Abstract
Dietary zinc deficiency in mice is accompanied by enhanced expression of the zinc uptake transporter Slc39a4 (Zip4) and repressed expression of Slc39a5 (Zip5) in tissues which regulate zinc homeostasis (intestine, pancreas and visceral yolk sac). Herein, mechanisms controlling this differential expression were investigated. The induction of Zip4 mRNA during zinc deficiency, and its repression in response to zinc repletion were found to reflect changes in Zip4 mRNA stability and not changes in the relative rate of transcription of this gene. During zinc deficiency, ZIP4 protein levels are increased and this protein is localized on the apical membranes. Administration of an oral gavage of zinc caused ZIP4 internalization and degradation in enterocytes and visceral endoderm cells. Similarly, ZIP4 is induced by zinc deficiency in cultured mouse Hepa cells and is rapidly degraded in response to added zinc. Zip5 mRNA abundance does not change in response to zinc, but the translation of this mRNA was found to be zinc-responsive. During zinc deficiency, Zip5 mRNA remains associated with polysomes, while the protein is internalized and degraded in enterocytes, acinar cells and endoderm cells. After zinc-gavage, ZIP5 is rapidly resynthesized and targeted to the basolateral membranes of these cell types.
Keywords: mRNA stability, post-transcriptional, protein stability, Slc39a4, Slc39a5, ZIP
Introduction
The storage, efflux and uptake of zinc are regulated in response to zinc availability. In mammals fed a diet with adequate zinc, this metal is absorbed by intestinal enterocytes, but excess zinc is released through the pancreas and back through the small intestine (King et al., 2000; Hambidge and Krebs, 2001). Under zinc-deficient conditions, zinc absorption by the small intestine increases, and zinc release from the pancreas and small intestine decreases (Hambidge and Krebs, 2001). During embryonic development of the mouse, the visceral endoderm of the yolk sac plays an important role in zinc homeostasis and also undergoes molecular adaptation to changes in maternal zinc status similar to those observed in the maternal intestine (Dufner-Beattie et al., 2004, 2007). We are exploring the mechanisms behind this adaptive response.
Two superfamilies of mammalian zinc transporters have been identified (Guerinot, 2000; Taylor and Nicholson, 2003; Palmiter and Huang, 2004). Solute carrier 30A (Slc30a) family members, named ZnTs, function in zinc efflux and compartmentalization (Palmiter and Huang, 2004), whereas the Slc39a family members, named ZIPs, function in the uptake of zinc and other metals into the cytoplasm. In mice and humans, 14 members of the ZIP family have been identified (Guerinot, 2000; Taylor and Nicholson, 2003). These proteins have eight predicted transmembrane domains and the ZIP superfamily can be subdivided into four subfamilies based on structural homology (Taylor and Nicholson, 2003). In mammals, most of the ZIP proteins fall into one of two subfamilies, named subfamily II (3 members) and LIV-1 (9 members). Studies of knockout mice revealed that the members of subfamily II are particularly important when dietary zinc becomes limiting (Dufner-Beattie et al., 2005, 2006; Peters et al., 2007). Less is known about the physiological functions of most LIV-1 family members. However, in humans, the importance of Zip4 is exemplified by the rare autosomal recessive disease, acrodermatitis enteropathica, which is caused by mutations in the Zip4 gene (Kury et al., 2002; Wang et al., 2002). This disease can be fatal if untreated by the administration of excess dietary zinc (Maverakis et al., 2007). The mouse Zip4 gene is essential early during development of the post-implantation embryo and heterozygosity hypersensitizes mice to the effects of dietary zinc deficiency (Dufner-Beattie et al., 2007). Although the physiological functions of ZIP5 are unknown, it is a closely related member of the LIV-1 subfamily that can also function as a zinc transporter (Dufner-Beattie et al., 2003b, 2004; Wang et al., 2004a) and is also expressed in tissues involved in zinc homeostasis (intestine, pancreas and visceral endoderm).
Our previous studies of ZIP4 and ZIP5 led to the suggestion that these proteins serve antagonistic functions in zinc homeostasis (Dufner-Beattie et al., 2004). Where ZIP4 is important for zinc acquisition from the diet (or mother), whereas ZIP5 functions to remove excess zinc from the blood when zinc is replete. Zip4 mRNA abundance is increased in the adult and neonatal intestine and embryonic visceral yolk sac during periods of dietary zinc deficiency (Dufner-Beattie et al., 2003b; Huang et al., 2006) leading to the intense localization of ZIP4 protein on the apical membranes of enterocytes and visceral endoderm cells and increased amounts of this protein (Huang et al., 2006). Studies of transfected cells revealed that zinc can stimulate the endocytosis and degradation of ZIP4 (Kim et al., 2004((•• a or b? ••)); Mao et al., 2007), and ZIP4 is removed from the apical surfaces of enterocytes and endoderm cells several hours after an injection of zinc into the peritoneum of zinc deficient mice (Dufner-Beattie et al., 2003b). Our recent studies of mouse ZIP5 revealed that this protein is localized on basolateral membranes of intestinal enterocytes, visceral endoderm cells and pancreatic acinar cells when dietary zinc is not limiting (Dufner-Beattie et al., 2004), but in contrast to ZIP4, this protein is removed from these basolateral membranes of each of these cell types during periods of dietary zinc deficiency. These changes in cellular localization were not accompanied by changes in Zip5 mRNA abundance (Dufner-Beattie et al., 2004).
The molecular mechanisms underlying the complex regulation of these zinc transporters in these specific cell types in response to dietary zinc is intriguing, yet poorly understood. Herein, we extended our previous studies of Zip4 and Zip5 regulation in vivo to include analyses of the effects of oral zinc repletion in zinc deficient mice and cultured cells and studies of the mechanisms involved in controlling these genes and proteins. These studies reveal that Zip4 and Zip5 expression is fine-tuned by several novel post-transcriptional, translational and post-translational mechanisms that each sense available zinc and reciprocally regulate these zinc transporters often within the same cell types and coordinately within the animal.
Results
Zinc regulation of Zip4 mRNA abundance requires new protein and RNA synthesis
Several mouse cell lines were examined to determine if they express and regulate the Zip4 gene in response to zinc. Zip4 mRNA was readily detectable by Northern blot hybridization of total RNA from mouse Hepa cells and the acinar-derived cell line 266-6, but not from mouse embryo fibroblasts, NIH3T3 cells or the mouse intestinal crypt-like cell line mIC-c12 (Figure 1A). In mouse Hepa cells, but not in 266-6 cells, zinc concentration in the culture medium modulated Zip4 mRNA abundance (Figure 1B,C) and as shown below, ZIP4 protein abundance in a manner that mimicked the regulation of Zip4 in mice (Figure 5D). Therefore, we used these cells as a model system in which to study regulation of the endogenous Zip4 gene. In the presence of 10% FBS in the culture medium, the addition of 100 to 150 μM zinc maximally repressed Zip4 mRNA (Dufner-Beattie et al., 2003b) and maximally induced metallothioneins-I (MT-I) mRNA in Hepa cells (Figure 1B,C). Expression of the mouse MT-I gene is regulated by the zinc-sensing transcription factor MTF-1 (Andrews et al., 2001) and MT-I functions to chelate excess zinc and provide a biologically important pool of labile zinc when zinc becomes limiting (Dalton et al., 1996).
Figure 1.
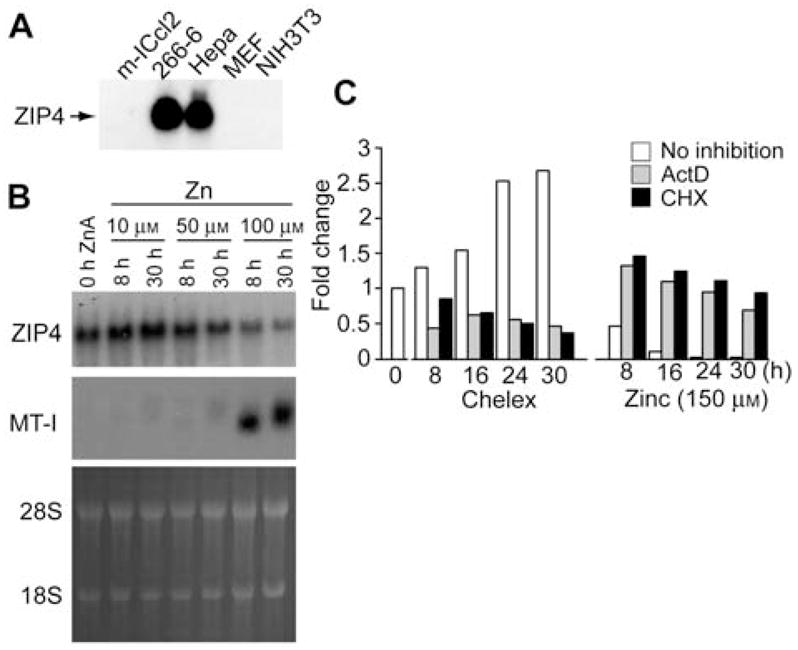
Effects of RNA and protein synthesis inhibitors on zinc regulation of Zip4 mRNA abundance.
