Abstract
The levels of Fibroblast Growth Factor (FGF) signaling play important roles in coordinating development of the mouse inner, middle and outer ears. Extracellular signal-regulated kinases (ERKs) are among the effectors that transduce the FGF signal to the nucleus and other cellular compartments. Attenuation of ERK activity by dephosphorylation is necessary to modulate the magnitude and duration of the FGF signal. Recently, we showed that inactivation of the ERK phosphatase, dual specificity phosphatase 6 (DUSP6) causes partially penetrant postnatal lethality, hearing loss and skeletal malformations. To determine whether other Dusps may function redundantly with Dusp6 during otic development, we surveyed the expression domains of the three ERK-specific DUSP transcripts, Dusp6, Dusp7 and Dusp9, in the embryonic mouse ear. We show that each is expressed in partially overlapping patterns that correspond to regions of active FGF signaling, implying combinatorial roles in negative regulation of this pathway during ear development.
Keywords: Fibroblast growth factor, cochlea, pillar cell, tympanic membrane, malleus, pinna, MAP kinase phosphatase (MKP), Mkp3/Pyst1, Mkp4/Pyst3, Mkp-X/Pyst2
Introduction
Normal development and function of the auditory-vestibular apparatus requires the coordinated deployment of signaling molecules to instruct appropriate transcriptional responses in the myriad tissues that contribute to the ear. Fibroblast growth factors (FGFs) comprise an important set of signals that are absolutely required for normal ear development. FGF signals are necessary at all stages of otic development, including for outer ear (pinna) morphogenesis, as well as for ossicle formation within the middle ear, and the initial induction, morphogenesis and sensory elaboration of the inner ear compartment (for reviews, see Mallo, 2003; Wright and Mansour, 2003b; Fritzsch et al., 2006; Schimmang, 2007).
The responses to FGF signals are dose-dependent and the magnitude of the signal must be precisely regulated to effect the appropriate outcome. Loss or reduction of FGF ligands causes sensorineural and/or conductive hearing deficits in several human syndromes (Rohmann et al., 2006; Gregory-Evans et al., 2007; Tekin et al., 2007), as well as defects of otic development and function in mice that are mutant for individual or multiple FGF ligands (Alvarez et al., 2003; Wright and Mansour, 2003a; Pirvola et al., 2004; Ladher et al., 2005; Hatch et al., 2007; Jacques et al., 2007; Zelarayan et al., 2007). FGF receptors 1, 2 and 3 are also required for normal otic development in both mice and humans (Colvin et al., 1996; Pirvola et al., 2000; Pirvola et al., 2002; Trokovic et al., 2003; Rohmann et al., 2006; Toydemir et al., 2006; Hayashi et al., 2007; Puligilla et al., 2007). Not surprisingly, over-activation of FGF signaling pathways due to gain-of-function mutations in FGF receptors (Hollway et al., 1998) or to reduced-function mutations in negative regulators of signaling are also detrimental to auditory function (Shim et al., 2005; Li et al., 2007). Far less, however, is understood regarding this latter class of activities, which make up an important modulatory component of the FGF signaling cascade.
FGF receptor signaling involves the integration of multi-protein complexes that function to transmit and amplify the signal intracellularly (Eswarakumar et al., 2005). Essential effector components in this cascade are the mitogen-activated protein kinases (MAPKs), of which the extracellular signal-regulated kinases (ERK) is one subset. Upon activation of ERK kinases by di-phosphorylation, substrates are recruited in an array of cellular compartments, including transcription factors in the nucleus that are, in turn, phosphorylated and activated to ultimately elicit a specific transcriptional response (Raman et al., 2007; Zhang and Dong, 2007). Most of the di-phosphorylated ERK in early mouse (E6.5-E10.5) and chick (HH10) embryos, including that found in the otic region, is dependent on FGF receptor activity (Corson et al., 2003; Lunn et al., 2007). This suggests that modulation of ERK activity may be a critical aspect of feedback for the FGF signaling pathway. All signaling systems require mechanisms to attenuate or terminate receptor activation, and this negative feedback can occur at numerous points throughout the cascade. The Sprouty genes encode proteins that inhibit MAPK signaling downstream of the receptor and upstream of ERK activation (Mason et al., 2006). The importance of these factors in otic development was demonstrated by analysis of Spry2 null mutant mice, which are hearing impaired (Shim et al., 2005). Spry2 is expressed in Deiters' cells in the organ of Corti. Spry2 mutants have an extra pillar cell likely due to a cell fate change from a Deiters to a pillar cell. This phenotype is suppressed and deafness is ameliorated, by removing one copy of Fgf8, implicating Spry2 in the negative regulation of FGF signaling.
