Summary
The Bambi (Bmp and activin membrane-bound inhibitor) gene encodes a transmembrane protein highly similar in amino acid sequence to TGF-beta type-I receptors. Unlike the TGF-beta receptors, however, the Bambi intracellular domain is short and lacks a serine/threonine-kinase domain that is essential for transducing TGF-beta signaling. Previous biochemical assays showed that Bambi interacts directly with BMP receptors and antagonizes BMP signaling. Interestingly, the expression of Bambi largely overlaps, both temporally and spatially, with that of Bmp4 during early embryonic development in Xenopus, zebrafish, and mice, which led to the hypothesis that Bambi may function to regulate BMP signaling during embryogenesis. To directly analyze the roles of Bambi during embryonic development, we generated mice carrying a conditional allele of Bambi, Bambiflox, with loxP sequences flanking the first exon that encodes the N-terminus and signal peptide region of the Bambi protein. Mice homozygous for this targeted conditional allele appear normal and fertile. We crossed the Bambiflox/+ mice to the EIIa-Cre transgenic mice and generated mice carrying deletion of the first exon of the Bambi gene. Surprisingly, mice homozygous for the deleted allele were viable, fertile and didn’t exhibit any discernible developmental defect. Our data exclude an essential role for Bambi in mouse embryonic development and postnatal survival.
Keywords: Bambi, BMP signaling, Conditional knockout, Cre/loxP
The transforming growth factor-beta (TGFβ) superfamily of secreted signaling molecules, including TGFβs, bone morphogenetic proteins (BMPs), and activins, plays essential roles throughout animal development. These molecules initiate cellular signaling by binding to and bringing together two types of receptor serine/threonine kinases. Upon ligand binding, the type-II receptor phosporylates and activates the type-I receptor kinase, which in turn phosphorylates and activates a set of transcriptional co-activators called Smad proteins and leads to their nuclear translocation and transcriptional activation of downstream target genes (reviewed in Shi and Massague, 2003; Nohe et al., 2004). In an expression screen for components involved in BMP4 signaling in Xenopus embryos, Onichtchouk et al. (1999) identified a gene encoding a transmembrane protein highly related to type-I TGFβ receptors. They named the gene product BAMBI (BMP and activin membrane-bound inhibitor) because it lacked an intracellular kinase domain and its overexpression antagonized BMP and activin signaling in Xenopus embryos (Onichtchouk et al., 1999). The Bambi gene is evolutionarily conserved and its homologs have been identified in human (Degen et al., 1996), Zebrafish (Tsang et al., 2000), mouse (Grotewold et al., 2001), and rat (Loveland et al., 2003).
Ectopic expression of Bambi mRNA in Xenopus and zebrafish embryos resulted in phenotypes resembling inhibition of BMP signaling (Onichtchouk et al., 1999; Tsang et al., 2000). In vitro biochemical assays showed that the Bambi protein associated with type-I and type-II BMP/TGFβ receptor complexes in the presence of ligands, suggesting that Bambi inhibits BMP/TGFβ signaling by interfering with receptor complex formation or phosphorylation of type-I receptors (Onichtchouk et al., 1999; Tsang et al., 2000). Interestingly, Bambi is coexpressed with Bmp4 in many developmental processes during embryogenesis in Xenopus (Onichtchouk et al., 1999), zebrafish (Tsang et al., 2000), and mouse (Grotewold et al., 2001). The co-expression and the ability of Bambi to antagonize BMP/TGFβ signaling suggest that Bambi may play important roles in the regulation of BMP4 signaling during embryonic development.
BMP4 signaling is essential for many developmental processes. Mouse embryos lacking BMP4 function die at midgestation with severe defects in mesoderm formation (Winnier et al., 1995). BMP4+/− heterozygous mice exhibit variable defects in craniofacial, eye, kidney, and limb development (Dunn et al., 1997). Moreover, mice lacking chordin and/or noggin, endogenous inhibitors of BMP signaling, have multiple developmental defects (Brunet et al., 1998; McMahon et al, 1998; Bachiller et al., 2000; Bachiller et al., 2003), indicating that the levels of BMP signaling are highly regulated during normal embryonic development. Since Bambi is a unique inhibitor of Bmp signaling at the receptor level and since Bambi mRNA is co-expressed with Bmp4 during craniofacial, eye, kidney, and limb development (Grotewold et al., 2001), it may play important roles in these developmental processes by fine-tuning BMP signaling.
