Abstract
Cells are known to be surrounded by nanoscale topography in their natural extracellular environment. The cell behavior, including morphology, proliferation, and motility of bovine pulmonary artery smooth muscle cells (SMC) were studied on poly(methyl methacrylate) (PMMA) and poly(dimethylsiloxane) (PDMS) surfaces comprising nanopatterned gratings with 350 nm linewidth, 700 nm pitch, and 350 nm depth. More than 90% of the cells aligned to the gratings, and were significantly elongated compared to the SMC cultured on non-patterned surfaces. The nuclei were also elongated and aligned. Proliferation of the cells was significantly reduced on the nanopatterned surfaces. The polarization of microtubule organizing centers (MTOC), which are associated with cell migration, of SMC cultured on nanopatterned surfaces showed a preference towards the axis of cell alignment in an in vitro wound healing assay. In contrast, the MTOC of SMC on non-patterned surfaces preferentially polarized towards the wound edge. It is proposed that this nanoimprinting technology will provide a valuable platform for studies in cell-substrate interactions and for development of medical devices with nanoscale features.
Keywords: Nanotopography, Smooth muscle cells, Bionanotechnology, Nanoimprinting, Nanomedicine, Cell-substrate interaction
1. Introduction
Cell-substratum interactions are central to many biological phenomena. Knowledge of these interactions is crucial to the understanding of many fundamental biological questions and to the design of medical devices. Tissue engineering is an example where control of these interactions is essential to the creation of functional engineered-tissues [1–6]. The performance of many implantable medical devices is also dependent on the desired device–tissue response [7]. Response of cells to topographical cues and the concept of contact guidance have been known for decades [8,9]. Various topographical features such as grooves, ridges, stops, pores, wells and nodes in micro- or nanoscale [10–12] have been presented to a wide variety of cells: fibroblasts [13–17], BHK cells [18], neuronal cells [19], macrophages [20,21], epithelial cells [22], endothelial cells, and smooth muscle cells (SMC) [23–26]. Topography can influence cellular responses from initial attachment and migration to differentiation and production of new tissue [10,12,27]. While the great majority of these cell-substrate interaction studies have been conducted on features in the micron range, recent findings underscore the phenomenon that mammalian cells do respond to nanoscale features on a synthetic surface [13,14,22–24,26]. Nanoscaled topography has been receiving increasing attention because of its resemblance to in vivo surroundings. Cells in their natural environment interact with extracellular matrix components in the nanometer scale. For example, the basement membrane of many tissues display features of pores, fibers, and ridges in the nanometer range [28]. Collagen fibers in connective tissues are also composed of tropocollagen molecules associating to form microfibrils with a periodicity, or the appearance of bands, of 66nm [10]. A better understanding of the cellular response to nanopatterns would therefore be of significance to the design and application of biomaterials.
A range of techniques can be used to create well-defined topographical and chemical cues for cell patterning. Many of these approaches rely on photolithography and reactive ion etching. This can be followed by anisotropic etching, or UV and glow discharge treatment [11]. Microcontact printing is also a popular technique. Other methods include inkjet printing and diamond cutting. These techniques are generally suitable only for micropatterning. To go down in size, photolithography is limited by diffraction limitations. Without the use of phase shift masks, its resolution is on the order of the wavelength of the light used for exposure (typically >200 nm). Electron-beam lithography can be used to produce nanoscale patterns, but it is expensive and time-consuming. A simple method has been reported to fabricate nanoislands of 13–95nm in height based on phase separation of polystyrene and poly(4-bromostyrene) spin-coated on silicon wafers [13,16]. The ability to produce nanofeatures of controlled size and geometry based on such phase separation phenomenon, however, is poor.
We have recently developed nanoimprinting techniques that can produce a wide range of nanostructures, including multilayer three-dimensional structures made of flexible polymers [29,30]. In this study we examined the behavior SMC cultured on poly(methyl methacrylate) (PMMA) and poly(dimethlsiloxane) (PDMS) surfaces, consisting of nanopatterned gratings with 350 nm linewidth, 700 nm pitch, and 350 nm depth. The morphology of cells grown on such gratings was characterized with respect to their elongation and alignment. In addition to proliferation, the cell motility was studied using the in vitro wound healing model, where the polarization of the microtubule organizing centers (MTOC) of the SMC during the healing was examined [31].
2. Materials and methods
2.1. Nanoimprinting
Nanoimprint lithography was used to generate gratings with 350 nm linewidth, 700 nm pitch, and 350 nm depth in a PMMA thin film that was spin-coated onto a Si substrate as previously described [30,32]. Briefly, a pre-patterned master SiO2 grating-mold was pressed onto the PMMA polymer film at a temperature of 180 °C and a pressure of 6MPa. Imprinting well above the glass transition temperature (Tg) of PMMA (Tg = 105 °C) results in a polymer that is in a viscous fluid state such that it is amenable to molding and flow. After 10 min of imprinting at elevated temperature and pressure, the SiO2 mold, PMMA polymer film, and Si substrate were allowed to cool to below the Tg of the PMMA, thereby producing a grating pattern in the polymer based on the master mold. Mold release from the polymer was facilitated by treating the mold surface, prior to imprinting, with 1H, 1H, 2H, 2H-perfluorodecyl-trichlorosilane (FDTS) in order to lower its surface energy.