(A) Northern blot detection of Zip4 mRNA in various mouse cell lines. Total RNA from the indicated mouse cell lines was fractionated by formaldehyde-agarose gel electrophoresis and transferred to nylon membranes. Membranes were hybridized with a mouse Zip4 cDNA probe and hybrids were detected by autoradiography (Dufner-Beattie et al., 2003b). (B) Northern blot detection of Zip4 mRNA in mouse Hepa cells incubated for the indicated times (0 h ZnA=0 time) in culture medium containing 10% FBS plus the indicated concentration of zinc (Zn). Total RNA was blotted and membranes hybridized with Zip4 and MT-I probes. Duplicate gels were stained with acridine orange (28S and 18S rRNAs are indicated) as a control for RNA integrity and loading. (C) Quantitation of Northern blots of Zip4 mRNA in mouse Hepa cells incubated for the indicated times in culture medium containing 10% Chelex-treated FBS (Chelex) ±150 μM ZnSO4 (Zinc), and ±10 μg/ml of actinomycin D (ActD), cycloheximide (CHX) or no inhibitor (no inhibition). The first unfilled bar (left) represents the untreated control (fold change=1) to which all other samples are compared. Total RNA was fractionated by electrophoresis, transferred to membranes and hybridized with the Zip4 probe and quantified by densitometry. Duplicate gels were stained with acridine orange as a control for RNA integrity and loading (not shown). Values (fold change) are expressed as the ratio of the experimental value to the 0 time untreated value.
Figure 5.
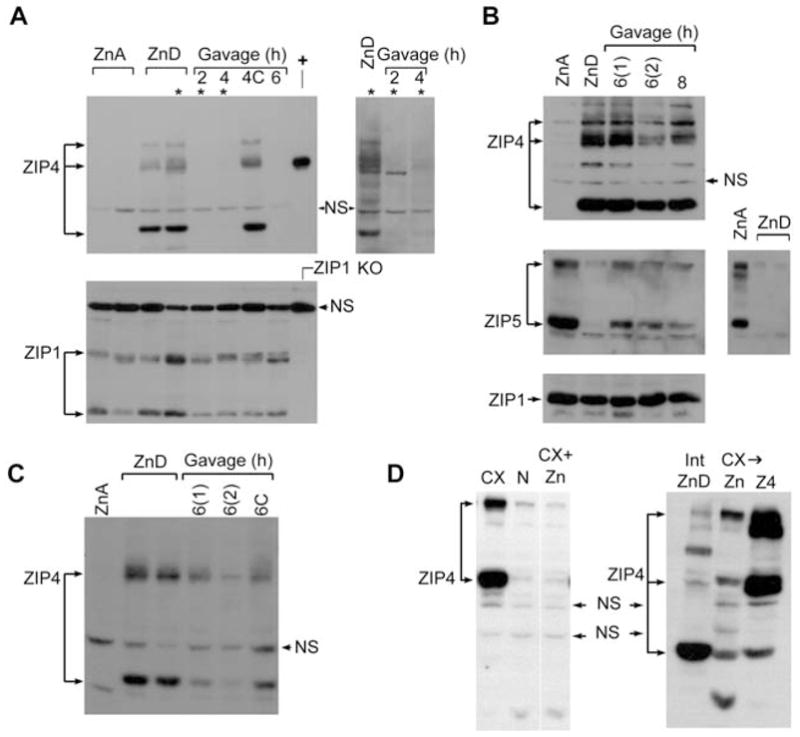
Effects of zinc on ZIP4 and ZIP5 protein abundance in the proximal small intestine (duodenum), visceral yolk sac (VYS) and cultured mouse Hepa cells.
(A) Western blot detection of ZIP4 (top panels) and ZIP1 (bottom panel) in membrane proteins from the proximal small intestine of mice fed a zinc-adequate (ZnA) or zinc-deficient (ZnD) diet for 10 days beginning on the day of weaning and then given an oral gavage of zinc or saline (4C), as detailed in the legend of Figure 4. Proximal small intestine was harvested at the indicated times after gavage (2, 4 or 6 h). Three prominent immunoreactive ZIP4 bands (ca. 150 kDa; ca. 73 kDa and 37 kDa) are indicated by arrows. Top panel: +, ZIP4 expressed in transfected cells (ca. 73 kDa); NS, non-specific band. Top right panel: a longer exposure of lanes marked (*) in the top left panel. Bottom panel: Zip1 KO, membrane proteins from intestine of homozygous ZIP1-knockout mice (Dufner-Beattie et al., 2006); NS, non-specific band. (B,C) Detection of ZIP4, ZIP5 and ZIP1 in VYS (B) or maternal small intestine (C). Pregnant mice were fed a ZnA or ZnD diet beginning on d8 of pregnancy. On d14, zinc-deficient mice were given an oral gavage of zinc or saline (6C), and VYS and maternal proximal small intestine were harvested at the indicated times (6 and 8 h) thereafter. Two separate experiments (n=5 each) are shown [6(1) and 6(2)]. Panel (B), right panel shows a ZIP5 Western blot of ZnA VYS and two additional and separate ZnD samples. Only the specific, ca. 35 kDa, band for ZIP1 is shown. (D) Detection of ZIP4 in Hepa cells or zinc-deficient intestine (Int ZnD). Hepa cells were cultured in medium containing either 10% normal FBS (N), 10% Chelex-treated FBS (CX), 10% Chelex-treated FBS to which 20 μM ZnSO4 had been added prior to culture (CX+Zn) or 10% Chelex-treated FBS towhich 20 μM ZnSO4 had been added for the last 2 h of the overnight culture (CX→N). In the right panel, Z4 refers to Hepa cells that were transfected with a ZIP4 expression vector.
The addition of 150 μM zinc to the culture medium reduced Zip4 mRNA abundance to low levels by 16 h (Figure 1C, Zinc). In contrast, Zip4 mRNA abundance increased in these cells cultured in medium containing 10% Chelex-treated FBS. This increase was clearly detectable by 8 h and was approximately 3-fold greater than control levels by 30 h of incubation (Figure 1C, Chelex). Similarly, incubation of these cells with the zinc chelator TPEN induced Zip4 mRNA (data not shown). The inclusion of actinomycin D or cycloheximide in the culture medium blocked the induction as well as repression of Zip4 mRNA by zinc (Figure 1C). Similar results were obtained using α-amanitin to inhibit mRNA synthesis (data not shown).
Zinc does not regulate transcription of the Zip4 gene
An in vitro nuclear run-on transcription assay was employed to directly examine the effects of zinc on Zip4 gene transcription (Figure 2). The effects of zinc on the relative rate of transcription of the MT-I gene was monitored as a positive control for zinc induction, and the rates of transcription of the GAPDH and β-actin genes were monitored as controls for housekeeping gene expression. When normalized to the transcription rate for the housekeeping genes, the relative rate of transcription of the Zip4 gene in Hepa cells incubated in medium containing 150 μM zinc or 10% Chelex-treated FBS remained constant for up to 48 h, long after Zip4 mRNA levels had increased. In contrast, the rate of MT-I gene transcription was rapidly and dramatically induced in response to zinc.
Figure 2.
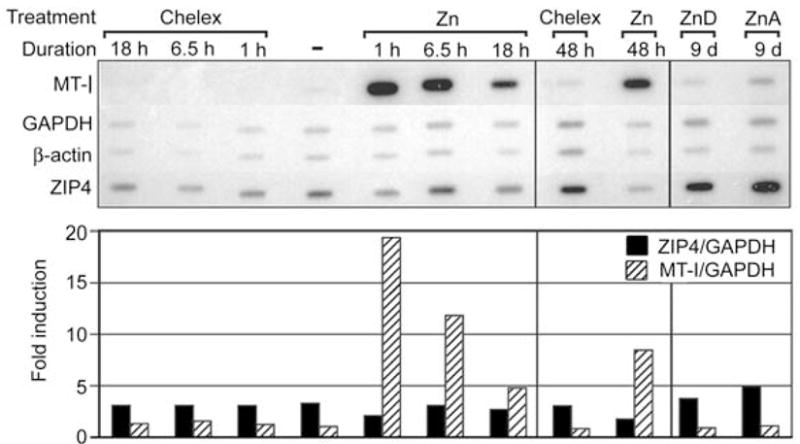
Effects of zinc on the relative rate of transcription of the Zip4 gene.
A nuclear run-on assay was employed to measure the relative rate of transcription of the mouse Zip4, MT-I, GAPDH and β-actin genes in Hepa cells and the mouse intestine (far right two lanes). Hepa cells were incubated for the indicated times [lane marked (−)=0 time] in culture medium containing 10% Chelex-treated FBS (Chelex) or in this medium also containing exogenous zinc (150 μM ZnSO4). The proximal small intestine (duodenum) was collected from pregnant mice (5 per group) fed a zinc-adequate (ZnA) or zinc-deficient (ZnD) diet for 24 h beginning on d8 of pregnancy, as described previously (Dufner-Beattie et al., 2003b, 2004). Nuclei isolated from cells and intestines were incubated in vitro in buffer containing [α-32P]UTP to label run-on transcripts. RNAs were isolated and equal counts of labeled run-on transcripts were hybridized to the indicated DNAs immobilized on nylon membrane strips. The cDNAs for mouse MT-I, GAPDH and β-actin and Zip4 genomic DNA were immobilized on the filter. The amount of labeled RNA hybridized was detected by autoradiography (top panel) and quantified by phosphorimaging. Values for Zip4 and MT-I hybrids are normalized to those of GAPDH and are expressed as fold-induction relative to the 0 time values (bottom panel).