To investigate the roles of Bambi in mammalian development, we generated mice carrying a conditional null allele of the Bambi gene, Bambiflox, through homologous recombination in mouse embryonic stem cells and subsequent production of chimeric mice (see MATERIALS AND METHODS, and FIG. 1a). Both Bambiflox/+ and Bambiflox/flox mice were viable, fertile and didn’t display any obvious phenotypic abnormality.
FIG. 1.
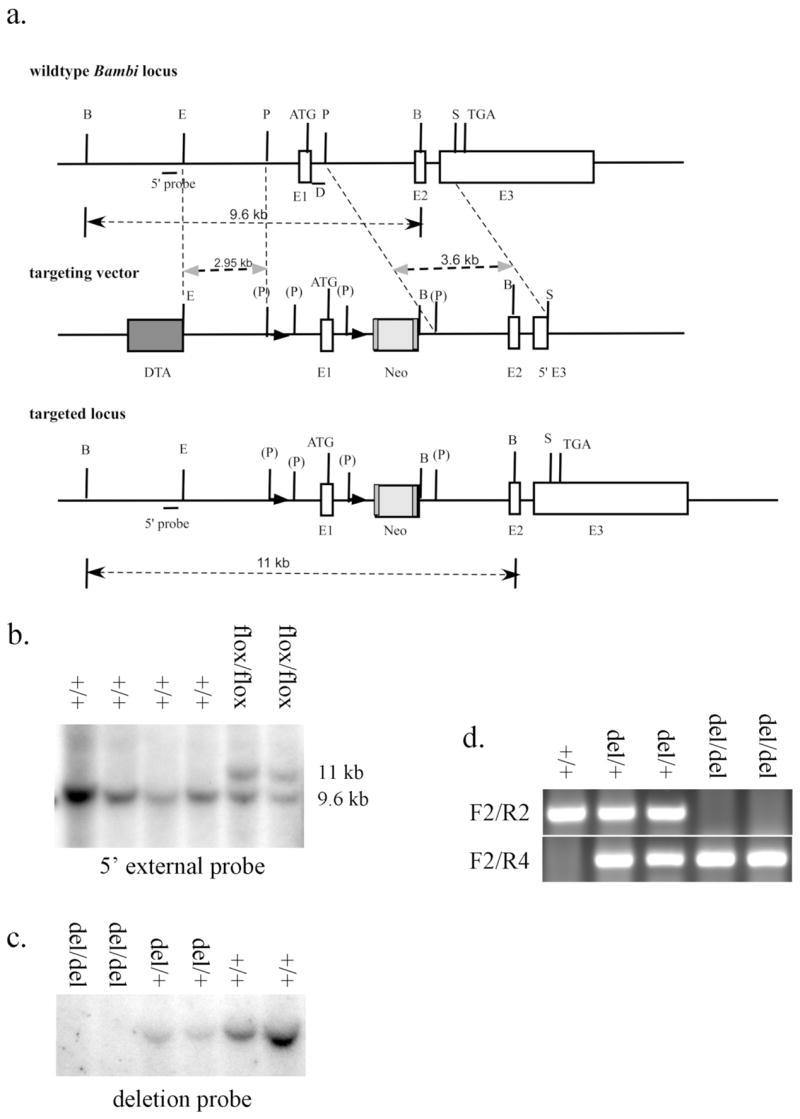
Generation of Bambiflox and Bambidel mice. (a) Schematic representation of the Bambi genomic locus, the gene targeting construct and the targeted (Bambiflox) allele. LoxP sequences are indicated by black triangles. D marks the deletion probe; DTA, diphtheria toxin A expression cassette; E1, exon 1; E2, exon 2; E3, exon 3; Neo, Frt-flanked PGK-Neo cassette. The restriction enzyme recognition sites are: B, Bgl I; E, EcoRI; P, PmlI; S, SacI. (P) marks PmlI site destroyed during subcloning. (b) Southern hybridization analysis of BglI-digested genomic DNA from representative G418-resistant ES clones using the 5′ external probe. The upper band (11 Kb) represents the targeted allele and the lower band (9.6 Kb) represents the wildtype allele. (c) Southern hybridization analysis of EcoRI-digested tail genomic DNA from Bambi+/+, Bambidel/+, and Bambidel/del mice using the deletion probe. A 7.7 Kb band was only detected in the wildtype and heterozyougs mouse DNA samples, but not in DNA from the homozygous Bambidel/del mice. (d) PCR analysis of tail DNA of Bambi+/+, Bambidel/+, and Bambidel/del mice using primers F2, R2 and R4. The upper fragment (215 bp) represents the wildtype allele, and the lower band (316 bp) represents the Bambidel allele.