2.2. Reproduction of nanograting with soft lithography
The nanopattern was also reproduced on poly(dimethylsiloxan) (PDMS) using soft lithography. The nanoimprinted PMMA-coated Si was used as a master sample for the replica molding. The PMMA-coated wafer was fluorinated with (tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane (United Chemical Technologies, Bristol, PA) for 2 h, washed with 0.01% Triton X (Sigma, St. Louis, MO) and cleaned by blowing with nitrogen gas. The PMMA-coated wafer was placed at the bottom of a medium-size weight boat (Fisher Scientific, Midland, MI) and 20 g of degassed PDMS solution with cross-linking reagent (Sylgard 184 Silicone Elastomer Kit, Dow Corning) was poured onto the wafer. The mold was baked at 50 °C overnight, cooled at room temperature and then separated from the master wafer. The surface morphology and fidelity of the nanogratings were inspected with scanning electron microscopy (SEM). Nanopatterned PDMS samples were plasma oxidized before cell culture in order to produce a hydrophilic surface.
2.3. Smooth muscle cell culture
Bovine pulmonary artery SMC were obtained from Cambrex (Walkersville, MD). The SMC were seeded on the pre-cut PMMA-coated Si wafer or PDMS replica at 50 × 103 cell/cm2. Cells were cultured with DMEM with 1 g/l of glucose and l-glutamine (Invitrogen, Frederick, MD) supplemented with 10% calf serum (Invitrogen) and 1% of penicillin and streptomycin sulphate (Invitrogen). Fresh medium was replenished every 2–3 days in all cultures. The morphology of the SMC on the PMMA-nanopatterns or PDMS replica was observed with SEM and confocal microscopy after staining. A glass cover slip (grade 1, Fisher Scientific), unpatterned PMMA, and unpatterned PDMS were used as controls.
2.4. Scanning electronic microscopy preparation
Samples with SMC cultured were fixed in 4% paraformaldehyde in PBS, washed in 0.1m sodium cacodylate, and post-fixed in 2% OsO4 in 0.1m Na cacodylate, pH 7.2. After post-fixation, the samples were stained in 2% uranyl acetate in water and dehydrated in a graded ethanol series. Critical point-dried samples were sputter-coated with a 5nm coating of chromium and viewed with a LEO FESEM (LEO 1550) (LEO Electron Microscopy Inc.) at 1 kV. PMMA-coated Si and PDMS samples with cell cultures were sputter-coated with 5nm of chromium without fixing before viewing.
2.5. Fluorescence staining of F-actin
Samples were fixed in 4% paraformaldehyde for at least 15 min and permeablized with 0.05% Triton-X and 50mm glycine solution for 20 min. The F-actin was stained with Oregon Green 488 phalloidin (Molecular Probes, Eugene, OR) diluted to 13nm in PBS, and the nucleus was stained with Hoechst for 30 min. Samples were thoroughly washed three times with PBS and mounted on glass cover slip with Gel/Mount (Abcam, Cambridge, MA) before inspection with confocal microscopy.
2.6. Elongation and alignment characterization
Five samples of each nanopatterned PMMA, unpatterned PMMA, nanopatterned PDMS, unpatterned PDMS, and glass cover slip seeded with SMC were fixed at day 3 and day 7. Eight separate regions of each sample were photographed (20 × or 40 ×). The images were analyzed with ImageJ NIH image processing software (Bethesda, MD).
The cells were characterized with respect to their elongation and alignment. The elongation (E) parameter describes the extent the equimomental elipse is lengthened or stretched out [22]. Thus, E is zero for a circle, and one for an ellipse with an axis ratio of 1:2. Alignment describes how well the long axis of an elongated cell, with E>4, is oriented with respect to the grating. Cells were considered aligned if the angle between the long axis and the grating was less than 15 °. The percentage of cell alignment and the E factor was measured. For each experiment an average of 300 cells was counted.
2.7. BrdU cell proliferation assay
Four hours prior to the fixation of samples with acid-alcohol, 70% ethanol and 1% acetic acid, 5-bromo-2-deoxyuridine, BrdU, (Amersham Biosciences, Piscataway, NJ) was added to the medium. The samples were stained with mouse anti-BrdU antibody (Amersham Biosciences), Alexa Fluor 546-conjugated anti-mouse antibody (Molecular Probes) as secondary antibody, goat serum (Sigma) as blocking medium, and Hoechst stain as a counter stain for the nucleus. Stained samples were inspected with confocal microscopy. Four samples of each patterned PMMA, unpatterned PMMA, patterned PDMS, and unpatterned PDMS were examined with an epifluorescence microscope at 40 × using IP lab and Nikon filter DAPI H365 01 for the Hoechst stain and Cy3/TRITC for Alexa Fluor 546. Thirty separate regions of each sample were photographed. The images were analyzed with ImageJ (NIH software).
2.8. Wound healing assay
Motility of the SMC on the patterned versus unpatterned surfaces was examined in terms of the polarization and reorientation of the MTOC after wounding [31]. When a confluent monolayer SMC on patterned PDMS and glass cover slip was observed, a wound was made parallel to the cell alignment by scraping the cell monolayer across the sample with the back of a surgical blade. The dimensions of the wound were about 0.5–1mm across and 5–7mm long.