A second class of MAPK inhibitors is a group of dual-specificity phosphatases encoded by the Dusp genes (also known as Mkps, for MAP kinase phosphatases). The mammalian genome contains at least 11 Dusp genes, many of whose protein products have been analyzed biochemically for substrate specificity. DUSP6 (MKP3/PYST1), DUSP7 (MKP-X/PYST2) and DUSP9 (MKP4/PYST3) comprise a structurally homologous subfamily of MAPK phosphatases that exhibit a preference for dpERK. Little is known about the physiological inducers of these Dusps, but we and others found that Dusp6 is expressed during embryonic development in a pattern that correlates with active centers of FGF signaling in the embryo, and that Dusp6 expression depends on FGF signaling (reviewed in, Alonso et al., 2004; Dickinson and Keyse, 2006; Owens and Keyse, 2007). Indeed, Dusp6 reporter constructs are proving useful to visualize areas of active FGF signaling in zebrafish embryos (Molina et al., 2007). Furthermore, analysis of Dusp6-deficient mice revealed incompletely penetrant phenotypes similar to those of activated FGF receptors, supporting a role for this phosphatase as a negative feedback regulator of FGF signaling (Li et al., 2007). Hearing loss associated with middle ear and otic capsule, but not cochlear, malformations were among the consequences of Dusp6 inactivation. The presence of homologous gene family members suggests the possibility of functional redundancy during peripheral auditory development.
Whether either Dusp7 or Dusp9 plays roles in otic development is unknown. Bypassing the placental requirement for X-linked Dusp9 via tetraploid rescue allowed the recovery of normal appearing, fertile Dusp9-deficient males, but the impact of this disruption on ear development or auditory function was not addressed (Christie et al., 2005). Dusp7 is expressed in a wide array of human cell lines, including those derived from cancers and lymphoblastoid cells (Levy-Nissenbaum et al., 2003), however nothing is known regarding the expression, or role, of this phosphatase during mouse development.
Here we examine the expression domains of mouse Dusp6, Dusp7 and Dusp9 between E9.0 and E18.5 in the tissues that give rise to the inner, middle and outer ear. We find that only Dusp9 has significant expression in the inner ear during mid-gestation, but all three genes are expressed in different epithelial and mesenchymal regions of the late gestation inner ear. There is significant overlap of the three genes in the mid-gestation precursors of the middle ear, and both Dusp6 and Dusp7 are expressed at different sites in the late gestation middle ear. Finally, all three genes were expressed during development of the outer ear. These studies set the stage for designing and analyzing the multi-genic conditional mutants that will be needed to understand the roles of these Dusp genes in all phases and compartments of otic development.
Results and Discussion
Dusp6, Dusp7 and Dusp9 are expressed in distinct, as well as overlapping, domains of the mid-gestation inner, middle and outer ear
To evaluate the potential for unique and redundant functions in the regulation of ERK signaling during otic development, we compared the mid-gestation expression patterns of Dusp6, Dusp7 and Dusp9 using whole mount in situ hybridization to E8.5-E11.5 embryos, followed by analysis of cryosections, and focused on defining expression in the otic region.