We crossed Bambiflox/+ mice to either EIIa-cre or CMV-cre transgenic mice, both of which could mediate deletion of loxP-flanked sequences in germ cells (Lakso et al., 1996; Dupe et al., 1997). Bambidel/+ mice were then intercrossed. The deletion of loxP-flanked sequence was confirmed by Southern hybridization analysis of EcoRI-digested genomic DNA using a probe corresponding to the deleted region (FIG. 1a). A 7.7 Kb band representing the wildtype allele was detected in both wildtype and Bambidel/+ heterozygous mice, but absent in Bambidel/del homozygotes (FIG. 1c). The presence of genomic DNA in all lanes was confirmed by hybridizing the same membrane to a probe corresponding to genomic DNA 5′ to the deletion (data not shown). The deletion was also verified by PCR genotyping. The wildtype allele gives a 215 bp PCR product using the primer pair F2/R2 and the deleted allele results in a 316 bp PCR product using the primers F2 and R4 (FIG. 1d). Furthermore, we performed RT-PCR analysis of total RNA from wildtype and Bambidel/del mutant mouse embryos using primers RF1 and RR1. As expected, a 329 bp product was amplified from wildtype but not homozygous mutant littermate RNA samples (FIG. 2b). Similarly, a 586 bp product was amplified from wildtype but not homozygous mutant littermate RNA samples using primers flanking the exons 1 and 2 (FIG. 2c); indicating that the loxP-flanked sequence had been successfully deleted by the Cre recombinase. Since the 1075 bp genomic deletion includes the Bambi gene promoter and the first exon encoding the signal peptide, the deleted allele should represent a null allele of Bambi.
FIG. 2.
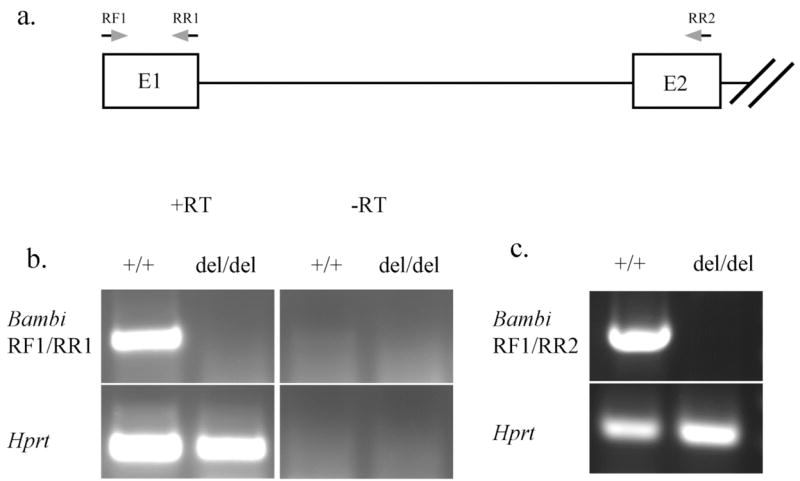
RT-PCR analysis of RNAs isolated from E10.5 wildtype and Bambi del/de embryos. (a) The approximate positions of primers used in (b) and (c) are indicated by arrows. E1, exon 1; E2, exon 2. (b) A 329 bp fragment corresponding to the entire exon 1 was only amplified from wildtype but not Bambi del/de embryo RNA samples. As internal control, RT-PCR product corresponding to the Hprt mRNA was amplified from both wildtype and Bambi del/de embryo RNA samples. RT, with reverse transcription; -RT, without reverse transcription. (c) A 586 bp fragment containing Exon 1 and Exon 2 sequences was amplified from wildtype but not mutant RNA samples using primers RF1 and RR2 as indicated in (a). RT-PCR amplification of a fragment from Hprt mRNA was used as internal control.