The cells were fixed at 0, 1, 2, 3 and 4 h after the wounding with ice-cold methanol for 5 min and then blocked with 33% goat serum (Sigma). The orientation of MTOC was assessed by immunofluorescence staining with rabbit anti-γ-tubulin antibody (Sigma) as primary antibody and Alexa Fluor 546-conjugated anti-mouse antibody (Molecular Probes) as secondary antibody. Hoechst stain was used as a counter stain for the nucleus. Four samples of each time point were examined with an epifluorescence microscope at 100 × using IP lab. Each cell was divided into four regions around the nucleus: front region facing the wound edge, the two sides, and the back region facing away from the wound edge. Diploid cells showing two MTOC were not counted. The number of cells with the MTOC located in each of the four regions was counted. On average, 150–200 cells were counted on each sample.
2.9. Data analysis
All data are presented as mean±SD. Student's t-test and ANOVA were used to evaluate the statistical significance where indicated. Significance level was set at p<0.01.
3. Results
3.1. Nanoimprinting on PMMA and replica molding of PDMS
The surface morphology and fidelity of the nanopatterns were examined with SEM (Fig. 1). The gratings were about 350 nm wide on both the nanoimprinted PMMA (Fig. 1A and B) and the replica PDMS sample (Fig. 1C), with no obvious defects observed. The surface topography was maintained even with a coating of collagen (bovine collagen I, Fig. 1D).
Fig. 1.
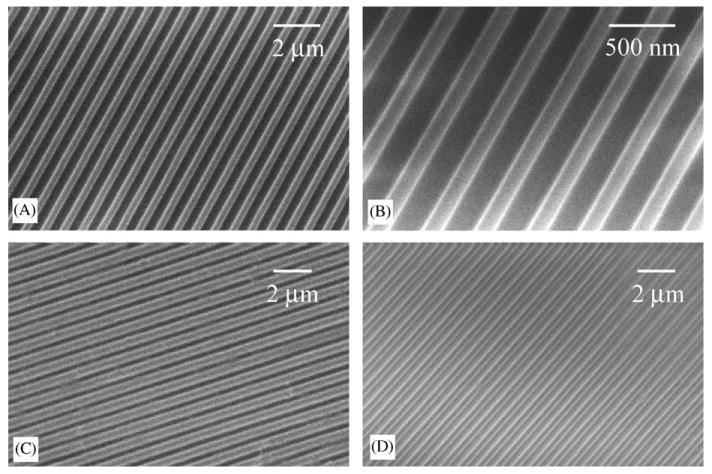
Scanning electron micrographs of (A and B) nano-imprinted gratins on PMMA coating on SiO2 wafer, (C) PDMS nanopatterned by replica molding and (D) collagen coated PDMS with nanopattern. Bar = 2 μm for A, C and D, bar = 500 nm for B.
3.2. SMC morphology
SMC cultured on the surface with nanogratings adapted with an elongated morphology and were mostly parallel to one another (Fig. 2A–D). In contrast, SMC cultured on unpatterned surfaces showed neither elongation nor orientation at both low and high cell densities (Fig. 2E and F). The orientation of the cells along the axis of the gratings could be seen more clearly under SEM (Fig. 2G), where they were randomly spread on unpatterned surfaces (Fig. 2H). The F-actin fiber was mostly stretched along the long axis of the cells. The nuclei of the cells were also elongated, and aligned to the long axis of the cell in most cases (Fig. 2A and B).
Fig. 2.
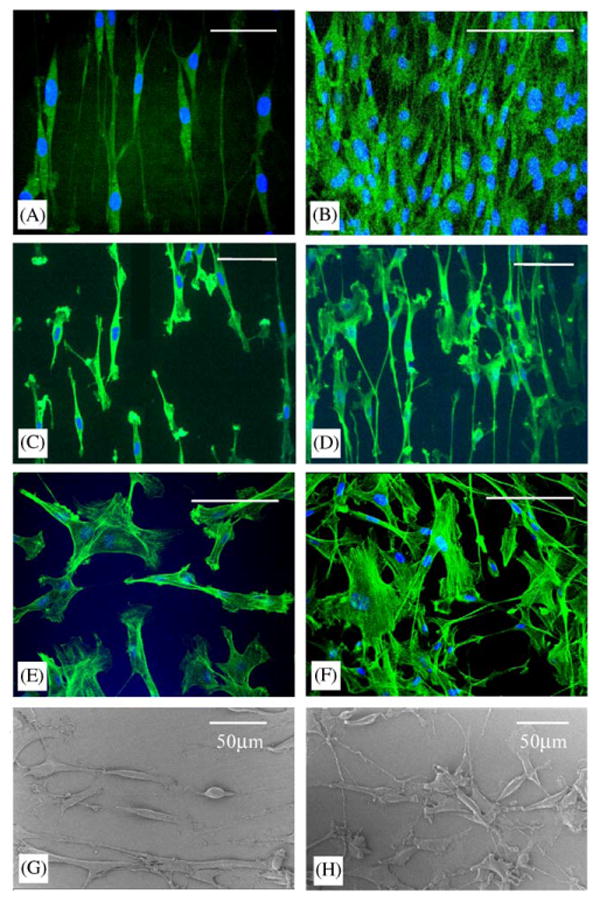
Confocal micrographs of F-actin stained SMC on (A) nano-imprinted PMMA at low cell density, (B) nano-imprinted PMMA at high cell density, (C) nanopatterned PDMS at low cell density, (D) nano-patterned PDMS at high cell density, (E) non-patterned PMMA and (F) glass cover slip. Scanning electron micrographs of SMC cultured on (G) nano-imprinted gratings on PMMA coated on SiO2 wafer and (H) non-patterned PMMA coated on SiO2 wafer. Bar = 50 μm for all except (B) Bar = 100 μm.