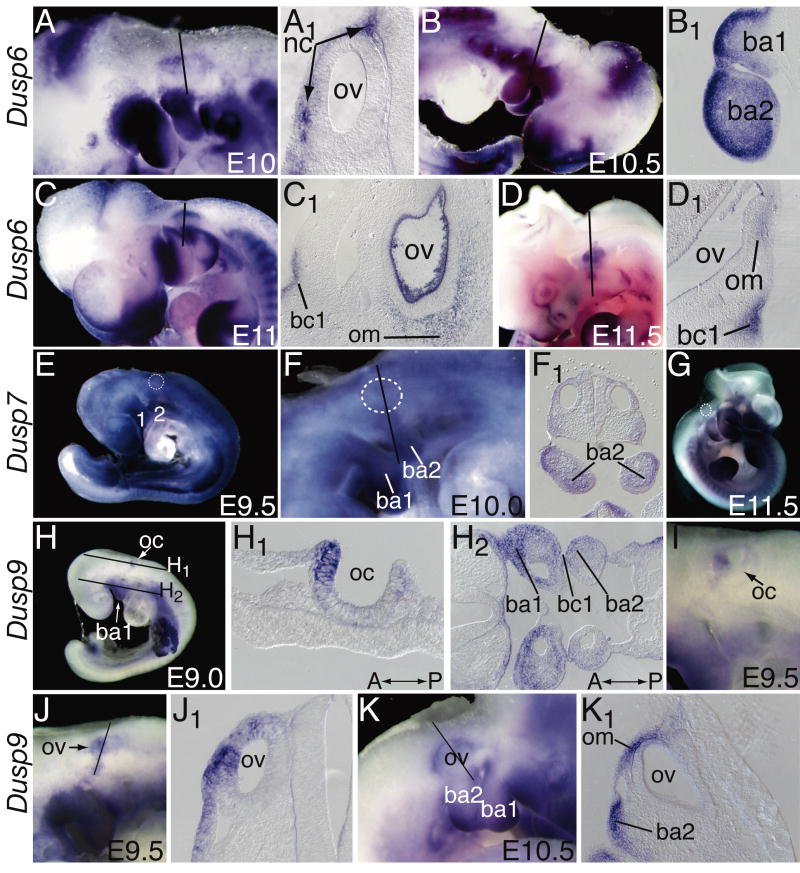
Dusp6 was seen in presumptive neural crest (nc) migrating lateral to the developing otic vesicles (ov) and was found from the earliest stages in the neural crest-derived mesenchyme of the branchial arches (ba1, ba2) (Figs. 1A,A1,B,B1,C). At E10.5 and E11, Dusp6 expression was also evident in mesenchyme adjacent to the otic vesicle as well as in first branchial cleft (bc1) ectoderm and immediately adjacent mesenchyme (Figs. 1C,C1,D,D1). Thus, Dusp6 was expressed in sites that could be responding to FGF signals controlling the development of the ossicles and outer ear (branchial arch mesenchyme and first branchial cleft, respectively), as well as the otic capsule (otic mesenchyme). Indeed, Dusp6 null animals have partially penetrant malformations of the ossicle and otic capsule (Li et al., 2007).
Figure 1. Expression of Dusp6, Dusp7 and Dusp9 transcripts in the otic region of mid-gestation (E9-E11.5) embryos.
(A-D1) Dusp6 expression, (E-G) Dusp7 expression and (H-K1) Dusp9 expression in whole embryos and sections (indicated by a subscript on the panel letter). Lines through whole embryos indicate the approximate plane of section of the corresponding section. All sections except H1 and H2 are in the transverse plane with dorsal up. Sections shown in H1 and H2 are oriented in the coronal plane with anterior (A) and posterior (P) indicated. Embryo stage is indicated in the lower right of each whole mount panel. The otic vesicle has been indicated with dashed white circles in panels E, F and G because it is difficult to see due to widespread Dusp7 staining. The blue precipitate inside the ov in C1 is caused by non-specific filling with probe of the acellular space inside the vesicle during the whole mount hybridization. Abbreviations: 1, branchial arch 1; 2, branchial arch 2, ba1, branchial arch1; ba2, branchial arch 2; bc1, branchial cleft 1, oc, otic cup; om, otic mesenchyme, ov, otic vesicle.
The Dusp7 expression pattern was not as discretely correlated with sites of FGF signaling, appearing rather widespread and diffuse (Figs. 1E,F,G). Indeed, there was no obvious change in Dusp7 expression in Fgfr1- or Fgfr2- deficient embryos, suggesting that it is not a target of FGF signals (data not shown). Nevertheless, sections of stained embryos revealed that like Dusp6, Dusp7 expression was concentrated in branchial arch mesenchyme (Figs. 1E,F,F1), but in contrast, was not highly enriched in outer ear or otic capsule progenitors (Fig. 1G and data not shown).