Transcription run-on assays were also performed using nuclei isolated from the mouse intestine obtained from pregnant mice fed a zinc-adequate (ZnA) or zinc-deficient (ZnD) deficient diet (Figure 2; 9d samples). In this experiment, pregnant mice were fed the ZnA or ZnD diet beginning on d8. Our previous studies showed that Zip4 mRNA is detectably increased in the dam’s intestine within 24 h under these experimental conditions (Dufner-Beattie et al., 2004). Therefore, the relative rate of transcription of Zip4 was determined 24 h after initiation of the ZnD diet. No difference in the transcription rate was noted between the ZnA and ZnD samples.
Zinc reciprocally regulates the cellular localization of ZIP4 and ZIP5 proteins in the absence of changes in mRNA abundance
To further examine the reciprocal regulation of ZIP4 and ZIP5 by dietary zinc, mice were fed the ZnD diet and then readministered zinc by oral gavage. Zip4 and Zip5 mRNA levels (Figure 3) and cellular localization (Figure 4) were monitored within the first few hours thereafter.
Figure 3.
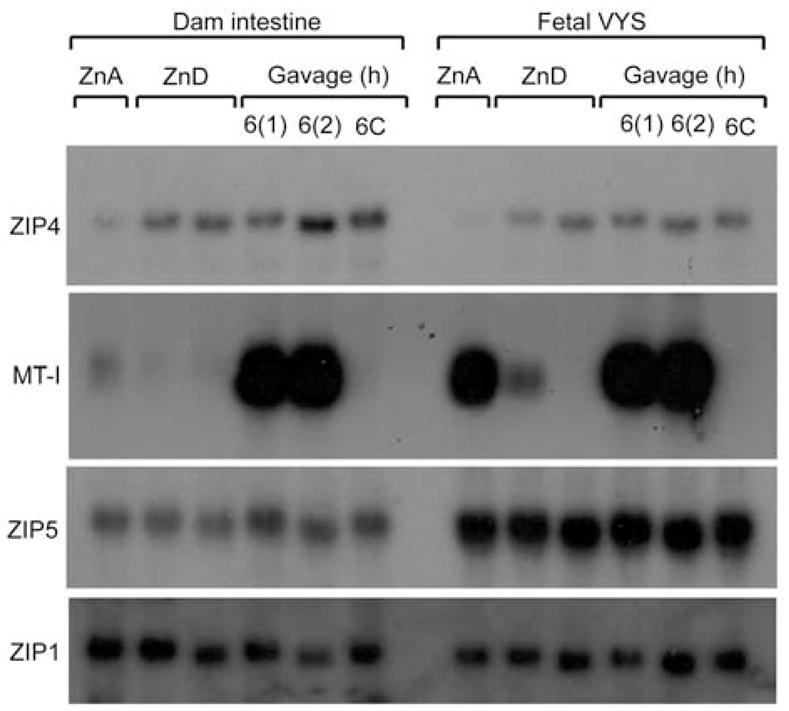
Effects of zinc repletion on Zip4 and Zip5 mRNA abundance in the proximal small intestine (duodenum) and visceral yolk sac (VYS).
Northern blot detection of Zip4, MT-I, Zip5 and Zip1 mRNAs in the maternal (dam) proximal small intestine (duodenum) and the fetal VYS is shown. Pregnant mice were fed a zinc-adequate (ZnA) or zinc-deficient (ZnD) diet for 6 days beginning on d8 of pregnancy (ending on d14). The zinc-deficient mice were then given an oral gavage of zinc or saline (6C), and tissues were harvested at the indicated times thereafter. Two independent groups of mice, labeled 6(1) and 6(2) were analyzed for the 6 h zinc-gavage time point. Specific transcripts were detected using total RNA, as described in the legend to Figure 1. Zip1 mRNA served as a good loading control, as it has equivalent expression amongst many tissues and is not zinc regulated.
Figure 4.
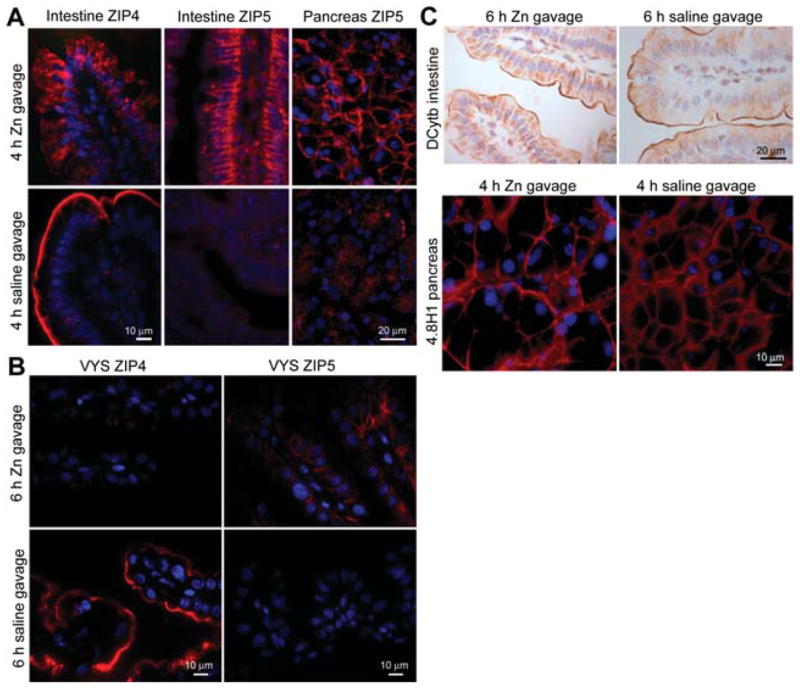
Effects of zinc repletion on the cellular localization of ZIP4 and ZIP5 proteins in the proximal small intestine (duodenum) and pancreas.
Two zinc deficiency models were used. Weanling mice were fed a zinc-deficient diet for 10 days beginning on the day of weaning. Pregnant mice were fed a zinc-deficient diet for 6 days beginning on d8 of pregnancy. On day 10 for weanlings and d14 for pregnant dams, the zinc-deficient mice were given an oral gavage of zinc (100 μmol ZnCl2/kg body weight) or saline, and tissues were harvested at the indicated times thereafter and fixed with paraformaldehyde in PBS. Cryosections of the indicated fixed tissues were prepared and ZIP4 and ZIP5 (A,B), or the indicated control proteins (C), were detected by immunofluorescence or immunohistochemistry. (A) Immunofluoresence localization of ZIP4 and ZIP5 in the proximal small intestine (duodenum) and pancreas. Zinc-deficient weanling mice were given an oral gavage of zinc or saline for the indicated times before harvest. Red: sites of antibody binding; blue: nuclei. (B) Immunofluoresence localization of ZIP4 and ZIP5 in the VYS. Zinc-deficient pregnant mice were given an oral gavage of zinc or saline, and fetal VYS was harvested 6 h later. (C) Immunohistochemical localization of the iron-regulated ferric reductase DCytb in the proximal small intestine and immunofluoresence localization of the basolateral membrane protein 4.8H1 in the pancreas from zinc-deficient weanling mice given an oral gavage of zinc or saline for the indicated time. Brown deposits: sites of antibody binding. Sections were counterstained with hematoxylin. The specificity of each of the antibodies used in this Figure has been previously demonstrated (De Lisle and Ziemer, 2000; McKie et al., 2001; Dufner-Beattie et al., 2003b, 2004).
Newly weaned mice grow rapidly and double in size within 2 or 3 weeks after weaning when fed the ZnA diet, but they fail to grow well when fed the ZnD diet and within 10 days they display overt signs of zinc deficiency, such as dermatitis, as well as stunted growth (Dufner-Beattie et al., 2005). Zinc demands also increase during pregnancy coincident with the rapid growth and differentiation of the embryo. Embryos in pregnant mice fed a ZnD diet beginning on d8 of pregnancy have a greatly increased risk of abnormal development (Dufner-Beattie et al., 2005, 2006). As noted previously (Dufner-Beattie et al., 2003b, 2004), Zip4 mRNA abundance was significantly increased in the maternal intestine and fetal VYS in mice fed the ZnD diet, while Zip5 and Zip1 mRNA abundance were unaltered (Figure 3). In a previous study, we noted that Zip4 mRNA levels in the VYS decline by 9 h after an intraperitoneal injection of zinc; earlier time points were not examined in that study. In both of the above experimental models of dietary zinc deficiency (newly weaned mice and pregnant mice), we found that an oral gavage of zinc, equivalent to the amount of zinc that might be eaten in 1 day by consuming the ZnA diet, did not cause a change in the abundance of Zip4, Zip5 or Zip1 mRNAs within 6 h in the intestine or VYS (Figure 3). However, MT-I mRNA was dramatically induced by a zinc gavage, but not a saline gavage.