3.3. Alignment and elongation characterization
The percentage of cell alignment on different surfaces is shown in Fig. 3. Cells were considered aligned if the angle between the long axis and the grating was less than 15°. The alignment of cells in nanoimprinted PMMA and nanopatterned PDMS was 96.1±2.7% and 89.7±5.3%, respectively.
Fig. 3.
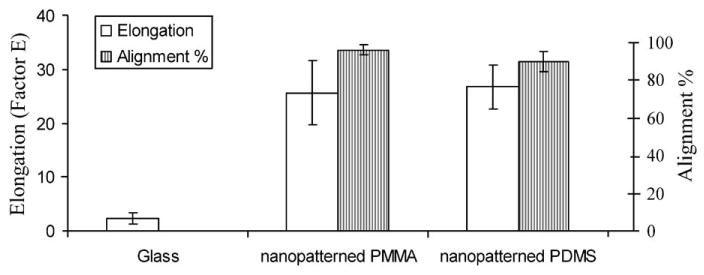
The percentage of cell alignment and the elongation factor of SMC cultured on glass cover slip, nano-imprinted PMMA and nanopatterned PDMS. Cells were considered as aligned if the angle between the long axis of the cells and the gratings was less then 15 °. Factor E = (long axis/short axis)−1.
Elongation of cells was assessed by measuring the length of the long axis and short axis, or the axis perpendicular to the long axis. The factor E equals to the long axis divided by the short axis minus one. The E factor of the SMC cultured on nanopatterned PDMS and PMMA was 26.7±10.5 and 25.7±6.9, respectively, while it was only 2.3±1.8 for SMC on glass cover slip (Fig. 3). The SMC elongation on the patterned surface was significantly higher than on glass cover slip.
3.4. BrdU cell proliferation assay
Cell proliferation was determined by counting the percentage of cells with incorporated BrdU during the 4 h of incubation (Fig. 4A–C). The values were 35.5±8.6% and 30.9±5.0% of SMC showing BrdU incorporation when cultured on nanopatterned PMMA and PDMS, versus 56.6±10.7% and 47.7±12.0% for cells cultured on unpatterned PMMA and PDMS surfaces, respectively (n = 4). The proliferation of SMC on the nanopattern was significantly lower (p<0.01) compared to surfaces without pattern.
Fig. 4.
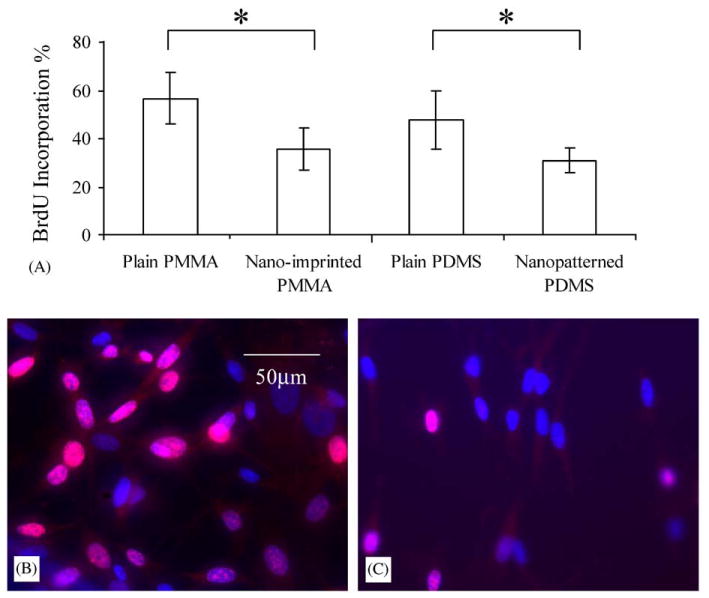
(A) The percentage of SMC incorporated BrdU during 4 h of incubation cultured on plain non-patterned PMMA, nano-imprinted PMMA, plain non-patterned PDMS and nanopatterned PDMS. Fluorescence images showing BrdU incorporation into the nucleus of SMC cultured on (B) non-patterned PMMA and (C) nano-imprinted PMMA after immuno-fluorescent staining. Bar = 50 μm.
3.5. Wound healing assay for cell motility
Cell migration behavior was examined by observing the polarization of MTOC at various time points after wounding. Previous studies show that microtubules (MT) are essential for the polarization of many cell types [31,33], and cell migration involves the MT and actin cytoskeletons. The MTOC would re-orientate towards the direction of migration. Figs. 5A and B illustrate the orientation of the MTOC towards either the wound edge or the wound axis (also the axis of gratings) for SMC cultured on glass cover slip and nanopatterned PDMS, respectively.
Fig. 5.