Dusp9 was the only family member showing strong and localized expression in the developing inner ear at these early stages. Expression initiated at the otic cup (oc) stage (E9.0, 14 somites, data not shown) and was confined to the anterior domain (Fig. 1H,H1,I), opposite to the expression domain of Fgf16 in the posterior otic cup (Wright et al., 2003, Hatch et al., manuscript in preparation). As the cup closed to form the otic vesicle (E9.5), Dusp9 was concentrated laterally, about midway between the dorsal and ventral otic poles (Figs. 1J,J1). Expression in the otic vesicle was no longer detectable after the cup closed entirely (E9.5, 21 somites), but could be seen subsequently in mesenchyme immediately adjacent to the dorsal half of the vesicle that was forming the vertical canal plate (E10.5, Figs. K,K1). This expression pattern suggests that Dusp9 could be involved in anterior-posterior patterning of the otic vesicle and/or outpouching of the lateral canal (Morsli et al., 1998; Wu et al., 1998).
Like both Dusp6 and Dusp7, Dusp9 was expressed strongly in branchial arch mesenchyme, although the highest levels became concentrated in the most distal reaches of the arches by the end of this period (Figs. 1H2,I,J,K and data not shown). Thus, Dusp7 and Dusp9 are reasonable candidates for redundancy with Dusp6 in middle ear and otic capsule formation (Li et al., 2007). In summary, all three Dusp genes could be regulating FGF signaling needed for ossicle and outer ear development, but only Dusp9 might have an early role in regulating responses of the otic vesicle to FGF signaling.
Dusp6, Dusp7 and Dusp9 are expressed in distinct, as well as overlapping, domains of the late gestation inner, middle and outer ear
To precisely determine the Dusp expression domains at later stages of embryonic development, transverse (E12.5, E14.5, E16.5) or sagittal (E18.5) paraffin sections taken through the otic region were subjected to in situ hybridization (Fig. 2).
Figure 2. Expression of Dusp6, Dusp7 and Dusp9 transcripts in the otic region of late gestation (E12.5-E18.5) embryos.
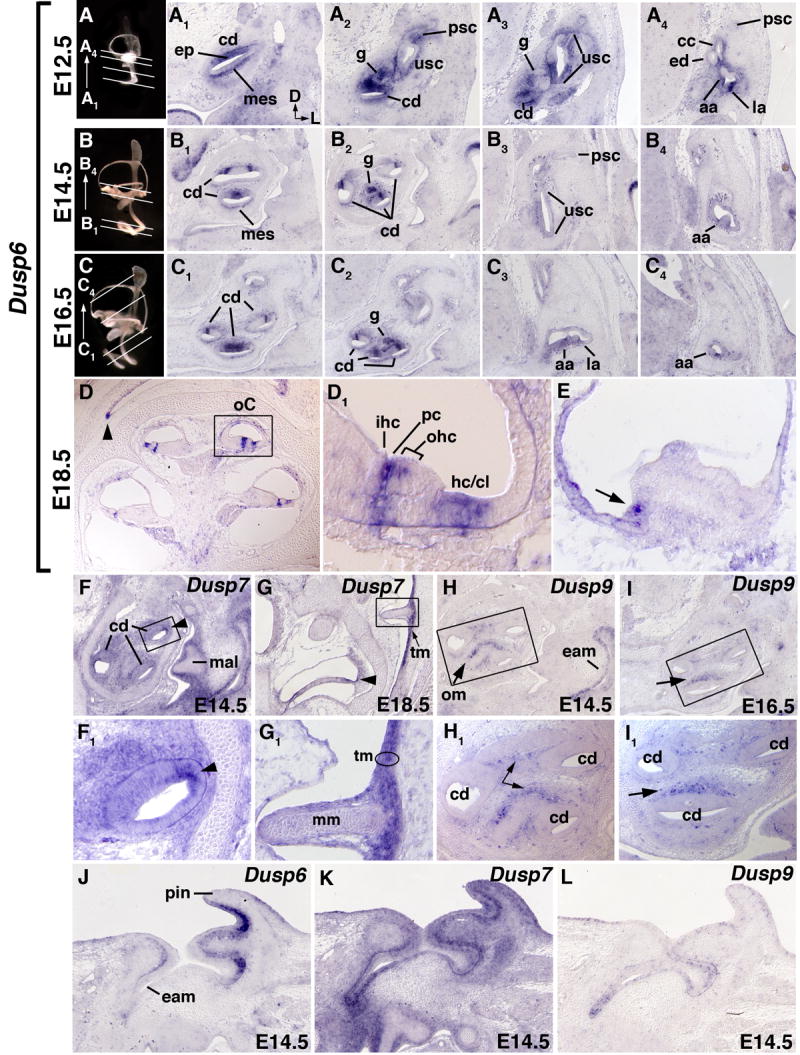
(A1-C4) Transverse sections of left ears hybridized with Dusp6 probe. The approximate planes of section are denoted by white lines through age-matched paint-filled inner ears (A, E12.5; B, E14.5; C, E16.5). Dorsal and lateral orientations for all panels are as indicated in A1. (D,D1,E) Sagittal sections of E18.5 heads hybridized with Dusp6 probe. The caret in D denotes expression at the margin of the tympanic