The rapid effects of zinc repletion on the localization of ZIP4 and ZIP5 were examined herein. ZIP4 was found to be removed from the apical membrane and internalized in enterocytes and visceral endoderm cells (Figure 4A,B), while ZIP5 was relocalized to the basolateral membrane of enterocytes, visceral endoderm cells (Figure 4A,B) and pancreatic acinar cells (Figure 4A) within a few (4 to 6 h) hours of administering an oral gavage of zinc. Administration of an oral gavage of saline did not elicit these changes. As an another important control for the specificity of these changes in membrane localization it was noted that zinc deficiency did not alter the apical localization of the iron-regulated DCytb on enterocytes nor the basolateral localization of the pancreatic protein 4.8H1 (Figure 4C). This control is lacking in all previously reported studies of ZIP4 and ZIP5 trafficking, but serves to confirm the specificity of the effects of zinc.
Zinc can regulate the turnover of ZIP4 and ZIP5 proteins
Western blot analysis was used to examine ZIP4 and ZIP5 proteins in response to zinc. ZIP4 protein was not detected in membranes prepared from the intestine (Figure 5A,C) or VYS (Figure 5B) taken from mice fed the ZnA diet. In contrast, this protein was abundant in the membranes prepared from mice fed the ZnD diet. Multiple species of ZIP4 were detected, which may reflect glycosylated forms of the protein (Kim et al., 2004((•• a or b? ••))). In addition, an approximately 37 kDa immuno-reactive band was detected in the ZnD samples. The identity of this peptide remains to be determined, but it appears to be ZIP4 specific and is present in the intestine and VYS only in zinc-deficient mice (Figure 5A).
An oral gavage of zinc resulted in the rapid loss of ZIP4 immunoreactivity in Western blots of intestinal membrane preparations (Figure 5A,C). By 2 h after the zinc gavage, little ZIP4 remained in the intestine membrane preparations from zinc-deficient newly weaned mice. In contrast, ZIP1 abundance was unaffected by dietary zinc or by an oral gavage of zinc. ZIP1 served as a good loading control for these membrane preparations. Western blot analysis of the maternal intestine from pregnant mice fed the ZnD diet and then given an oral gavage of zinc also revealed a significant loss of ZIP4 immunoreactivity (Figure 5C). Although this reduction was not as dramatic as that noted in the intestine of newly weaned mice, it was significant and reproducible. In contrast, Western blot analysis of membrane proteins from the VYS revealed moderately reduced abundance of ZIP4 by 8 h after the zinc gavage (Figure 5B). Taken together, the immunolocalization (Figure 4) and Western blot analyses (Figure 5) suggest that zinc stimulates the endocytosis of ZIP4 in vivo in enterocytes and visceral endoderm cells, as was previously noted in transfected cultured cells (Kim et al., 2004((•• a or b? ••))), and that endocytosis can, but may not necessarily, lead to rapid degradation of the protein.
Hepa cells upregulated ZIP4 protein when cultured in zinc-deficient medium (Figure 5D, left panel). This induction was blocked when zinc-deficient medium was supplemented with zinc during the incubation (Figure 5D, left panel). Similar to intestine, repletion of zinc to zinc-deficient Hepa cells resulted in the rapid loss of ZIP4 immunoreactivity (Figure 5D, right panel).
ZIP5 could not be detected by Western blot analysis of intestine membrane preparations, but this protein was readily detectable in membrane preparations from the VYS and from the pancreas. ZIP5 was detected in VYS membranes from mice fed the ZnA diet, but was dramatically and reproducibly reduced or absent in VYS membranes from mice fed the ZnD diet (Figure 5B). However, ZIP5 was rapidly induced, in response to an oral gavage of zinc, in the embryonic VYS (Figure 5). In a previous report, we concluded that ZIP5 protein levels do not change in the VYS as a result of zinc deficiency (Dufner-Beattie et al., 2004). We reanalyzed the original data and discovered that we were misled by a non-specific band in those Western blots. Apparently, ZIP5 protein aggregated in those samples leaving a non-specific band at the approximate molecular mass expected for ZIP5 which confounded our interpretation. Herein, we repeated the results in the VYS multiple times (Figures 5B and 6) and then extended these studies to include analyses of the pancreas. During zinc deficiency, ZIP5 was degraded in the maternal pancreas as well as in the VYS (Figure 6A; note the induction of ZIP4 in VYS collected from those dams). To further validate these findings, we immunoprecipitated ZIP5 from the VYS and examined the precipitates by Western blot analysis. This approach increased the level of sensitivity of detection and reduced background (Figure 6B). The results confirm the conclusion that ZIP5 is dramatically reduced or absent in the VYS during zinc deficiency.
Figure 6.
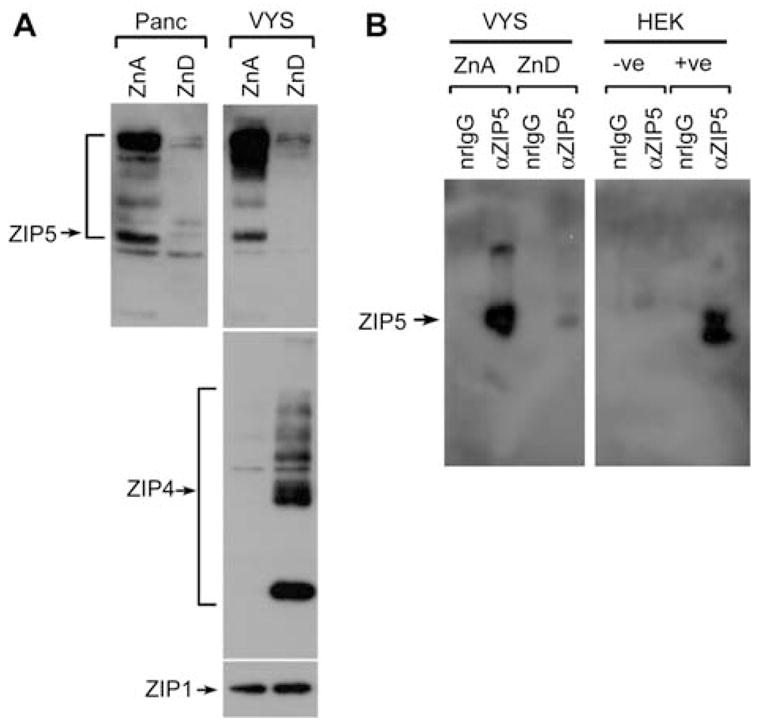
Effect of zinc deficiency on ZIP5 protein abundance in the pancreas and visceral yolk sac (VYS).
(A) Western blot detection of ZIP5 (top panels), ZIP4 (middle panel) and ZIP1 (bottom panel) in membrane proteins prepared from pancreas (top left) and VYS (top right) from pregnant dams fed a zinc-adequate (ZnA) or zinc-deficient (ZnD) diet for 6 days beginning on d8 of pregnancy. Note that ZIP4 and ZIP1 are not expressed in pancreatic acinar cells. (B) Immunoprecipitation and Western blot detection of ZIP5 in VYS and HEK 293 cells transfected with empty vector (−ve) or with (+ve) a ZIP5 expression vector. VYS sac and HEK 293 cells were sonicated in IP buffer and samples were precipitated overnight using normal rabbit IgG (nrIgG) or ZIP5 antiserum (αZIP5). Immune complexes were subjected to Western blot analysis using ZIP5 antiserum.
ZIP5 protein synthesis is controlled by a zinc-regulated translational mechanism
The rapid accumulation of ZIP5 synthesized from pre-existing mRNA could reflect the recruitment of this mRNA back into actively translating polysomes in response to zinc, the activation of translation of stalled polysomal mRNA or the inhibition of rapid turnover of this protein. To begin to address these possibilities, we first determined whether the association of Zip5 mRNA with polysomes was altered during zinc deficiency (Figure 7). VYS polysomes (membrane-bound and free) were prepared from mice fed the ZnA or ZnD diet during pregnancy (d8–d14). Polysomes were fractionated by sucrose density gradient sedimentation and the distribution of Zip4 and Zip5 mRNAs in the gradient was determined by reverse transcription-polymerase chain reaction (RT-PCR) (Figure 7B). The results revealed that Zip5 mRNA is predominantly associated with the polysome fractions, regardless of dietary zinc status. Zip5 mRNA apparently does not appear to cycle on and off polysomes in response to zinc.
Figure 7.
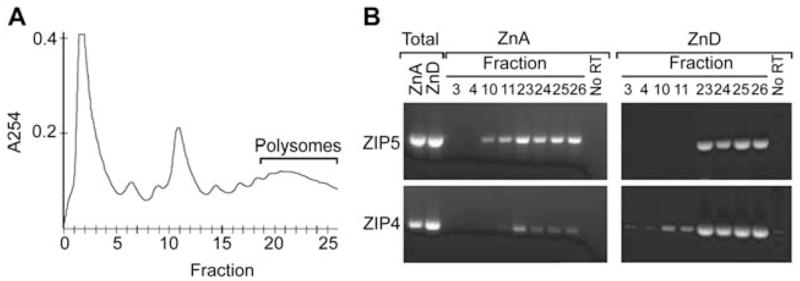
Effects of dietary zinc on the association of Zip4 and Zip5 mRNAs with polysomes.
