||||||||||||||||||||||
| Chen S | Int J Gynecol Obstet (1999) | Australia | Fresh biopsies | My09/11 | 186 | 153/33 | 91.9 | 53.8 | 17.2 | 4.8 | |||||||||||||||
| Thompson CH | Gynecol Oncol (1994) | Australia | Fresh/fixed biopsies | pU-1M/pU-2R | 103 | 103/0 | 86.4 | 65.0 | 18.4 | 2.9 | 0.0 | 0.0 | |||||||||||||
| Duggan MA | Human Pathol (1995) | Canada | Fixed biopsies | L1C1/C2 of DBH neg samples only | 76 | 0/76 | 69.7 | 35.6 | 39.5 | 0.0 | 2.6 | 0.0 | 0.0 | 0.0 | |||||||||||
| Bosch FX | JNCI (1995) | USA, Canada | Fresh biopsies | My09/11 | 57 | 57/0 | 93.0 | 57.9 | 15.8 | 14.0 | 5.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.5 | 0.0 | 0.0 | 1.8 | 0.0 | 0.0 | |||
| Burger RA | JNCI (1996) | USA | Fresh biopsies | My09/11 | 401 | 297/104 | 84.8 | 51.4 | 19.7 | 2.2 | 2.5 | 1.0 | 0.5 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.2 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 |
| Burnett AF | Gynecol Oncol (1992) | USA | Fixed biopsies | My09/11 | 21 | 18/3 | 100.0 | 71.4 | 28.6 | 42.9 | 4.8 | 19.0 | 0.0 | ||||||||||||
| Ferguson AW | Mod Pathol (1998) | USA | Fixed biopsies | MY09/11 | 27 | 0/27 | 59.3 | 25.9 | 25.9 | 7.4 | |||||||||||||||
| Paquette RL | Cancer (1993) | USA | Fresh biopsies | MY09/11 | 45 | 28/17 | 93.3 | 48.9 | 40.0 | 4.4 | 0.0 | ||||||||||||||
| Schwartz SM | J Clin Oncol (2001) | USA | Fixed biopsies | MY09/11 | 465 | 354/111 | 88.2 | 58.5 | 23.2 | 1.3 | 2.4 | 3.4 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | 0.2 | 0.0 | 0.2 | 0.2 | 0.0 |
| Sebbelov AM | Microbes Infect (2000) | USA | Fixed biopsies | GP5/6 of TS-PCR neg samples only | 53 | 53/0 | 98.1 | 77.4 | 3.8 | 0.0 | 20.8 | 30.2 | 0.0 | ||||||||||||
| Wistuba II | Cancer Res (1997) | USA | Fixed biopsies | SPF10 | 20 | 20/0 | 90.0 | 60.0 | 20.0 | 5.0 | 5.0 | ||||||||||||||
| Resnick RM | JNCI (1990) | USA (+Holland) | Fixed biopsies | My09/11, WD72/76 | 33 | 29/4 | 100.0 | 75.8 | 15.2 | 3.0 | 3.0 | 3.0 | 0.0 | ||||||||||||
| Pirog EC | Am J Pathol (2000) | USA (+Poland) | Fixed biopsies | SPF10 | 76 | 0/75 | 89.5 | 39.5 | 38.2 | 10.5 | 1.3 | 1.3 | 0.0 | 2.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |||
| Region subtotal | 1562 | 1112/450 | 87.5 | 54.9 | 22.1 | 3.3 | 3.9 | 3.2 | 0.2 | 0.6 | 0.4 | 0.0 | 0.2 | 0.0 | 1.1 | 0.2 | 0.1 | 0.0 | 0.2 | 0.1 | 0.0 | ||||
|
South and Central America | |||||||||||||||||||||||||
| Bosch FX | JNCI (1995) | Argentina,Bolivia,Brazil,Chile,Colombia,Cuba,Panama,Paraguay | Fresh biopsies | MY09/11 | 505 | 505/0 | 92.9 | 50.5 | 9.5 | 7.3 | 6.9 | 3.6 | 2.2 | 3.2 | 2.0 | 2.8 | 0.6 | 1.0 | 0.2 | 0.4 | 0.2 | ||||
| Alonio LV | MEDICINA (2000) | Argentina | Biopsies | GP 5+/6+ | 30 | 30/0 | 93.3 | 46.7 | 20.0 | 3.3 | 3.3 | 6.7 | |||||||||||||
| Eluf-Neto J | Br J Cancer (1994) | Brazil | Exfol. cells | GP 5/6 | 186 | 186/0 | 84.4 | 53.8 | 9.7 | 2.2 | 3.2 | 0.0 | |||||||||||||
| Lorenzato F | Int J Gynecol Cancer (2000) | Brazil | Exfol. cells | MY09/11 | 59 | 59/0 | 89.8 | 59.3 | 38.5 | 0.0 | 11.9 | 5.1 | 3.4 | 1.7 | 3.4 | 0.0 | |||||||||
| Rabelo-Santos SH | International Papillomavirus Conference Proceedings (2000) | Brazil | Fixed biopsies | GP 5+/6+ | 56 | 51/5 | 80.4 | 57.1 | 5.4 | 1.8 | 8.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |||||||||
| Munoz N | Int J Cancer (1992) | Colombia | Exfol. cells | MY09/11 | 87 | 87/0 | 72.4 | 50.6 | 5.7 | 5.7 | 0.0 | 3.4 | 0.0 | ||||||||||||
| Herrero R | JNCI (2000) | Costa Rica | Exfol. cells | MY09/11 +HMB01 | 34 | 34/0 | 88.0 | 47.0 | 15.0 | 0.0 | 5.9 | 8.8 | 12.0 | 0.0 | 2.9 | 2.9 | 2.9 | 2.9 | 2.9 | 5.9 | 0.0 | 0.0 | 2.9 | 0.0 | |