recess. D1 shows a higher magnification view of the organ of Corti (oC), indicated by the boxed region in D, Panel E shows a representative section through the sensory region (crista) at the base of a semicircular canal. An arrow indicates the expression at the margin of the crista. (F,F1,G,G1) Dusp7 expression at E14.5 and E18.5. F1 and G1 show enlargements of the boxed regions in panels F and G, respectively. A caret indicates the margin of the cochlear duct. (H,H1,I,I1) Dusp9 expression at E14.5 and E16.5. H1 and I1 show enlargements of the boxed regions in panels H and I, respectively. Dusp6 (J), Dusp7 (K), and Dusp9 (L) expression in transverse sections of the E14.5 outer ear. Abbreviations: cd, cochlear duct; ep, cochlear epithelium; mes, mesenchyme; ed, endolymphatic duct; cc, common crus; usc, utriculosaccular chamber; g, spiral ganglion; psc, posterior semicircular canal; aa, anterior semicircular canal ampulla; la, lateral semicircular canal ampulla; oC, organ of Corti; ihc, inner hair cell; pc, pillar cells; ohc, outer hair cells; hc/cl, Hensen cells/Claudius cells; mal, malleus; mm, manubrium of the malleus; tm, tympanic membrane; om, otic mesenchyme; pin, pinna; eam, external acoustic meatus.
Inner ear expression
Dusp6 exhibited robust expression in the presumptive sensory epithelium (ep) of the cochlear duct (cd) at all stages examined (Figs. 2A1,A2,B1,B2,C1,C2,D,D1). Transcripts were not detected at E11.5 in the inner ear epithelium (Fig. 1D1), thus the presumed onset of cochlear Dusp6 expression was between E11.5 and E12.5, corresponding both to the time when the sensory domain becomes specified, and to the location of Fgfr1 transcripts (Pirvola et al., 2002; Barald and Kelley, 2004). As the cochlear epithelium matured, the Dusp6 domain was increasingly restricted (Figs. 2B1,B2,C1,C2), initially resembling the combination of Fgfr1 and Fgfr3 patterns (Pirvola et al., 2002; Barald and Kelley, 2004; Hayashi et al., 2007). By E18.5 (Figs. 2D,D1), Dusp6 expression was restricted to the presumptive pillar cells (pc), which separate the inner (ihc) and outer hair cells (ohc), and also express Fgfr3 (Hayashi et al., 2007), and to the more lateral Hensen/Claudius (hc/cl) cell domain, in which Fgfr1 and Fgfr2 expression have been detected (O. Bermingham-McDonogh, personal communication). Thus, DUSP6 could potentially regulate the FGF signaling required for FGFR1-mediated cellular proliferation and/or FGFR3-mediated pillar cell development (Pirvola et al., 2002; Hayashi et al., 2007; Puligilla et al., 2007).