Of 125 pups from heterozygous intercross genotyped at two weeks of age, 30 (24.0%) were of wildtype, 59 (47.2%) were heterozygous, and 36 (28.8%) were homozygous for the Bambidel allele. Chi-square analysis indicated that the distribution of these three genotypes was not significantly different from the expected Mendelian ratio. Morphological and histological examination of Bambidel/del homozygous mice didn’t detect any overt defects, indicating that the Bambi gene was not essential for normal mouse embryogenesis and postnatal survival.
To assess whether loss of Bambi function has any effect on postnatal growth, the body weights of homozygous mutant, heterozygous and wild type littermates were measured at regular internals up to two months after birth (FIG. 3). Independent student’s t-test indicated that the body weights of Bambidel/del female mice were not significantly different from those of wildtype and heterozygous littermates before weaning (pups were usually weaned at 21 – 22 days after birth). However, the Bambidel/del homozygous mutant female mice exhibited a small (9%–10%) but statistically significant (P<0.05) reduction in body weight, compared to their wildtype littermates. The Bambidel/+ heterozygous female mice had body weights in between and not statistically significant from the wildtype and homozygous mutant littermates. For male mice, body weights of Bambidel/del homozygous mutants were not significantly different from those of wildtype and heterozygous littermates at all the stages examined.
FIG. 3.
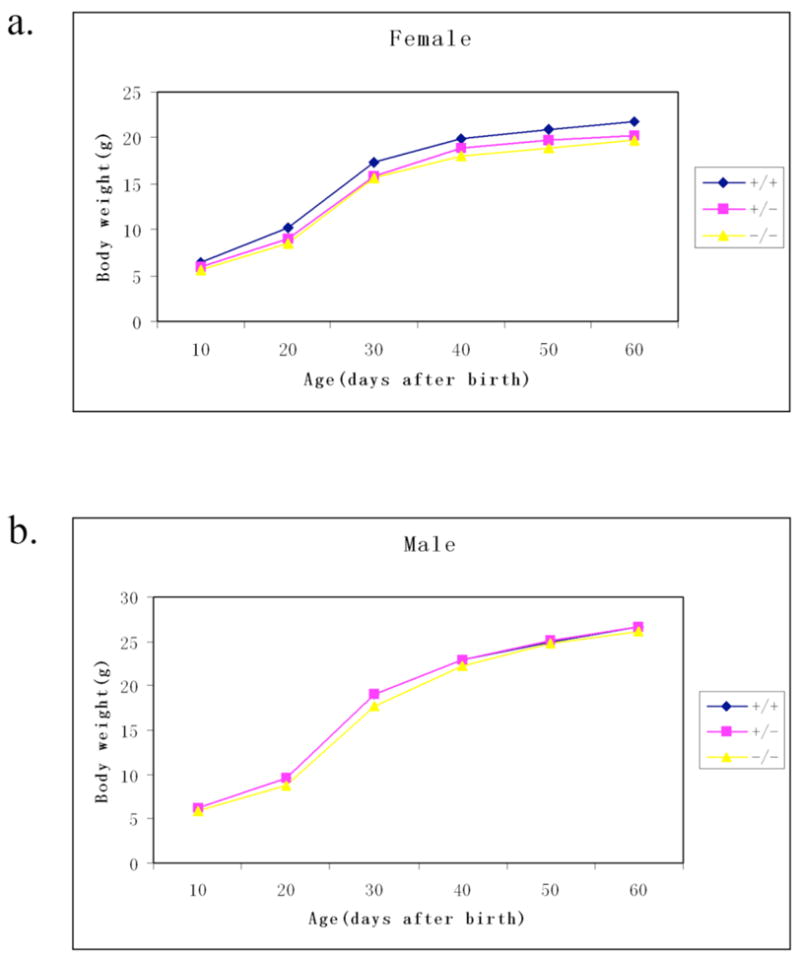
Growth curves of wild type mice, Bambidel/+ heterozygous, and Bambidel/del homozygous mice. (a) Growth curves of female wildtype (n=7), Bambidel/+ heterozygous (n=12), and Bambidel/del homozygous (n=7) mice. From 30 days onwards, the body weights of the Bambidel/del female mice were 9%–10% lower than those of wildtype mice (P<0.05). (b) Growth curve shows the average body weights of male wildtype (n=9), Bambidel/+ heterozygous (n=18), and Bambidel/del homozygous (n=10) mice taken on the days indicated for a period of 60 days. At all the stages examined, body weights of Bambidel/del male mice were not significantly different from those of wildtype and heterozygous male mice.