Fetal VYS was harvested on d14 from dams fed the ZnA or the ZnD diet beginning on d8 of pregnancy. Membrane-bound and free ribosomes were solubilized and resolved on 10–50% linear sucrose gradients. (A) Fractions from the sucrose gradient were monitored for absorbance at 254 nm (A254) beginning at the top of the gradient (fraction 0). Fractions 0–5 contained the smallest particles, whereas fraction 6, 9 and 11 contained 40S ribosomal subunits, 60S ribosomal subunits and 80S monosomes, respectively. Fractions 20–26 contained polysomes. (B) RNA was isolated from the indicated fractions and amplified by RT-PCR using primers specific for Zip4 and Zip5 mRNAs. Total ZnA and ZnD: RT-PCR products from total extract RNA isolated before sucrose gradient fractionation. No RT: negative controls where the reverse transcriptase enzyme was omitted.
ZIP5 protein is underrepresented in total methionine and cysteine residues which, coupled with its relative low abundance, precluded unequivocal detection of the pulse-labeled protein. However, the effects of the cell-permeable proteasomal inhibitors, epoxomicin, MG132 and clasto-lactacystin β-lactone, and the specific lysosomal inhibitor, bafilomycin A1 on ZIP5 protein abundance were examined in VYS explant cultures. VYS harvested from pregnant mice fed the ZnD diet, were incubated in medium containing lysosomal inhibitor or proteasome inhibitors either individually (Figure 8A) or in cocktails with or without MG132 (Figure 8B). The latter inhibitor has been reported to transiently stall translation (Cowan and Morley, 2004). Protein synthesis was monitored in these explant cultures and no difference in total [35S]-methionine and cysteine incorporation was found (not shown). IP analysis of ZIP5 in membrane preparations revealed that neither the proteosomal nor lysosomal inhibitors caused the accumulation of ZIP5 protein in the VYS, even after 12 h of incubation (Figure 8A,B). In contrast, the inclusion of exogenous zinc in the VYS explant culture resulted in the internalization of ZIP4 and the reappearance of ZIP5 on basolateral membranes by 12 h (Figure 8C).
Figure 8.
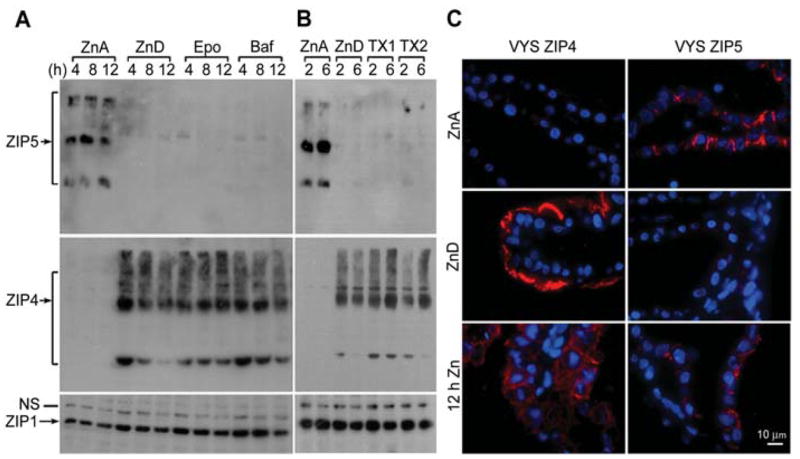
Effects of proteosome and lysosome pathway inhibitors or zinc on ZIP5 and ZIP4 protein abundance and localization in the visceral yolk sac (VYS) explant cultures.
VYS were harvested from d14 fetuses carried in pregnant females that had been fed a zinc-adequate (ZnA) diet or a zinc-deficient (Zn-D) diet beginning on d8 of pregnancy. VYS were then cultured in vitro under the conditions described below. (A) VYS from zinc-adequate dams were incubated in zinc-adequate medium (DMEM containing 10% FBS; ZnA). VYS from zinc-deficient dams were incubated in zinc-deficient medium (DMEM containing 10% Chelex-treated FBS with DMSO vehicle; ZnD), or ZnD medium also containing 50 nM synthetic epoxomicin (Epo) or 100 nM bafilomycin A1 (Baf) for the indicated times (up to 12 h). ZIP5 was immunoprecipitated (top left panel) and detected as described in the legend of Figure 6B. An equal amount of total membrane protein from each sample was examined by Western blot analysis for ZIP4 (middle panels) and ZIP1 as a loading control (bottom panels). (B) VYS from zinc-adequate dams were incubated in ZnA medium. VYS from zinc-deficient dams were incubated in ZnD medium plus or minus a mixture of 1 μM epoxomicin, 50 μM MG132 and 50 μM clasto-lactacystin β-lactone (TX1) or 1 μM epoxomicin and 50 μM clasto-lactacystin β-lactone (TX2) for the indicated times. ZIP5, ZIP4 and ZIP1 detected by Western blot analysis as described in (A). (C) VYS from zinc-adequate dams were incubated for 12 h in ZnA medium, as above. VYS from zinc-deficient dams were incubated for 12 h in ZnD medium ±100 μM ZnCl2 (12 h Zn). VYS were fixed and processed, and ZIP4 and ZIP5 were detected by immunofluorescence as described in the legend of Figure 4. Blue: nuclei; red: sites of antibody binding.
These results suggest that ZIP5 is not being synthesized and rapidly degraded in the VYS during zinc deficiency, but instead are consistent with the concept that zinc deficiency causes the translation of Zip5 mRNA to stall on the polysomes and that a zinc-sensing mechanism reactivates its translation when zinc is repleted. This zinc-sensing mechanism appears to be cell autonomous in the VYS endoderm.
Discussion
How ZIP4 and ZIP5 proteins are upregulated and down-regulated, respectively, in response to zinc deficiency (Dufner-Beattie et al., 2004), in the same cell types and coordinately in several organs is unknown (Cousins et al., 2006). Herein, we have continued to address these issues. The results of this study reveal that several novel zinc-sensing pathways orchestrate these changes via diverse post-transcriptional, translational and post-translational mechanisms in vivo (see Figure 9). Studies of ZIP4 in transfected cells suggested that zinc regulates the trafficking of this protein (Kim et al., 2004((•• a or b? ••)); Wang et al., 2004b) and a recent study identified a histidine-rich region of the human ZIP4 protein that is essential for its ubiquitination and degradation following exposure to a high concentration of zinc (Mao et al., 2007). Nothing is known about the trafficking of ZIP5 in transfected cells, although it is targeted to the basolateral membrane in polarized cells (Wang et al., 2004a). We extended these studies in vivo to examine the physiological relevance of these findings in transfected cells. It is of paramount importance to understand how these zinc transporters are regulated in vivo, although these studies are technically challenging.
Figure 9.
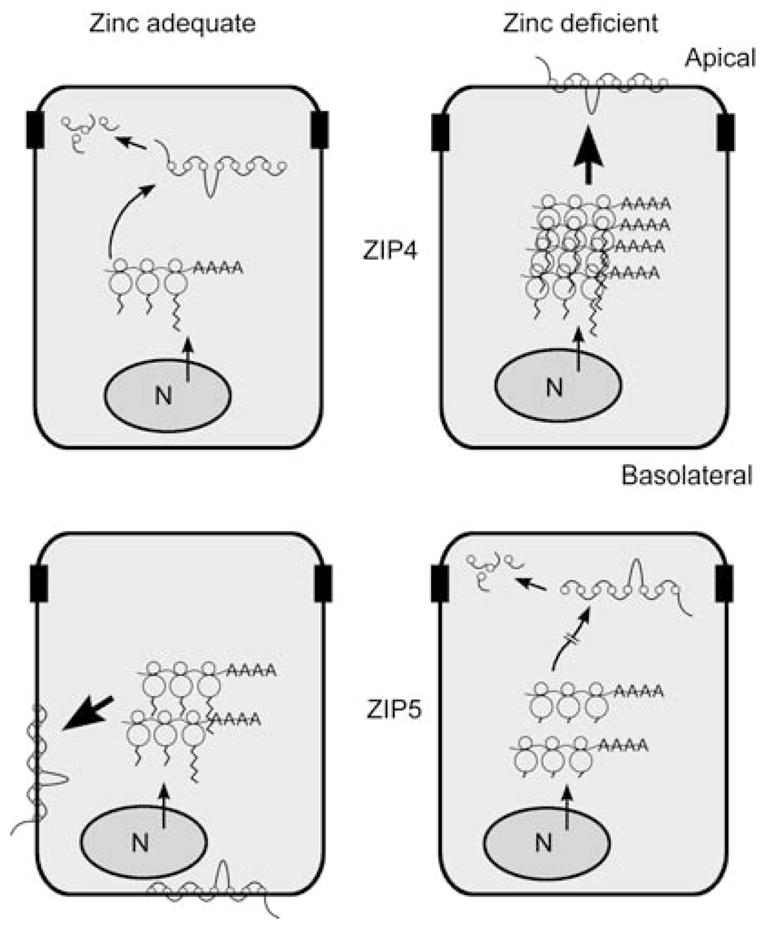
Cartoon summarizing mechanisms by which ZIP4 and ZIP5 are reciprocally regulated by zinc in the same cell. The mouse Zip4 and Zip5 genes are actively expressed in the intestine and fetal visceral yolk sac (VYS). Zip5 is also actively expressed in pancreatic acinar cells. This cartoon depicts the polarized intestinal enterocyte. N, nucleus. Polysomal mRNAs are indicated as lines ending in AAAA and bound by three ribosomes, with the protein tails extending from the ribosomes. ZIP4 and ZIP5 proteins are indicated as lines connected by eight circles representing the eight transmembrane domains. The degraded proteins are indicated as individual circles and lines inside the cells. Zinc apparently does not alter the rate of transcription of either Zip4 (top) or Zip5 (bottom), as indicated by the arrows on the nuclei in each panel. Large bold arrows indicate the most active expression of each of these zinc transporters. Top left: when zinc is adequate, Zip4 mRNA is unstable and ZIP4 protein is not detected on the apical membrane, but is degraded. Top right: when zinc becomes deficient, Zip4 mRNA is stabilized, leading to increased accumulation of Zip4 mRNA and ZIP4 protein and the localization of ZIP4 at the apical membrane. Repletion of zinc causes the rapid endocytosis and ultimate degradation of ZIP4 and destabilization of Zip4 mRNA. Bottom left: Zip5 mRNA abundance remains unchanged in response to zinc. When zinc is adequate, Zip5 mRNA is translated and ZIP5 protein accumulates in the basolateral membrane. Bottom right: when zinc becomes deficient, Zip5 mRNA translation is arrested (hatched arrow), but this mRNA remains associated with polysomes. ZIP5 protein is removed from the membrane and degraded. Repletion of zinc causes the rapid resynthesis and relocalization of this protein to the basolateral membrane.