Dusp6 was also expressed strongly in the spiral ganglion (g), at least in the more basally located sections, starting at E12.5 (Fig. 2A2) and continuing through E16.5 (Figs. 2B2,C2). Since the ganglion neurons become post-mitotic in a basal-to-apical gradient between E11.5 and E16 (Ruben, 1967), the pattern of Dusp6 could reflect a role for FGF signaling in this process. In the vestibular system, Dusp6 transcripts were seen in an anterior domain of the utriculosaccular chamber (usc) that presumably gives rise to the saccule (Fig. 2A3). This expression was only prominent at E12.5. Expression of Dusp6 was also apparent in the developing cristae of each semicircular canal, narrowing to the most lateral margin as development proceeded (Figs. 2A4,B4,C4,E).
Other sites of inner ear Dusp6 expression included mesenchyme immediately adjacent to the developing Reissner's membrane (Fig. 2A1, B1) and also that near the semicircular canal ducts (Fig 2A4), which is fated to be resorbed during perilymphatic duct (scala vestibuli) formation. The former expression domain is adjacent to Fgf9 expression in Reissner's membrane and the semicircular canal ducts, and coincides with expression of Fgfr1(IIIc) and Fgfr2(IIIc) (Pirvola et al., 2004). These data suggest that Dusp6 could regulate the FGF9 signaling that influences formation of the otic capsule and scala vestibuli (Pirvola et al., 2004).
The only discrete Dusp7 expression domain in the inner ear epithelium was found at the lateral edge of the cochlear duct (cd) (Figs. 2F,F1,G, indicated by carets), which will eventually contribute to the stria vascularis. Dusp7 transcripts were found quite widely, but not especially strongly, throughout the head mesenchyme (data not shown). There was also diffuse expression of Dusp7 throughout the cochlear mesenchyme (Fig. 2F,F1, data not shown for E12.5 and E16.5) that diminished by E18.5 (Fig. 2G). The significance of this expression pattern may become apparent when the gene is mutated, but it is interesting to note that the few lateral cochlear duct cells that express Dusp7 are either adjacent to or coincident with cells that express Fgf16 (E. Hatch, LDU and SLM, unpublished data).
In contrast to Dusp6 and Dusp7, we did not detect Dusp9 in cochlear epithelium after E10.5, but expression was clearly apparent at E14.5 and E16.5 in otic mesenchyme (om) adjacent to the developing sensory epithelium (Fig. 2H,H1,I,I1, indicated by arrows). This mesenchyme abuts Fgf3 and Fgf10-expressing cells in the developing sensory domain (Wilkinson et al., 1989; Pirvola et al., 2000; Pauley et al., 2003), the Fgf8-expressing inner hair cell progenitors (Shim et al., 2005; Hayashi et al., 2007; Jacques et al., 2007) and Fgf10-expressing cells in the cochlear ganglion (Pirvola et al., 2000; Pauley et al., 2003) and is eventually cleared during formation of the scala tympani. FGF3 and FGF10 are each required for normal inner ear morphogenesis (Mansour et al., 1993; Pauley et al., 2003; Ohuchi et al., 2005; Hatch et al., 2007), but their receptor, FGFR2(IIIb), is present mainly in non-sensory cochlear epithelium and Fgfr2b mutants form a perilymphatic space, albeit abnormally shaped (Pirvola et al., 2000). FGF8 is apparently required for pillar cell differentiation, with no effects reported on otic morphogenesis (Jacques et al., 2007; Zelarayan et al., 2007). These considerations suggest the possibility of additional FGFR or other RTK signaling events that DUSP9 might regulate in otic mesenchyme.
Middle ear
Dusp6 and Dusp7, but not Dusp9, were expressed in the late gestation middle ear. Dusp6 transcripts were found in the margin of the tympanic cavity at E18.5 (Fig. 2D, caret). Dusp7 was found in mesenchyme surrounding the chondrifying ossicles at E14.5 (Fig. 2F, malleus) and at E18.5 it was strongly induced in all three layers of the tympanic membrane (tm); the epithelium of the invaginated external acoustic meatus (eam), the pharyngeal endoderm-derived epithelium, and the interstitial branchial mesenchyme (Fig. 2G,G1). It has been proposed that signals arising from the EAM are required to coordinate the development of the malleus and its insertion point into the tympanic membrane (Mallo et al., 2000). Both Fgf4 and Fgf9 are expressed in the developing EAM (Mallo et al., 2000), so it is tempting to suggest that Dusp7 might participate in this process and/or contribute to development of the tympanic membrane. Furthermore, loss of Dusp6 expression from the margins of the tympanic cavity might account for the variably penetrant defects found in the shape of the tympanic opening in Dusp6 mutants (Li et al., 2007).