We further investigated whether loss of Bambi function affected male and female fertility. Bambidel/del homozygous males and females were intercrossed and littersizes were monitored. Pups were born with normal litter sizes and were normally nursed by their mothers, suggesting that deletion of Bambi has no significant effect on fertility. These data also exclude any maternal or paternal effect on the Bambi gene.
Since BMP signaling plays important roles in skeletal development, and genetic inactivation of other endogenous Bmp inhibitors resulted in skeletal defects (e.g., Balemans et al., 2002), we examined skeletal preparations of newborn pups. Compared with wildtype littermates, Bambidel/del mutant mice didn’t exhibit any obvious defect in the skeleton (data not shown).
Bambi is co-expressed with Bmp4 during craniofacial and limb development (Grotewold et al., 2001). We carefully examined these structures in newborn mice by histological and skeletal preparations. Bambidel/del mutant mice didn’t display any obvious craniofacial or limb defects (Fig. 4).
FIG. 4.
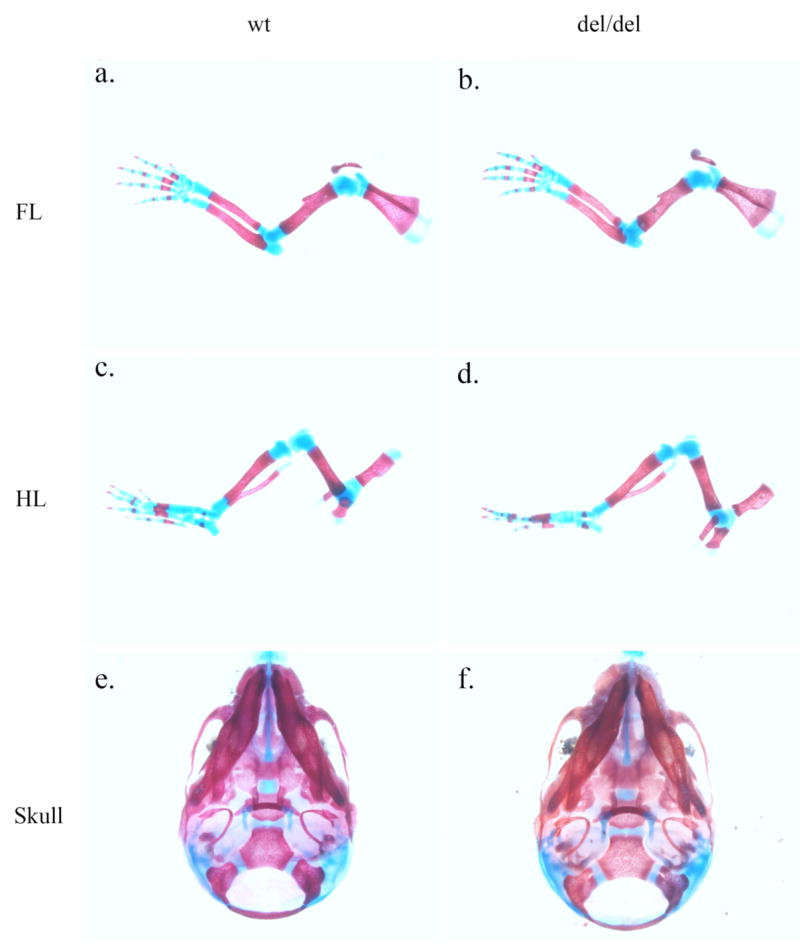
Bambi is not essential for limb and crnaiofacial development. Skeletal preparations of the forelimb (a, b), hindlimb (c, d), and skull (e, f) were compared between wildtype (a, c, e) and Bambidel/del homozygous (b, d, f) newborn littermates. No significant difference was found. (e) and (f) show the ventral view of the skulls.