| Ferrera A | Int J Cancer (1999) | Honduras | Exfol. cells | MY09/11 | 104 | 99/5 | 80.6 | 47.1 | 13.5 | 5.8 | 3.8 | 9.6 | 6.7 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Torroella-Kouri M | Gynecol Oncol (1998) | Mexico | Fresh biopsies+Exfol. Cells | MY09/11 +HMB01 | 69 | 59/10 | 87.0 | 42.0 | 14.5 | 10.1 | 2.9 | 0.0 | 4.3 | 2.9 | 0.0 | 4.3 | 1.4 | 0.0 | 0.0 | 1.4 | 4.3 | 1.4 | 0.0 | 1.4 | 1.4 |
| Meyer T | J Infect Dis (1998) | Mexico | Fresh biopsies | MY09/11 | 60 | 60/0 | 96.7 | 43.3 | 16.7 | 3.3 | 23.3 | 6.7 | 0.0 | 1.7 | 1.7 | 0.0 | 1.7 | 0.0 | 3.3 | 0.0 | 1.7 | 3.3 | 0.0 | 0.0 | 0.0 |
| Illades-Aguiar B | International Papillomavirus Conference Proceedings (2001) | Mexico | Fresh biopsies | MY09/11 | 74 | 66/8 | 97.3 | 60.8 | 10.8 | 0.0 | 10.8 | 1.4 | 4.1 | 2.7 | 0.0 | 0.0 | 0.0 | 0.0 | 1.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Santos C | Br J Cancer (2001) | Peru | Fresh biopsies | GP5+/6+ | 196 | 171/25 | 94.9 | 56.1 | 12.2 | 4.6 | 9.7 | 4.1 | 2.0 | 7.7 | 3.6 | 0.5 | 1.0 | 1.0 | 0.0 | 0.0 | 2.6 | 0.0 | 0.0 | 1.0 | 0.5 |
| Lo KWK | Int J Cancer (2002) | China | Fresh biopsies | MY09/11, GP5+/6+ | 809 | 731/78 | 83.7 | 66.9 | 6.3 | 0.4 | 0.6 | 1.5 | 3.2 | 2.2 | |||||||||||
| Mortazavi SH | Asian Pacific J Cancer Prev (2002) | Iran | Fixed biopsies | TS-PCR only | 69 | 61/8 | 85.5 | 73.9 | 11.6 | 1.2 | |||||||||||||||
| Ishikawa H | Cancer (2001) | Japan | Fixed biopsies | TS-PCR only | 52 | 52/0 | 76.9 | 53.8 | 23.1 | 1.9 | |||||||||||||||
| Dybikowska A | Oncol Rep (2002) | Poland | Exfol. cells | MY09/11 | 53 | 53/0 | 53.8 | 48.1 | 0.0 | 1.9 | 1.9 | 1.9 | 0.0 | ||||||||||||
| Pegoraro RJ | Int J Gynecol Cancer (2002) | South Africa | Fresh biopsies | MY09/11 | 190 | 190/0 | 98.4 | 46.8 | 14.2 | 4.7 | 10 | 1.1 | 1.6 | 1.6 | 0.0 | 0.5 | |||||||||
| Cuzick J | Br J Cancer (2000) | UK | Exfol. cells | MY09/11 | 116 | 85/31 | 94 | 65.5 | 11.2 | 0.0 | 11.2 | 6.9 | 1.7 | 0.0 | 1.7 | 0.0 | 1.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Region subtotal | 1460 | 1407/53 | 89.3 | 51.7 | 10.6 | 5.5 | 7.0 | 4.0 | 2.9 | 3.3 | 2.0 | 1.9 | 0.7 | 0.7 | 0.5 | 0.5 | 1.7 | 0.4 | 0.0 | 0.4 | 0.4 | ||||
| Total | 10 058 | 8550/2725 | 86.0 | 51.0 | 16.3 | 4.4 | 3.9 | 3.8 | 2.7 | 2.3 | 0.9 | 0.8 | 0.6 | 0.5 | 0.6 | 0.5 | 0.4 | 0.2 | 0.2 | 0.2 | 0.2 | ||||
Given that HPV is considered a virtually necessary cause of ICC, we further examined results restricted to HPV DNA-positive cases. This increased type-specific prevalence by a factor of ≈1.1 consistently for each HPV type, with no impact on the relative distribution of HPV types. However, given that many of the PCR systems used by the included studies amplify only a subset of HPV types, many HPV-‘negative’ cases may actually be infected with other, unascertained, HPV types. Thus, we did not consider it appropriate to restrict to HPV-positive cases when comparing type-specific prevalence across studies where PCR methodology differed considerably.
However, in order to make a broad overall estimate if one does assume that, HPV DNA should be detectable in 100% of ICC and that the distribution of undetected types in HPV-negative cases is similar to that in positive cases, this meta-analysis suggests that vaccinating against HPV16 and 18 should prevent over 70% of worldwide ICC. However, a worldwide vaccine against only HPV16/18 may prevent a larger proportion of ICC in Europe, North America and Australia (≈75%), than in Africa, Central and South America and Asia (59–64%), where a larger proportion of ICC cases were associated with other HPV types. Although this study identifies at least 18 HPV types associated with ICC from around the world, the most important type after HPV16 and 18 appears to be HPV45, followed by types 31, 33, 58 and 52, the relative importances of which vary by region.