Outer ear
Each of the three Dusp genes was also expressed at different levels and in contiguous domains in the developing outer ear. Dusp6 was strongly expressed in the ectodermal layer of the pinna (pin) (Fig. 2J), whereas Dusp7 expression, although widely expressed throughout the pinna and EAM, was found most strongly in a region of chondrifying mesenchyme adjacent to Dusp6 (Fig. 2K). In contrast, Dusp9 expression was confined to dispersed cells in the ectodermal layer of the pinna and EAM (Fig. 2L). Although FGF signaling involving Fgf8, Fgf4 and Fgfr1 clearly plays roles in pinna development (Partanen et al., 1998; Abu-Issa et al., 2002; Frank et al., 2002; Boulet et al., 2004), it is not clear when these signaling components are required, nor whether these particular ligands and receptors are expressed in the late gestation pinna. Thus, it is difficult to propose specific roles in pinna development for the Dusp genes at this stage, beyond the suggestion that they could be involved in regulating epithelial-mesenchymal signaling. Expression of Dusps in the developing EAM is intriguing as this ectodermal tissue is thought to require signals from the developing tympanic ring to develop normally (Mallo and Gridley, 1996).
In summary, only Dusp6 and Dusp7 were found in late gestation otic epithelium, but since their patterns were not overlapping, Dusp7 is not a good candidate for redundancy with Dusp6, suggesting that perhaps other negative regulators of ERK signaling could provide this function. All three Dusp genes were expressed in cochlear mesenchyme and could have roles in development of the perilymphatic spaces. Only Dusp7 showed expression in the developing middle ear at this stage and it was found in locations suggesting roles in coordination of insertion of the malleus into the tympanic membrane. All three Dusps were found in partially overlapping and adjacent tissue layers in the developing pinna and EAM, suggesting roles in the regulation of FGF-directed outer ear morphogenesis.
Methods
In situ hybridization
Whole mount in situ hybridization of digoxigenin-labeled cRNA probes to early CD-1 (Charles River) embryos, followed by cryosectioning to evaluate cellular detail has been described previously (Wright and Mansour, 2003a). In situ hybridization of the probes to paraffin sections of older embryos was performed essentially as described by Hayashi et al. (2007) with some modifications. Briefly, E12.5-E16.5 embryos were decapitated and the heads submerged in Modified Carnoy's fixative at 4°C overnight. E17 and older embryos were decapitated, the heads were bisected mid-sagittally, the brain was removed, and the remainder fixed as above. Slides containing 10 μm sections of paraffin-embedded heads were dewaxed and incubated overnight at 68°C in a hybridization buffer containing 50% formamide and 1 μg/ml digoxigenin-labeled anti-sense RNA. After the non-specifically bound probe was removed, slides were incubated in alkaline phosphatase-conjugated anti-digoxigenin antibody (1:2000, Roche) and developed in BM Purple Substrate (Roche). Full details are available upon request.
Probes
The Dusp6 probe (5′ UTR) has been described (Li et al., 2007). The Dusp7 probe was generated from a clone containing a 498 bp segment of the 3′ UTR (2357-2855bp of GenBank accession NM_153479). The Dusp9 probe was generated from a clone containing a 544bp segment of the 3′ UTR (1640-2184 bp, GenBank accession NM_029352).
Acknowledgments
We thank Dr. Olivia Bermingham-McDonogh for discussions of FGF ligand and receptor expression data, and her and Dr. Toshinori Hayashi for helpful advice regarding in situ hybridization on tissue sections. This work was supported by NIH/NIDCD grant R01DC002043 to SLM.