We previously reported that Zip4 mRNA abundance increases in vivo in enterocytes and visceral endoderm cells during periods of zinc deficiency (Dufner-Beattie et al., 2003b). This regulation can be mimicked in cultured mouse Hepa cells that express Zip4 by manipulating zinc in the culture medium. This finding strongly supports the concept that changes in zinc availability directly regulate Zip4 expression. However, the induction of Zip4 mRNA occurs relatively slowly in response to zinc deficiency in vivo and in vitro and is not accompanied by an increase in the relative rate of Zip4 transcription, but rather reflects increased mRNA stability. It is interesting to note that the Zip4 gene has no metal response element, no identifiable TATA box or Inr sequence in the proximal promoter region, and it has two transcription start sites producing an A form and a smaller, much less abundant B form of the mRNA (Dufner-Beattie et al., 2003b). In addition, 5′ end mapping experiments suggest that the transcription start site of the abundant and zinc inducible A form of Zip4 mRNA is also imprecise (J. Dufner-Beattie, unpublished results). Taken together, these results are mostly consistent with the concept that the Zip4 gene is not inducible, but its tissue-specific expression is tightly controlled, as is the stability of its mRNA.
How zinc regulates the stability of Zip4 mRNA remains to be determined. These studies suggest the possibility that zinc may repress the expression of a protein that specifically stabilizes this mRNA. But, it was also noted that excess zinc destabilizes Zip4 mRNA in a process that requires new protein synthesis. Zinc may reciprocally regulate genes that stabilize and those that destabilize Zip4 mRNA. This mRNA does not contain an AU-rich sequence in the 3′-untranslated region which would be typical of an mRNA with a very short half-life (Wilkie et al., 2003). Details of the zinc-sensing mechanism(s) that regulate(s) Zip4 mRNA abundance remain to be delineated.
ZIP4 protein localization and turnover were also shown to be regulated by zinc availability in vivo. This was particularly obvious in the intestines of zinc-deficient mice given a bolus of zinc directly into the stomach. Previous findings of transfected cells showed internalization of ZIP4 in response zinc (Kim et al., 2004((•• a or b? ••))). We now report that zinc repletion in vivo caused the rapid endocytosis of ZIP4 and degradation of ZIP4 in the intestine. During the course of our studies, it was reported that human ZIP4 in transfected cells can be internalized and degraded in response zinc and it was noted that the extent of degradation was influenced by the zinc concentration and involved both proteasomal and lysosomal degradation pathways (Mao et al., 2007). Our studies are consistent with the concept that internalization and subsequent degradation can both be regulated by zinc. ZIP4 in the intestine of newly weaned mice was rapidly internalized and degraded in response to a bolus of oral zinc, whereas ZIP4 in the VYS was internalized but not necessarily degraded. This may reflect a dose-response for zinc. The concentration of zinc in the gut after an oral gavage of zinc would be expected to be significantly greater than that seen by the embryonic VYS after a maternal zinc gavage. Perhaps, ZIP4 can be recycled to the membrane under some physiological conditions. As an important tool for the future study of ZIP4 protein regulation, we have also shown that Hepa cells regulate ZIP4 protein in a manner similar to that observed in vivo with its upregulation during zinc deficiency and rapid degradation following zinc repletion.
The synthesis and targeting of ZIP5 to basolateral membranes in vivo are both modulated by zinc. This was documented to occur in the VYS and pancreas by Western blot analysis, but we assume this also occurs in the intestine, based on the lack of a significant immunofluorescence signal for this protein from zinc-deficient mice and the rapid reappearance of the signal after zinc-repletion. Remarkably, zinc stimulated the rapid accumulation of this protein in the absence of changes in mRNA. This represents the first demonstration of translational control of a zinc transporter in response to zinc. Translational control mechanisms are known to be operative in iron homeostasis. The regulation of the translation of ferritin and ferroportin 1 by iron (McKie et al., 2000; Crichton et al., 2002) and the arrest of ceruloplasmin translation in macrophages exposed to IFN-γ (Mazumder et al., 2005) reflect changes in the initiation of translation and release of the non-translating mRNAs from polysomes. Zip5 mRNA remains associated with polysomes during zinc deficiency and inhibitors of the proteasome and lysosome did not result in the reaccumulation of ZIP5 protein in VYS explant cultures. Taken together, these data support a translational stall mechanism for ZIP5 when dietary zinc is limiting. When phosphorylated, the fragile X mental retardation protein associates with polysomes that appear to be stalled in the act of translation (Ceman et al., 2003), and recent studies have suggested important roles for microRNAs in this (Jin et al., 2004) and other translational control mechanisms (Kim et al., 2004((•• a or b? ••))). Further studies are required to delineate the mechanisms which control ZIP5 translation.
In summary, these studies provide evidence that ZIP4 and ZIP5 are both dynamically regulated by several post-transcriptional, translational and post-translational mechanisms. Interestingly, distinct zinc-sensing pathways coordinately regulate these zinc transporters in such a way that ZIP4 is removed from the apical membrane and can be degraded, while ZIP5 is resynthesized and targeted to the basolateral membrane in response to increased zinc availability. In two specialized cell types (enterocytes and visceral endoderm cells), these processes occur within the same cell. In addition, this coordinate regulation of ZIP5 extends to pancreatic acinar cells. Given the importance of ZIP4 in zinc homeostasis in humans and mice, our findings suggest that ZIP5 may also be a critically important zinc transporter, as evidenced by these intricate regulatory mechanisms. Our studies reveal for the first time the existence of several novel zinc-sensing mechanisms in mammals. Unraveling the details of these molecular mechanisms will require further investigation.
Materials and methods
Animal care and use
All experiments involving mice were conducted in accordance with the National Institutes of Health guidelines for the care and use of experimental animals and were approved by the Institutional Animal Care and Use Committee. CD-1 mice (weanlings and 6–8-week-old females) were purchased from Charles River Breeding Laboratories (www.criver.com). Mouse diets were purchased from Harlan Teklad (www.teklad.com) and have been described previously (Dalton et al., 1996; Dufner-Beattie et al., 2003a,b). Zinc levels in the diets were as follows: ZnD, 1 ppm zinc; ZnA, 50 ppm zinc.
Dietary zinc manipulation
Two experimental designs of dietary zinc deficiency were employed. In experimental design one, pregnant mice (d1=vaginal plug) were fed the ZnA or ZnD diet beginning on d8 of pregnancy, under conditions described in detail previously (Dufner-Beattie et al., 2003b, 2004, 2006), and dams and fetuses were analyzed on d14. In experimental design two, mice weaned for 2–3 days (weaning 21 days after birth) were then fed the ZnA diet or ZnD diet for 10 days and then analyzed. In both of these experimental designs, zinc was readministered by oral gavage of 100 μmol ZnCl2/kg body weight (approximately equal to daily dietary intake in ca. 100 μl) or saline as a control. Mice (n=5–6 per group) were killed and tissues snap frozen for RNA and protein assays or placed in freshly prepared 4% paraformaldehyde in phosphate-buffered saline (PBS) and processed for immunohistochemistry and immunofluorescence. All of the above dietary and gavage manipulations were replicated in two independent experiments to ensure reproducibility of results.
RNA extraction and Northern blot hybridization
RNA was isolated as described in detail previously (Langmade et al., 2000; Andrews et al., 2001). Total RNA (3–6 μg) was size-fractionated by agarose-formaldehyde gel electrophoresis, transferred and UV cross-linked to the nylon membrane. Northern blot membranes were hybridized and washed under stringent conditions as previously described (Dalton et al., 1996). Hybrids were detected by autoradiography at −80°C. Duplicate gels were stained with acridine orange to ensure equivalent loading and integrity of total RNA.