Acknowledgments
The work reported in this paper was undertaken by Dr Gary Clifford during the tenure of an IARC Postdoctoral Fellowship from the International Agency for Research on Cancer. We thank the Vaccine Development team (VAD) of the Department of Vaccines and Biologicals of the World Health Organization, Geneva and Swiss Bridge (Award 2001) who supported and contributed to the funding of this work. We are grateful to Dr Teresa Aguado, Coordinator, VAD and Dr Sonia Pagliusi, Scientist, VAD for their critical comments during the preparation of the manuscript, as well as to Dr Rolando Herrero. We also thank those authors who made additional data available from their published studies.
Appendix A
A comparison of alternative regional groupings, HPV prevalence is presented by the study in Table AI
References
- Andersson S, Rylander E, Larsson B, Strand A, Silfversvard C, Wilander E (2001) The role of human papillomavirus in cervical adenocarcinoma carcinogenesis. Eur J Cancer 37: 246–250 [DOI] [PubMed] [Google Scholar]
- Barbosa MS, Schlegel R (1989) The E6 and E7 genes of HPV-18 are sufficient for inducing two-stage in vitro transformation of human keratinocytes. Oncogene 4: 1529–1532 [PubMed] [Google Scholar]
- Bernard HU, Chan SY, Manos MM, Ong CK, Villa LL, Delius H, Peyton CL, Bauer HM, Wheeler CM (1994) Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis 170: 1077–1085 [DOI] [PubMed] [Google Scholar]
- Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV (1995) Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst 87: 796–802 [DOI] [PubMed] [Google Scholar]
- Breslow NE, Day NE (1980) Statistical methods in cancer research. Vol. I – The analysis of case–control studies. IARC Sci Publ 32: 5–338 [PubMed] [Google Scholar]
- Burger RA, Monk BJ, Kurosaki T, Anton-Culver H, Vasilev SA, Berman ML, Wilczynski SP (1996) Human papillomavirus type 18: association with poor prognosis in early stage cervical cancer. J Natl Cancer Inst 88: 1361–1368 [DOI] [PubMed] [Google Scholar]
- Chaouki N, Bosch FX, Munoz N, Meijer CJ, El Gueddari B, El Ghazi A, Deacon J, Castellsague X, Walboomers JM (1998) The viral origin of cervical cancer in Rabat, Morocco. Int J Cancer 75: 546–554 [DOI] [PubMed] [Google Scholar]
- Chan SY, Delius H, Halpern AL, Bernard HU (1995) Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol 69: 3074–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, O'Sullivan H, Tabrizi SN, Fairley CK, Quinn MA, Garland SM, (1999) Prevalence and genotyping of HPV in cervical cancer among Australian women. Int J Gynaecol Obstet 67: 163–168 [DOI] [PubMed] [Google Scholar]
- Combita AL, Bravo M-M, Touze A, Coursaget P (2002) Serologic response to human oncogenic papillomavirus types 16,18,31,33,39,58 and 59 virus-like particles in Columbian women with invasive cervical cancer. Int J Cancer 97: 796–803 [DOI] [PubMed] [Google Scholar]
- Davies P, Kornegay J, Iftner T (2001) Current methods of testing for human papillomavirus. Best Pract Res Clin Obstet Gynaecol 15: 677–700 [DOI] [PubMed] [Google Scholar]