In conclusion, we have generated mice carrying a floxed conditional allele of Bambi. These mice were crossed to Cre-transgenic mice and mice carrying the deleted null Bambi allele were generated. Analysis of Bambidel/del mutant mice indicated that Bambi function is not essential for embryonic development and postnatal survival although it has some minor effect on the growth of female mice after weaning.
MATERIALS AND METHODS
Generation of Bambiflox and Bambidel mice
A BAC clone containing the Bambi genomic region was obtained by screening the RPCI-22 129/SvEvTac mouse BAC library (BACPAC Resources, Children’s Hospital of Oakland, Oakland, CA). An 7.7 Kb EcoRI fragment containing exons 1 and 2 as well as an 13.5 Kb BamHI fragment containing the entire gene were subcloned into pBluescript plasmid vectors for construction of the targeting vector. The targeting vector contained the following fragments in the 5′ to 3′ order: (1) a diphtheria toxin (DTA) expression cassette, (2) an 2.9 Kb EcoRI/PmlI fragment from 5′ to the Bambi gene promoter as the 5′ homologous arm, (3) a loxP sequence, (4) the 1.07 Kb PmlI fragment containing the Bambi gene promoter and first exon, (5) another loxP sequence followed by an FRT-flanked PGKneo expression cassette, and (6) the 3.6 Kb PmlI-SacI fragment from Intron1 to exon3 region as the 3′ homology arm (FIG. 1a). The final targeting vector was linearized at a unique KpnI restriction site 3′ to the 3′ homology arm and electroporated into CJ7 mouse embryonic stem (ES) cells.
Following electroporation, ES cells were subjected to G418 selection as described previously (Swiatek and Gridley, 1993). G418-resistant ES clones were screened by Southern hybridization analysis of BglI-digested ES genomic DNA using a 5′ external probe. Three independent targeted ES clones were microinjected into C57BL/6J blastocysts that were subsequently transferred into the uterus of pseudopregnant C57BL/6J female mice to generate chimeric mice. Chimeric mice from two of the three ES clones gave germ-line transmission of the targeted allele and were crossed to C57BL/6J female mice to generate Bambiflox/+ heterozygous mice, which were than intercrossed to generate and test whether there is any obvious defects in the Bambiflox/flox mice.
To generate mice with loss of Bambi gene function, Bambiflox/+ heterozygous mice were crossed to either EIIa-Cre or CMV-Cre transgenic mice (purchased from The Jackson Laboratory, Bar Harbor, ME). Mice carrying the targeted allele and the Cre transgene were backcrossed to C57BL/6J mice to generate mice heterozygous for the deletion of the floxed first exon (Bambidel/+) but without the Cre transgene. The Bambidel/+ heterozygous males and females were intercrossed for generation and analysis of Bambidel/del homozygous mutant mice.
All animal protocols were approved by the University of Rochester Committee on Animal Resources in accordance with National Institute of Health guidelines.