Probes for mouse MT-I, Zip1, Zip4, and Zip5 mRNAs were as described previously (Dufner-Beattie et al., 2003a, 2004). Probes were used at 2×106 cpm/ml of hybridization solution.
Cell culture
Mouse Hepa cells were maintained at 37°C in a humidified 5% CO2 incubator in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U penicillin/ml, and 100 μg streptomycin/ml. To generate zinc-deficient conditions in culture, heat-inactivated FBS was treated with Chelex 100 resin, as described previously (Kim et al., 2004((•• a or b? ••))). Essential metals except zinc were repleted by the DMEM which was adjusted to 10% Chelex-treated FBS.
Stock solutions of 10 mg/ml actinomycin D (Sigma-Aldrich, www.sigma.com) and 10 mg/ml cycloheximide (Sigma-Aldrich) were prepared in methanol and 100% ethanol, respectively.
Mouse 266-6 cells were obtained from the American Type Culture Collection and cultured according to their suggested protocol, and the intestinal cell line mIC-c12 (Bens et al., 1996) was a generous gift from Dr. A. Vandewalle (Paris, France) and was cultured as they described. These cell lines were assayed for expression of the Zip4 gene, but were not used in further experiments.
Visceral yolk sac explant culture and immunoprecipitation of ZIP5
Pregnant dams were fed ZnA or ZnD diet as described above. Visceral yolk sacs (VYS; n=5 per treatment) were collected on d14 into prewarmed DMEM, blotted to remove excess medium and transferred to prewarmed DMEM containing: 10% FBS; 10% Chelex-treated FBS with dimethylsulfoxide (DMSO) vehicle; 10% Chelex-treated FBS plus 100 μM epoxomicin ZnCl2, 10% Chelex-treated FBS containing 50 nM (Calbiochem; www.calbiochem.com), or 100 nM bafilomycin A1 (Calbiochem) or 1 μM epoxomicin plus 50 μM MG132 (Sigma-Aldrich) and 50 μM clasto-lactacystin β-lactone (Sigma-Aldrich) or 1 μM epoxomicin and 50 μM clasto-lactacystin β-lactone (Sigma-Aldrich). VYS (1 per 2 ml medium) were incubated at 37°C for the indicated times and snap frozen in liquid nitrogen. Samples were thawed by sonication (5 VYS per ml) in ice-cold lysis buffer [20 mM Tris-base pH 7.4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM NaF, 5 mM Na3VO4 and complete protease inhibitor cocktail (Roche; www.roche-applied-science.com)], and membranes were prepared as described below. Membranes were resuspended in 300 μl of immunoprecipitation buffer [50 mM Tris base, pH 8.0, 150 mM NaCl, 1% NP-40, 0.2% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate and Roche protease inhibitor cocktail] by sonication. An equivalent amount of total protein from each sample was used for Western blot analysis of ZIP4 and ZIP1, as described below.
For each immunoprecipitation (IP) reaction, 1 mg of VYS protein was adjusted to 0.5 ml with IP buffer, precleared with EZview red protein A affinity gel (Sigma-Aldrich), and then immunoprecipitated using normal rabbit IgG or ZIP5 antibody overnight at 4°C. Samples were transferred to fresh tubes and incubated with EZview red protein A affinity gel at 4°C for 4 h to capture immune complexes. Immune complexes were collected by centrifugation and washed 5 times with 10 bed volumes of IP buffer per wash. ZIP5 protein was eluted from each complex with 10 μg of ZIP5 antigenic peptide, as described previously (Dufner-Beattie et al., 2004), into a small volume of IP buffer at room temperature. A portion of the eluted volume was carefully transferred to a fresh tube containing SDS-sample buffer and freshly prepared dithiothreitol and heated at 42°C for 30 min. Samples were analyzed using an 8% polyacrylamide gel and transferred to PVDF ((•• please clarify in ful ••)) membranes and analyzed by Western blot detection as described below.
Nuclear run-on assay
Run-on assays were performed according to previously published methods (McKnight and Palmiter, 1979; Derman et al., 1981). Nuclei were purified from cultured Hepa cells, as well as the proximal small intestine from pregnant dams, as described previously (Derman et al., 1981). Mice were fed the ZnA or ZnD diet for 1 day beginning on d8 of pregnancy (Dufner-Beattie et al., 2003b, 2004). Nuclei were labeled and RNA isolated, as described previously (McKnight and Palmiter, 1979).
DNA was denatured and then immobilized on nylon membranes by slot blotting, as described previously (Dufner-Beattie et al., 2001). The cDNA (3 μg) for mouse MT-I, β-actin and GAPDH or the genomic mouse Zip4 gene (Dufner-Beattie et al., 2003b) were denatured in 10 mM NaOH, neutralized by dilution into 6× SSC (350 μl total volume) and loaded via slot blot onto a well on strips of nylon membrane. Each well was rinsed with 500 μl of 6× SSC and the filter was exposed to UV light to cross-link the DNA.
Labeled run-on transcripts were hybridized (2.5×106 cpm/ml hybrid buffer) (Church and Gilbert, 1984) at 65°C for 36 h with membrane strips containing immobilized DNA. After hybridization, the membranes were washed successively as follows: 5 min at 25°C in 2× SSC; 30 min at 25°C in 2× SSC containing 10 μg/ml RNase A; 10 min at 65°C in 2× SSC, 0.1% SDS; 10 min at 65°C in 0.1× SSC, 0.1% SDS. Hybrids were detected by autoradiography and quantified by radioanalytic analysis using ImageQuant software version 5.0 ((••Manufacturer? city? state? country? ••)) on a Typhoon 9410 phosphorimager ((••Manufacturer? city? state? country? ••)).
Preparation of membrane proteins and Western blot analysis
Membrane proteins were prepared and analyzed as described previously (Dufner-Beattie et al., 2004; Huang et al., 2006). Membrane proteins (20 to 25 μg) were resolved on 8% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Membranes were blocked overnight and then incubated with primary antibody in blocking solution at the appropriate dilution (ZIP4 antiserum 1:800, ZIP5 antiserum 1:600, and ZIP1 antiserum 1:800) for 2 h at room temperature. After extensive washing, membranes were incubated with goat anti-rabbit horseradish peroxidase-conjugated secondary antibody and the blot was developed using an ECL Plus reagent (Amersham; www.amershambiosciences.com), according to the manufacturer’s instructions, and detected using Kodak BioMax MS film (www.kodak.com).
Immunohistochemistry/immunofluorescence
These methods were essentially as described in detail previously (Dufner-Beattie et al., 2003b, 2004), with modifications. Antiserum against duodenal cytochrome b (DCytb) was previously described and kindly provided by Dr. Andrew McKie (King’s College, London, UK) (McKie et al., 2001). Cell culture supernatant containing rat monoclonal antibody against the epithelial marker 4.8H1 was previously described (De Lisle and Ziemer, 2000) and kindly provided by Dr. Robert DeLisle (University of Kansas Medical Center, Kansas City, KS, USA). Immunohistochemistry was performed using Zymed Histostain-SP kits (www.zymed.com) and DAB staining. Immunofluorescene was performed using QDot 655 streptavidin conjugate (Invitrogen; www.invitrogen.com) to detect antibody binding sites. For ZIP5 visualization in the pancreas and intestine, tyramide signal amplification (Perkin-Elmer; www.perkinelmer.com) was used to enhance the specific fluorescence signal. Sections were mounted with 90% glycerol containing DAPI for subsequent imaging using a Leica DM 4000B fluorescence microscope ((• • Manufacturer? city? state? country? • •)) and Adobe Photoshop image capture software ((•• Manufacturer? city? state? country? ••)).
Sucrose density gradient fractionation of polysomes and RT-PCR detection of Zip4 and Zip5 transcript distribution
VYS were collected on d14 of pregnancy from dams fed the ZnD and ZnA diet, as described in detail above. Freshly collected tissues were homogenized in 40 mM HEPES (pH 7.4), 100 mM KCl, 5 mM MgCl2, 100μg/ml cycloheximide, and 400 U/ml of RNAse Block (Stratagene; www.stratagene.com). The homogenate was centrifuged at 10 000 g at 4°C for 30 min. The supernatant was adjusted to 0.05% sodium deoxycholate, 2% Triton X-100 and an aliquot was collected for extraction of total input RNA. Polysomes were separated on a linear 10–50% sucrose gradient by centrifugation at 40 000 g at 4°C for 2.5 h. Gradients were collected from the top and fractions were monitored for absorbance at 254 nm. A total of 26 fractions were collected into Trizol, E. coli rRNA (16S and 23S; Roche) was added to make the total RNA amount in each fraction approximately equivalent. RNA was isolated, dissolved in 5 mM EDTA (pH 8.0) and precipitated with 3 vol of 4 M ammonium acetate (pH 7.0) at 0°C for 3 h. The precipitate was collected by centrifugation, redissovled and ethanol precipitated. RNA was collected by centrifugation, dried under vacuum and dissolved in 22 μl of RNAse-free water. Volume equivalents of the fractions were then used for comparison by RT-PCR.