- Eluf-Neto J, Booth M, Munoz N, Bosch FX, Meijer CJ, Walboomers JM (1994) Human papillomavirus and invasive cervical cancer in Brazil. Br J Cancer 69: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravitt P, Hakenewerth A, Stoerker J (1991) A direct comparison of methods proposed for use in widespread screening of human papillomavirus infections. Mol Cell Probes 5: 65–72 [DOI] [PubMed] [Google Scholar]
- Guerrero E, Daniel RW, Bosch FX, Castellsague X, Munoz N, Gili M, Viladiu P, Navarro C, Zubiri ML, Ascunce N, Gonzalez LC, Tafur L, Izarzugaza I, Shah KV (1992) Comparison of ViraPap, Southern hybridization, and polymerase chain reaction methods for human papillomavirus identification in an epidemiological investigation of cervical cancer. J Clin Microbiol 30: 2951–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harima Y, Sawada S, Nagata K, Sougawa M, Ohnishi T (2002) Human papillomavirus (HPV) DNA associated with prognosis of cervical cancer after radiotherapy. Int J Radiat Oncol Biol Phys 52: 1345–1351 [DOI] [PubMed] [Google Scholar]
- Hildesheim A, Hadjimichael O, Schwartz PE, Wheeler CM, Barnes W, Lowell DM, Willett J, Schiffman M (1999) Risk factors for rapid-onset cervical cancer. Am J Obstet Gynecol 180: 571–577 [DOI] [PubMed] [Google Scholar]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (1995) Monographs on the evaluation of the carcinogenic risks to humans, Vol. 64, Human papillomaviruses. International Agency for Research on Cancer: LLyon: 64: 96–102 [Google Scholar]
- Illades-Aguiar B, Leyva-Vazquez MA, Alarcon-Romero LC, Alfaro-Flores E, Antonio-Veyar V, Lopez-Zamudio N, Teran-Porcayo MA (2000) Human papillomavirus infection in Mexican women with cervical cancer, squamous intraepithelial lesion and cytologically normal. Proceedings of the 18th International Papillomavirus Conference, Paris
- Koutsky LA, Ault KA, Wheeler CM, Brown DR, Barr E, Alvarez FB, Chiacchierini LM, Jansen KU (2002) A controlled trial of a human papillomavirus type 16 vaccine. N Engl J Med 347: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Kleter B, van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, ter Schegget J, Lindeman J, ter Harmsel B, Burger M, Quint W (1999) Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol 37: 2508–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw KL, Hildesheim A, Burk RD, Gravitt P, Wacholder S, Manos MM, Scott DR, Sherman ME, Kurman RJ, Glass AG, Anderson SM, Schiffman M (2001) A prospective study of human papillomavirus (HPV) type 16 DNA detection by polymerase chain reaction and its association with acquisition and persistence of other HPV types. J Infect Dis 183: 8–15 [DOI] [PubMed] [Google Scholar]
- Lin P, Koutsky LA, Critchlow CW, Apple RJ, Hawes SE, Hughes JP, Toure P, Dembele A, Kiviat NB (2001) HLA Class II DR-DQ and increased risk of cervical cancer among Senegalese women. Cancer Epidemiol Biomarkers Prev 10: 1037–1045 [PubMed] [Google Scholar]
- Meyer T, Arndt R, Christophers E, Beckmann ER, Schroder S, Gissmann L, Stockfleth E, Gonzalez LC, Kaldor JM, Guerrero E, Lorincz A, Santamaria M, Alonso de Ruiz P, Aristizabal N, Shah KV (1998) Association of rare human papillomavirus types with genital premalignant and malignant lesions. J Infect Dis 178: 252–255 [DOI] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Tafur L, Izarzugaza I, Gili M, Viladiu P, Navarro C, Martos C, Ascunce N, Gonzalez LC, Kaldor JM, Guerrero E, Lorincz A, Santamaria M, Alonso de Ruiz P, Aristizabal N, Shah KV (1992) The causal link between human papillomavirus and invasive cervical cancer: a population-based case–control study in Colombia and Spain. Int J Cancer 52: 743–749 [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Yoshikawa H, Onda T, Kawana T, Iwamoto A, Taketani Y (1996) Type of human papillomavirus is related to clinical features of cervical carcinoma. Cancer 78: 1935–1941 [PubMed] [Google Scholar]