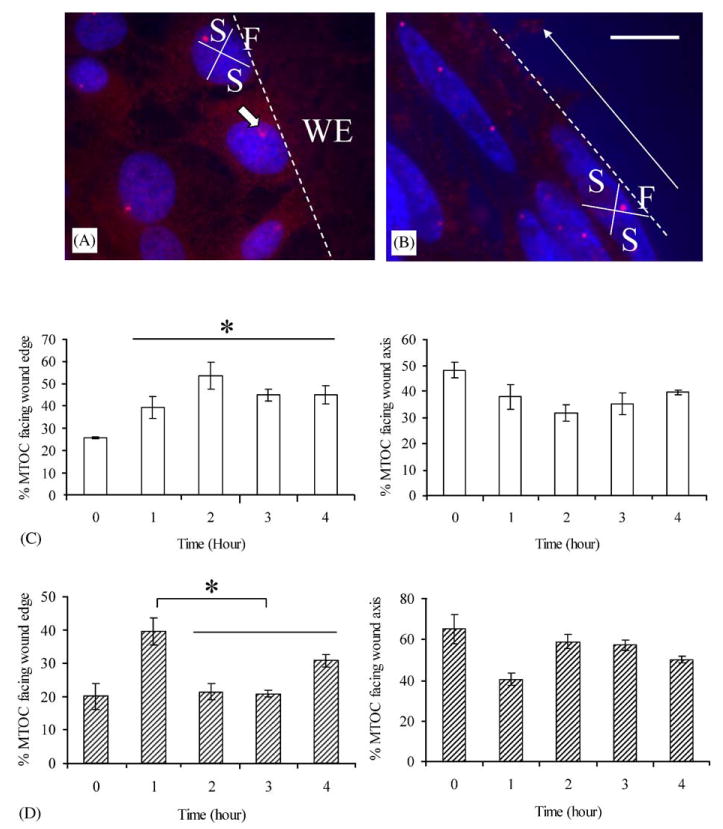
MTOC polarization over the time course of wound closure for SMC. Fluorescent labeling of the MTOC and nucleus on the wounded SMC monolayer on (A) glass cover slip and (B) nanopatterned PDMS 1-h post wounding. Each cell at the wound edge was divided into four regions as shown in (A) and (B). The location of MTOC of one of the cell is indicated with a block white arrow in (A). The white dotted line outlines the wound edge. The direction of cell alignment, hence the gratings on the nanopatterned PDMS, is indicated with a white arrow in (B). The percentage of wound-edge cells with an MTOC orientated towards the wound edge and sideway of the edge at 0, 1, 2, 3 and 4 h post wounding for SMC cultured on (C) glass cover slip and (D) nanopatterned PDMS gratings. Bar = 20 μm. WE = wound edge, F = front quadrant, S = side quadrants.
For SMC cultured on glass cover slip (Fig. 5A and C), the MTOC polarization was nearly random at time zero before wounding. At 1, 2, 3 and 4 h after wounding, the wound-edge cells had a polarized MTOC. The polarization peaked at 2-h after wounding. The orientation of the MTOC towards the wound edge and hence the direction of SMC migration is similar to the behavior of other types such as endothelial cells and fibroblasts.
For SMC cultured on nanopattern PDMS (Fig. 5B and D), a higher percentage of MTOC of the cells oriented toward the two sides, which was the direction of cell alignment and grating axis before wounding, compared to the random orientation observed in SMC cultured on a glass cover slip. At 1 h after wounding, a significantly higher percentage of MTOC was polarized to the wound edge; meanwhile, the percentage of MTOC polarized along the grating axis decreased. However, after 2 h, the percentage of MTOC polarized toward the wound edge decreased, but the percentage of MTOC polarized toward the grating axis increased, returning to the state before wounding. The polarization of MTOC towards the grating axis suggests that the cells re-orientated themselves along the grating axis while migrating. This strong preference of MTOC polarization along the axis of the grating indicates the strong migration cue presented by the surface topography.
4. Discussion
In this study, we observed that the bovine pulmonary artery SMC cultured on nanoscale ridge and groove patterns, fabricated by nanoimprinting, exhibited an elongated morphology and alignment parallel to the axis of the gratings. Besides elongation of cell bodies, alignment and elongation of nuclei were also observed. On the nanopatterned PMMA and PDMS surfaces the proliferation rate of the SMC was significantly reduced to 35.5±8.6% and 30.9±5.0% of BrdU incorporation in 4 h, compared to 56.6±10.7% and 47.7±12.0% for SMC on the unpatterned reference surfaces, respectively. In the motility study, the MTOC of the SMC cultured on glass cover slip were polarized toward the wound edge starting from 1 h after wounding. On the other hand, the MTOC of SMC cultured on nanogratings initially polarized briefly to the wound edge 1 h after wounding, then polarized towards the direction of the alignment axis instead starting from 2 h after wounding. Collectively these observations highlight the significant influence of nanograting topography on SMC behavior.
Cell alignment and elongation of different cell types have been studied on micro- [15,17,18,21,25,34–40] or nanoscale gratings [22]. Cell alignment, F-actin alignment and elongation are usually observed on the edges of micro-ridges. The alignment would also be depth dependent. Our study was conducted on a pattern in which the dimensions of the gratings in terms of width, periodicity, and depth are all sub-micrometer in size. The findings show that this nanopattern would exert qualitatively similar influence on cellular morphology as other nanogratings. Work is underway to define the threshold of size features that would still command a significant topographical influence.