Genotyping
For genotyping, DNA was isolated from either tail or embryonic yolk sac samples. To distinguish wildtype from the Bambiflox allele, forward primer F2 (GGGATCCGGGTCTTGGAGAC) and reverse primer R2 (TCGGAGTTTGCTACGAGACC) were used for PCR, amplifying an 215 bp product from the wildtype allele and an 336 bp product from the Bambiflox allele. The Bambidel allele was detected by PCR using the forward primer F2 and reverse primer R4 (TTCGGAATAGGAACTTCGTCG), which yielded an 316 bp product. The program used for PCR genotyping reactions is as follows: 1 cycle of 94°C for 4 minutes; followed by 40 cycles of 30 seconds at 94°C, 30 seconds at 62°C, and 40 seconds at 72°C. PCR products were resolved on 1.5% agarose gels.
RT-PCR
Total RNAs were extracted from E10.5 whole embryos using Trizol reagents (Invitrogen), treated with RNase-free DNase I (Promega) to remove any residual genomic DNA, and purified further using the RNAeasy kit (QIAGEN). 3 μg of each purified total RNA sample was reverse-transcribed to generated first-strand cDNAs using the Superscript II reverse transcription kit (Invitrogen). The following primers were used for RT-PCR analysis of Bambi: RF1, GTGTAGGCGTTGCTCTCTGT; RR1, CTTTGGTGAGCAGCACAGCC; and RR2, CGTCATGCAGTCCTCGATAA. The following PCR program was used for the PCR reactions: 1 cycle of 94°C for 4 min; followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and 40 seconds at 72°C.
Skeletal Preparation, Growth Curve and Statistical Analysis
Skeletal preparation was performed as described previously (Martin et al., 1995). For growth curve analysis, the body weights of mice were measured every 10 days for a period of 60 days starting from the 10th day after birth. Male and female growth curves were generated separately. For statistical analysis, independent Student’s t test was used, and a P value less than 0.05 was regarded as significant.
Chi-square test was used for determining whether distribution of genotypes of progeny from Bambidel/+ heterozygous intercrosses reflected the expected Mendelian ratio, and P value less than 0.05 was considered significant.
Acknowledgments
We thank Tom Gridley for the CJ7 ES cells, Phil Soriano and Anne Moon for plasmids for construction of the gene targeting vector. This work was supported by NIH/NIDCR grants DE015207 and DE16215 (subcontract from the University of Iowa) to RJ. JOB was supported by the NIH training grant DE07202.
LITERATURE CITED
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Shneyder N, Tran U, Anderson R, Rossant J, De Robertis EM. The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development. 2003;130:3567–3578. doi: 10.1242/dev.00581. [DOI] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- Degen WG, Weterman MA, van Groningen JJ, Cornelissen IM, Lemmers JP, Agterbos MA, Geurts van Kessel A, Swart GW, Bloemers HP. Expression of nma, a novel gene, inversely correlates with the metastatic potential of human melanoma cell lines and xenografts. Int J Cancer. 1996;65:460–465. doi: 10.1002/(SICI)1097-0215(19960208)65:4<460::AID-IJC12>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Dunn NR, Winnier GE, Hargett LK, Schrick JJ, Fogo AB, Hogan BL. Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev Biol. 1997;188:235–247. doi: 10.1006/dbio.1997.8664. [DOI] [PubMed] [Google Scholar]
- Dupe V, Davenne M, Brocard J, Dolle P, Mark M, Dierich A, Chambon P, Rijli FM. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- Grotewold L, Plum M, Dildrop R, Peters T, Ruther U. Bambi is coexpressed with Bmp-4 during mouse embryogenesis. Mech Dev. 2001;100:327–330. doi: 10.1016/s0925-4773(00)00524-4. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland KL, Bakker M, Meehan T, Christy E, von Schonfeldt V, Drummond A, de Kretser D. Expression of Bambi is widespread in juvenile and adult rat tissues and is regulated in male germ cells. Endocrinology. 2003;144:4180–4186. doi: 10.1210/en.2002-0124. [DOI] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeobox gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 2004;16:291–299. doi: 10.1016/j.cellsig.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401(6752):480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Swiatek P, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Tsang M, Kim R, de Caestecker MP, Kudoh T, Roberts AB, Dawid IB. Zebrafish nma is involved in TGFbeta family signaling. Genesis. 2000;28:47–57. doi: 10.1002/1526-968x(200010)28:2<47::aid-gene20>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]