RT was carried out using StrataScript reverse transcriptase, according to the manufacturer’s instruction (Stratagene), with random primers. Samples were amplified by PCR for 35 cycles using Taq polymerase (New England Biolabs; www.NEB.com) and the following primers: Zip4 416 bp amplimer, sense primer 5′-GCC TGG TCT TGA CTG CCT TGC T-3′, antisense primer 5′-AGG CTC CAC TTA GCT GCT GCT G-3′; Zip5 524 bp amplimer, sense primer 5′-TGG GCC CAG CTG AAC AGG AAC A-3′, antisense primer 5′-GTG CTG GAG CTG GGG TTT TGA-3′. PCR products were resolved on 2% agarose-TAE gels containing 0.5 μg/ml ethidium bromide and imaged on an Alpha Innotech FluorChem 8900 ((•• Manufacturer? city? state? country? ••)).
Acknowledgments
This work was funded, in part, by a National Institutes of Health grant DK063975 to G.K.A. B.P.W. was supported, in part, by a KU Medical Center Biomedical Research Training Program Scholarship. T.K. was supported, in part, by fellowships from the Uehara Memorial Foundation and the Naito Foundation. We wish to thank Jim Geiser for invaluable technical assistance and Eileen Roach for digital graphics. We also wish to acknowledge the generous gift of several antibodies: Dr. Robert De Lisle (anti-4.8H1), Dr. Andrew McKie (anti-DCytb), Drs. Yien-Ming Kuo and Jane Gitschier (anti-ZIP4 and anti-ZIP5).
References
- Andrews GK, Lee DK, Ravindra R, Lichtlen P, Sirito M, Sawadogo M, Schaffner W. The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 2001;20:1114–1122. doi: 10.1093/emboj/20.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bens M, Bogdanova A, Cluzeaud F, Miquerol L, Kerneis S, Kraehenbuhl JP, Kahn A, Pringault E, Vandewalle A. Transimmortalized mouse intestinal cells (m-ICc12) that maintain a crypt phenotype. Am J Physiol. 1996;270:C1666–C1674. doi: 10.1152/ajpcell.1996.270.6.C1666. [DOI] [PubMed] [Google Scholar]
- Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. J Biol Chem. 2006;281:24085–24089. doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- Cowan JL, Morley SJ. The proteasome inhibitor, MG132, promotes the reprogramming of translation in C2C12 myoblasts and facilitates the association of hsp25 with the eIF4F complex. Eur J Biochem. 2004;271:3596–3611. doi: 10.1111/j.0014-2956.2004.04306.x. [DOI] [PubMed] [Google Scholar]
- Crichton RR, Wilmet S, Legssyer R, Ward RJ. Molecular and cellular mechanisms of iron homeostasis and toxicity in mammalian cells. J Inorg Biochem. 2002;91:9–18. doi: 10.1016/s0162-0134(02)00461-0. [DOI] [PubMed] [Google Scholar]
- Dalton TP, Fu K, Palmiter RD, Andrews GK. Transgenic mice that over-express metallothionein-I resist dietary zinc deficiency. J Nutr. 1996;126:825–833. doi: 10.1093/jn/126.4.825. [DOI] [PubMed] [Google Scholar]
- De Lisle RC, Ziemer D. Processing of pro-Muclin and divergent targeting of its products to zymogen granules and the apical plasma membrane of pancreatic acinar cells. Eur J Cell Biol. 2000;79:892–904. doi: 10.1078/0171-9335-00121. [DOI] [PubMed] [Google Scholar]
- Derman E, Krauter K, Walling L, Weinberger C, Ray M, Darnell JE., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981;23:731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Lemon RS, Thornburn A. Retinoic acid-induced expression of autotaxin in N-myc-amplified neuroblastoma cells. Mol Carcinog. 2001;30:181–189. doi: 10.1002/mc.1028. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Langmade SJ, Wang F, Eide D, Andrews GK. Structure, function, and regulation of a subfamily of mouse zinc transporter genes. J Biol Chem. 2003a;278:50142–50150. doi: 10.1074/jbc.M304163200. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Wang F, Kuo YM, Gitschier J, Eide D, Andrews GK. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J Biol Chem. 2003b;278:33474–33481. doi: 10.1074/jbc.M305000200. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Kuo YM, Gitschier J, Andrews GK. The adaptive response to dietary zinc in mice involves the differential cellular localization and zinc-regulation of the zinc transporters ZIP4 and ZIP5. J Biol Chem. 2004;279:49082–49090. doi: 10.1074/jbc.M409962200. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Huang ZL, Geiser J, Xu W, Andrews GK. Generation and characterization of mice lacking the zinc uptake transporter ZIP3. Mol Cell Biol. 2005;25:5607–5615. doi: 10.1128/MCB.25.13.5607-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufner-Beattie J, Huang ZL, Geiser J, Xu W, Andrews GK. Mouse ZIP1 and ZIP3 genes together are essential for adaptation to dietary zinc deficiency during pregnancy. Genesis. 2006;44:239–251. doi: 10.1002/dvg.20211. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Weaver BP, Geiser J, Bilgen M, Larson M, Xu W, Andrews GK. The mouse acrodermatitis gene Slc39a4 (ZIP4) is essential for development and heterozygosity causes hypersensitivity to zinc deficiency. Hum Mol Genet. 2007;16:1391–1399. doi: 10.1093/hmg/ddm088. [DOI] [PubMed] [Google Scholar]
- Guerinot ML. The ZIP family of metal transporters. Biochim Biophys Acta. 2000;1465:190–198. doi: 10.1016/s0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- Hambidge M, Krebs NF. Interrelationships of key variables of human zinc homeostasis: relevance to dietary zinc requirements. Annu Rev Nutr. 2001;21:429–452. doi: 10.1146/annurev.nutr.21.1.429. [DOI] [PubMed] [Google Scholar]
- Huang ZL, Dufner-Beattie J, Andrews GK. Expression and regulation of SLC39A family zinc transporters in the developing mouse intestine. Dev Biol. 2006;295:571–579. doi: 10.1016/j.ydbio.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kim BE, Wang FD, Dufner-Beattie J, Andrews GK, Eide DJ, Petris MJ. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J Biol Chem. 2004a;279:4523–4530. doi: 10.1074/jbc.M310799200. [DOI] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004b;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Shames DM, Woodhouse LR. Zinc homeostasis in humans. J Nutr. 2000;130(Suppl):1360S–1366S. doi: 10.1093/jn/130.5.1360S. [DOI] [PubMed] [Google Scholar]
- Kury S, Dreno B, Bezieau S, Giraudet S, Kharfi M, Kamoun R, Moisan JP. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat Genet. 2002;31:239–240. doi: 10.1038/ng913. [DOI] [PubMed] [Google Scholar]
- Langmade SJ, Ravindra R, Daniels PJ, Andrews GK. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J Biol Chem. 2000;275:34803–34809. doi: 10.1074/jbc.M007339200. [DOI] [PubMed] [Google Scholar]
- Mao X, Kim BE, Wang F, Eide DJ, Petris MJ. A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. J Biol Chem. 2007;282:6992–7000. doi: 10.1074/jbc.M610552200. [DOI] [PubMed] [Google Scholar]
- Maverakis E, Fung MA, Lynch PJ, Draznin M, Michael DJ, Ruben B, Fazel N. Acrodermatitis enteropathica and an overview of zinc metabolism. J Am Acad Dermatol. 2007;56:116–124. doi: 10.1016/j.jaad.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Fox PL. Regulation of macrophage ceruloplasmin gene expression: one paradigm of 3′-UTR-mediated translational control. Mol Cells. 2005;20:167–172. [PubMed] [Google Scholar]
- McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- McKnight GS, Palmiter RD. Transcriptional regulation of the ovalbumin and conalbumin genes by steroid hormones in chick oviduct. J Biol Chem. 1979;254:9050–9058. [PubMed] [Google Scholar]
- Palmiter RD, Huang L. Efflux and compartmentalization of zinc by members of the SLC30 family of solute carriers. Eur J Physiol. 2004;447:744–751. doi: 10.1007/s00424-003-1070-7. [DOI] [PubMed] [Google Scholar]
- Peters JL, Dufner-Beattie J, Xu W, Geiser J, Lahner B, Salt DE, Andrews GK. Targeting of the mouse Slc39a2 (Zip2) gene reveals highly cell-specific patterns of expression, and unique functions in zinc, iron and calcium homeostasis. Genesis. 2007;45:339–352. doi: 10.1002/dvg.20297. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Nicholson RI. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta. 2003;1611:16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhou B, Kuo YM, Zemansky J, Gitschier J. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am J Hum Genet. 2002;71:66–73. doi: 10.1086/341125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kim BE, Petris MJ, Eide DJ. The mammalian ZIP5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J Biol Chem. 2004a;279:51433–51441. doi: 10.1074/jbc.M408361200. [DOI] [PubMed] [Google Scholar]
- Wang FD, Kim BE, Dufner-Beattie J, Petris MJ, Andrews G, Eide DJ. Acrodermatitis enteropathica mutations affect transport activity, localization and zinc-responsive trafficking of the mouse ZIP4 zinc transporter. Hum Mol Genet. 2004b;13:563–571. doi: 10.1093/hmg/ddh049. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]