The decrease of proliferation rate is also in agreement with the literature. As Thakar et al. indicated in their micro-pattern study, the decrease of SMC proliferation could be due to the decrease in cell spreading and change in mechanical force exerted on the cells [25]. The decreased proliferation rate of SMC is similar to that observed in vivo, where SMC proliferate slowly [41]. In adult blood vessels, SMC are in the “contractile” state where the rate of proliferation, migration and production of extracellular matrix are low. During vascular development and in response to vascular injury, the SMC dramatically increase their rate of proliferation, migration as well as synthetic capacity in order to play a critical role in vascular repair. The elongated morphology and alignment, which resemble the natural state of SMC in vivo, suggest that the nanopatterned surface would be a more favorable substrate for the culture of these cells.
The motility behavior indicates that the tendency for SMC to move along the axis of contact guidance is very strong compared to their tendency to migrate forward in wound healing. The MTOC were significantly polarized toward the wound edge for SMC cultured on non-patterned surfaces after wounding. In contrast, the MTOC polarization toward the direction of cell alignment was significantly higher on the nanopatterned surface than on the non-patterned surface for all time points after the wound healing except at the 1-h time point. The change in the MTOC polarization indicates that the migration rate of SMC would be altered on nanopatterned surfaces.
Nucleus elongation and alignment were observed in this study. The elongation of cells and nucleus have been correlated with changes in gene expression profile and cell differentiation in other studies [15,42–44]. The nucleus is mechanically integrated with the physical entity of the cell. Forces are transferred to the nucleus through actin-intermediate filament system during changes in cell shape [45]. The mechanical tension causing alignment of cells can rearrange the centromere through deformation of the nucleus [15]. The tensile force can also increase microtubule mass in the cell by mechano-transduction via integrin–cytoskeleton coupling, affecting the cellular structure and phenotypes in SMC [46]. Meanwhile, the mechano-sensitive Ca2+ channel in the nuclear membrane would induce release of Ca2+ from the perinuclear space [42], which in turn would regulate gene expression. Relan et al. has shown that cell elongation induces expression of laminin α2 (LM2) chain in mouse embryonic mesenchymal smooth muscle precursor cells [44]. Similarly, various studies have shown that distortion of nucleus would result in changes of gene and protein expression [15,43]. The nanoimprinted pattern can therefore be a valuable tool to study cell physiology and the relationship between nucleus distortion and gene regulation of various cell types.
Another interesting observation in this study is the close agreement of cellular behavior on both the nanopatterned PMMA and PDMS surfaces. It suggests that the strong topographical cue can overcome the influence of polymer surface chemistry. PDMS is considerably more compliant than PMMA. This would also suggest the dominance of this topographical cue over the stiffness of the substrate. In our ongoing study, it was observed that a collagen coating on the patterned PDMS did not mask the topographical influence of the nanopatterns (data not shown). Previous studies showed that the effect of topographic cue is stronger than the biochemical cue for BHK cell [18] if the depth of the groove is more than 2 μm. In this study, we showed that even with a depth of 350 nm, the difference in surface chemistry is not obvious for SMC.
Approaches taken in this study may provide insight to pathological differentiation of SMC in vascular injury or atherosclerosis. They may also offer input for design of tissue engineering scaffolds. For instance, SMC orientation is essential in providing a strong mechanical property for tissue-engineered vascular grafts. The nanoimprinting techniques reported in this study can produce scaffolds with topographical features to direct the SMC orientation. The imprinting techniques used in this study are applicable to a wide range of polymers, including the ones popular for tissue engineering scaffolds, such as polylactide and polycaprolactone. Moreover, they can be applied to non-planar surfaces and generate multi-layered patterns. With the knowledge that much of the cellular environment in vivo involves nanoscale features, this three-dimensional nanoimprinting technology offers interesting opportunities to study the interaction of cells with topographical features comparable to the size of ECM components, and on materials practical for biomedical applications.
5. Conclusion
Bovine pulmonary artery smooth muscle cells (SMC) cultured on poly(methyl methacrylate) (PMMA) and poly(dimethylsiloxane) (PDMS) surfaces comprising nanopatterned gratings with 350 nm linewidth, 700 nm pitch, and 350 nm depth showed significant elongation and alignment, both in cytoskeleton and nuclei. In addition to a reduced proliferation compared to cells cultured on non-patterned controls, the polarization of microtubule organizing centers (MTOC) of the SMC cultured on nanopatterned surfaces showed a preference towards the axis of cell alignment in an in vitro wound healing assay. Applicable to a wide range of biomedical polymers and on non-planar surfaces, this nanoimprinting technology has great potential in producing structures for studying cell physiology in response to nanotopography and in developing medical devices with nanoscale features.
Acknowledgments
The authors would like to acknowledge NIH for funding support (R21EB003203) and S. Raghavan for technical advice on MTOC staining.