- Nindl I, Arndt R, Schneede P, Enno C, Eggert S, Meyer T (2001) Low oncogenic potential of HPV53 examined in patients with normal cytology, condylomata acuminata, CIN and cervical cancer. Proceedings of the 19th International Papillomavirus Conference, Paris, Vol. 20
- O'Leary JJ, Landers RJ, Crowley M, Healy I, O'Donovan M, Healy V, Kealy WF, Hogan J, Doyle CT (1998) Human papillomavirus and mixed epithelial tumors of the endometrium. Hum Pathol 29: 383–389 [DOI] [PubMed] [Google Scholar]
- Parkin DM, Whelan SL, Ferlay J, Raymond L, Young J (1997) Cancer Incidence in Five Continents, Vol. VII. IARC Scientific Publications No. 143. Inteernationall Agency for Research on Cancer: Lyon
- Parkin DM, Pisani P, Ferlay J (1999) Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer 80: 827–841 [DOI] [PubMed] [Google Scholar]
- Pirog EC, Kleter B, Olgac S, Bobkiewicz P, Lindeman J, Quint WG, Richart RM, Isacson C (2000) Prevalence of human papillomavirus DNA in different histological subtypes of cervical adenocarcinoma. Am J Pathol 157: 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, Burk RD (1997) PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol 35: 1304–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabelo-Santos SH, Magalhaes AV, Amaral RG (2001) Human papillomavirus DNA prevalence among women with CIN III and invasive cancer. Proceedings of the 19th International Papillomavirus Conference
- Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ (1995) The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol 76: 1057–1062 [DOI] [PubMed] [Google Scholar]
- Sasieni P, Adams J (2001) Changing rates of adenocarcinoma and adenosquamous carcinoma of the cervix in England. Lancet 357: 1490–1493 [DOI] [PubMed] [Google Scholar]
- Schiffman MH, Bauer HM, Lorincz AT, Manos MM, Byrne JC, Glass AG, Cadell DM, Howley PM (1991) Comparison of Southern blot hybridisation and polymerase chain reaction methods for the detection of human papillomavirus DNA. J Clin Microbiol 29: 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SM, Daling JR, Shera KA, Madeleine MM, McKnight B, Galloway DA, Porter PL, McDougall JK (2001) Human papillomavirus and prognosis of invasive cervical cancer: a population-based study. J Clin Oncol 19: 1906–1915 [DOI] [PubMed] [Google Scholar]
- Sebbelov AM, Davidson M, Kruger KS, Jensen H, Gregoire L, Hawkins I, Parkinson AJ, Norrild B (2000) Comparison of human papillomavirus genotypes in archival cervical cancer specimens from Alaska natives, Greenland natives and Danish Caucasians. Microbes Infect 2: 121–126 [DOI] [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189: 12–19 [DOI] [PubMed] [Google Scholar]
- White H (1980) A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica 48: 817–830 [Google Scholar]
- Wong YF, Cheung TH, Chan MK, Poon CS, Chung TK (2000) Prevalence and genotype of human papillomavirus in cervical carcinoma: a nationwide perspective in China. Proceedings of the 18th International Papillomavirus Conference, Paris
- Yang YC, Shen J, Tate JE, Wang KG, Su TH, Wang KL, Jeng CJ, Chen HS, Chiang S, Crum CP (1997) Cervical cancer in young women in Taiwan: prognosis is independent of papillomavirus or tumor cell type. Gynecol Oncol 64: 59–63 [DOI] [PubMed] [Google Scholar]