References
- 1.Nerem RM. Tissue engineering: confronting the transplantation crisis. Adv Exp Med Biol. 2003;534:1–9. doi: 10.1007/978-1-4615-0063-6_1. [DOI] [PubMed] [Google Scholar]
- 2.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260(5110):920–6. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 3.Patel N, Padera R, Sanders GH, Cannizzaro SM, Davies MC, Langer R, et al. Spatially controlled cell engineering on biodegradable polymer surfaces. Faseb J. 1998;12(14):1447–54. doi: 10.1096/fasebj.12.14.1447. [DOI] [PubMed] [Google Scholar]
- 4.Boyan BD, Hummert TW, Dean DD, Schwartz Z. Role of material surfaces in regulating bone and cartilage cell response. Biomaterials. 1996;17(2):137–46. doi: 10.1016/0142-9612(96)85758-9. [DOI] [PubMed] [Google Scholar]
- 5.Aucoin L, Griffith CM, Pleizier G, Deslandes Y, Sheardown H. Interactions of corneal epithelial cells and surfaces modified with cell adhesion peptide combinations. J Biomater Sci Polym Ed. 2002;13(4):447–62. doi: 10.1163/156856202320253956. [DOI] [PubMed] [Google Scholar]
- 6.Boateng SY, Hartman TJ, Ahluwalia N, Vidula H, Desai TA, Russell B. Inhibition of fibroblast proliferation in cardiac myocyte cultures by surface microtopography. Am J Physiol Cell Physiol. 2003;285(1):C171–82. doi: 10.1152/ajpcell.00013.2003. [DOI] [PubMed] [Google Scholar]
- 7.Wisniewski N, Reichert M. Methods for reducing biosensor membrane biofouling. Colloids Surf B Biointerfaces. 2000;18(3–4):197–219. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 8.Curtis AS, Varde M. Control of cell behavior: topological factors. J Natl Cancer Inst. 1964;33:15–26. [PubMed] [Google Scholar]
- 9.Das GD, Lammert GL, McAllister JP. Contact guidance and migratory cells in the developing cerebellum. Brain Res. 1974;69(1):13–29. doi: 10.1016/0006-8993(74)90366-7. [DOI] [PubMed] [Google Scholar]
- 10.Curtis A, Wilkinson C. Nanotechniques and approaches in biotechnology. Trends Biotechnol. 2001;19(3):97–101. doi: 10.1016/s0167-7799(00)01536-5. [DOI] [PubMed] [Google Scholar]
- 11.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20(6):573–88. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson CDW. Nanostructures in biology. Microelectronic Engineering. 1995;27(1–4):61–5. [Google Scholar]
- 13.Dalby MJ, Gadegaard N, Riehle MO, Wilkinson CD, Curtis AS. Investigating filopodia sensing using arrays of defined nano-pits down to 35nm diameter in size. Int J Biochem Cell Biol. 2004;36(10):2015–25. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Dalby MJ, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Polymer-demixed nanotopography: control of fibroblast spreading and proliferation. Tissue Eng. 2002;8(6):1099–108. doi: 10.1089/107632702320934191. [DOI] [PubMed] [Google Scholar]
- 15.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284(2):274–82. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 16.Dalby MJ, Yarwood SJ, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp Cell Res. 2002;276(1):1–9. doi: 10.1006/excr.2002.5498. [DOI] [PubMed] [Google Scholar]
- 17.Wojciak-Stothard B, Curtis AS, Monaghan W, McGrath M, Sommer I, Wilkinson CD. Role of the cytoskeleton in the reaction of fibroblasts to multiple grooved substrata. Cell Motil Cytoskeleton. 1995;31(2):147–58. doi: 10.1002/cm.970310207. [DOI] [PubMed] [Google Scholar]
- 18.Britland S, Morgan H, Wojiak-Stodart B, Riehle M, Curtis A, Wilkinson C. Synergistic and hierarchical adhesive and topographic guidance of BHK cells. Exp Cell Res. 1996;228(2):313–25. doi: 10.1006/excr.1996.0331. [DOI] [PubMed] [Google Scholar]
- 19.Corey JM, Feldman EL. Substrate patterning: an emerging technology for the study of neuronal behavior. Exp Neurol. 2003;184 1:S89–96. doi: 10.1016/s0014-4886(03)00392-3. [DOI] [PubMed] [Google Scholar]
- 20.Wojciak-Stothard B, Curtis A, Monaghan W, MacDonald K, Wilkinson C. Guidance and activation of murine macrophages by nanometric scale topography. Exp Cell Res. 1996;223(2):426–35. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- 21.Wojciak-Stothard B, Madeja Z, Korohoda W, Curtis A, Wilkinson C. Activation of macrophage-like cells by multiple grooved substrata. Topographical control of cell behaviour. Cell Biol Int. 1995;19(6):485–90. doi: 10.1006/cbir.1995.1092. [DOI] [PubMed] [Google Scholar]
- 22.Andersson AS, Backhed F, von Euler A, Richter-Dahlfors A, Sutherland D, Kasemo B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials. 2003;24(20):3427–36. doi: 10.1016/s0142-9612(03)00208-4. [DOI] [PubMed] [Google Scholar]
- 23.Dalby MJ, Riehle MO, Johnstone H, Affrossman S, Curtis AS. In vitro reaction of endothelial cells to polymer demixed nanotopography. Biomaterials. 2002;23(14):2945–54. doi: 10.1016/s0142-9612(01)00424-0. [DOI] [PubMed] [Google Scholar]
- 24.Miller DC, Thapa A, Haberstroh KM, Webster TJ. Endothelial and vascular smooth muscle cell function on poly(lactic-co-glycolic acid) with nano-structured surface features. Biomaterials. 2004;25(1):53–61. doi: 10.1016/s0142-9612(03)00471-x. [DOI] [PubMed] [Google Scholar]
- 25.Thakar RG, Ho F, Huang NF, Liepmann D, Li S. Regulation of vascular smooth muscle cells by micropatterning. Biochem Biophys Res Commun. 2003;307(4):883–90. doi: 10.1016/s0006-291x(03)01285-3. [DOI] [PubMed] [Google Scholar]
- 26.Thapa A, Webster TJ, Haberstroh KM. Polymers with nano-dimensional surface features enhance bladder smooth muscle cell adhesion. J Biomed Mater Res. 2003;67A(4):1374–83. doi: 10.1002/jbm.a.20037. [DOI] [PubMed] [Google Scholar]
- 27.Curtis AS, Wilkinson CD. Reactions of cells to topography. J Biomater Sci Polym Ed. 1998;9(12):1313–29. doi: 10.1163/156856298x00415. [DOI] [PubMed] [Google Scholar]
- 28.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell Tissue Res. 2000;299(1):39–46. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 29.Huang XD, Bao LR, Cheng X, Guo LJ, Pang SW, Yee AF. Reversal imprinting by transferring polymer from mold to substrate. J Vac Sci Technol B. 2002;20(6):2872–6. [Google Scholar]
- 30.Bao LR, Cheng X, Huang XD, Guo LJ, Pang SW, Yee AF. Nanoimprinting over topography and multilayer three-dimensional printing. J Vac Sci Technol B. 2002;20(6):2881–6. [Google Scholar]
- 31.Magdalena J, Millard TH, Machesky LM. Microtubule involvement in NIH 3T3 Golgi and MTOC polarity establishment. J Cell Sci. 2003;116(Pt 4):743–56. doi: 10.1242/jcs.00288. [DOI] [PubMed] [Google Scholar]
- 32.Tan L, Kong YP, Bao LR, Huang XD, Guo LJ, Pang SW, et al. Imprinting polymer film on patterned substrates. J Vac Sci Technol B. 2003;21(6):2742–8. [Google Scholar]
- 33.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106(4):489–98. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 34.Clark P, Dunn GA, Knibbs A, Peckham M. Alignment of myoblasts on ultrafine gratings inhibits fusion in vitro. Int J Biochem Cell Biol. 2002;34(7):816–25. doi: 10.1016/s1357-2725(01)00180-7. [DOI] [PubMed] [Google Scholar]
- 35.Dow JA, Clark P, Connolly P, Curtis AS, Wilkinson CD. Novel methods for the guidance and monitoring of single cells and simple networks in culture. J Cell Sci. 1987;8(Suppl):55–79. doi: 10.1242/jcs.1987.supplement_8.4. [DOI] [PubMed] [Google Scholar]
- 36.Jung DR, Kapur R, Adams T, Giuliano KA, Mrksich M, Craighead HG, et al. Topographical and physicochemical modification of material surface to enable patterning of living cells. Crit Rev Biotechnol. 2001;21(2):111–54. doi: 10.1080/20013891081700. [DOI] [PubMed] [Google Scholar]
- 37.Kam L, Boxer SG. Cell adhesion to protein-micropatterned-supported lipid bilayer membranes. J Biomed Mater Res. 2001;55(4):487–95. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 38.Recknor JB, Recknor JC, Sakaguchi DS, Mallapragada SK. Oriented astroglial cell growth on micropatterned polystyrene substrates. Biomaterials. 2004;25(14):2753–67. doi: 10.1016/j.biomaterials.2003.11.045. [DOI] [PubMed] [Google Scholar]
- 39.Webb A, Clark P, Skepper J, Compston A, Wood A. Guidance of oligodendrocytes and their progenitors by substratum topography. J Cell Sci. 1995;108(Pt 8):2747–60. doi: 10.1242/jcs.108.8.2747. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson CDW, Riehle M, Wood M, Gallagher J, Curtis ASG. The use of materials patterned on a nano- and micro-metric scale in cellular engineering. Mater Sci Eng C-Biomimetic and Supramolecular Systems. 2002;19(1–2):263–9. [Google Scholar]
- 41.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84(3):767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 42.Itano N, Okamoto S, Zhang D, Lipton SA, Ruoslahti E. Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc Natl Acad Sci USA. 2003;100(9):5181–6. doi: 10.1073/pnas.0531397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99(4):1972–7. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Relan NK, Yang Y, Beqaj S, Miner JH, Schuger L. Cell elongation induces laminin alpha2 chain expression in mouse embryonic mesenchymal cells: role in visceral myogenesis. J Cell Biol. 1999;147(6):1341–50. doi: 10.1083/jcb.147.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94(3):849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putnam AJ, Schultz K, Mooney DJ. Control of microtubule assembly by extracellular matrix and externally applied strain. Am J Physiol Cell Physiol. 2001;280(3):C556–64. doi: 10.1152/ajpcell.2001.280.3.C556. [DOI] [PubMed] [Google Scholar]