Grant Sponsor: NIH/NIDCD Grant number: R01DC002043
References
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, Schimmang T. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131:4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol. 2004;273:361–372. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Christie GR, Williams DJ, Macisaac F, Dickinson RJ, Rosewell I, Keyse SM. The dual-specificity protein phosphatase DUSP9/MKP-4 is essential for placental function but is not required for normal embryonic development. Mol Cell Biol. 2005;25:8323–8333. doi: 10.1128/MCB.25.18.8323-8333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Dickinson RJ, Keyse SM. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci. 2006;119:4607–4615. doi: 10.1242/jcs.03266. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Beisel KW. Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res. 2006;1091:151–171. doi: 10.1016/j.brainres.2006.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory-Evans CY, Moosajee M, Hodges MD, Mackay DS, Game L, Vargesson N, Bloch-Zupan A, Ruschendorf F, Santos-Pinto L, Wackens G, Gregory-Evans K. SNP genome scanning localises oto-dental syndrome to chromosome 11q13 and microdeletions at this locus implicate FGF3 in dental and inner ear disease and FADD in ocular coloboma. Hum Mol Genet. 2007;16:3482–3493. doi: 10.1093/hmg/ddm204. [DOI] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, Mansour SL. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development. 2007;134:3615–3625. doi: 10.1242/dev.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- Hollway GE, Suthers GK, Battese KM, Turner AM, David DJ, Mulley JC. Deafness due to Pro250Arg mutation of FGFR3. Lancet. 1998;351:877–878. doi: 10.1016/S0140-6736(98)24012-8. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Nissenbaum O, Sagi-Assif O, Kapon D, Hantisteanu S, Burg T, Raanani P, Avigdor A, Ben-Bassat I, Witz IP. Dual-specificity phosphatase Pyst2-L is constitutively highly expressed in myeloid leukemia and other malignant cells. Oncogene. 2003;22:7649–7660. doi: 10.1038/sj.onc.1206971. [DOI] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JS, Fishwick KJ, Halley PA, Storey KG. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol. 2007;302:536–552. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Mallo M. Formation of the outer and middle ear, molecular mechanisms. Curr Top Dev Biol. 2003;57:85–113. doi: 10.1016/s0070-2153(03)57003-x. [DOI] [PubMed] [Google Scholar]
- Mallo M, Gridley T. Development of the mammalian ear: coordinate regulation of formation of the tympanic ring and the external acoustic meatus. Development. 1996;122:173–179. doi: 10.1242/dev.122.1.173. [DOI] [PubMed] [Google Scholar]
- Mallo M, Schrewe H, Martin JF, Olson EN, Ohnemus S. Assembling a functional tympanic membrane: signals from the external acoustic meatus coordinate development of the malleal manubrium. Development. 2000;127:4127–4136. doi: 10.1242/dev.127.19.4127. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Molina GA, Watkins SC, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol. 2007;7:62. doi: 10.1186/1471-213X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Yasue A, Ono K, Sasaoka S, Tomonari S, Takagi A, Itakura M, Moriyama K, Noji S, Nohno T. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn. 2005;233:177–187. doi: 10.1002/dvdy.20319. [DOI] [PubMed] [Google Scholar]
- Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- Partanen J, Schwartz L, Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. 1998;12:2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev Biol. 2004;273:350–360. doi: 10.1016/j.ydbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, Fritzsch B, Kelley MW. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Rohmann E, Brunner HG, Kayserili H, Uyguner O, Nurnberg G, Lew ED, Dobbie A, Eswarakumar VP, Uzumcu A, Ulubil-Emeroglu M, Leroy JG, Li Y, Becker C, Lehnerdt K, Cremers CW, Yuksel-Apak M, Nurnberg P, Kubisch C, Schlessinger J, van Bokhoven H, Wollnik B. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 1967;220(Suppl):221–244. [PubMed] [Google Scholar]
- Schimmang T. Expression and Functions of FGF Ligands during Early Otic Development. Int J Dev Biol. 2007;51:473–481. doi: 10.1387/ijdb.072334ts. [DOI] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Tekin M, Hismi BO, Fitoz S, Ozdag H, Cengiz FB, Sirmaci A, Aslan I, Inceoglu B, Yuksel-Konuk EB, Yilmaz ST, Yasun O, Akar N. Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am J Hum Genet. 2007;80:338–344. doi: 10.1086/510920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toydemir RM, Brassington AE, Bayrak-Toydemir P, Krakowiak PA, Jorde LB, Whitby FG, Longo N, Viskochil DH, Carey JC, Bamshad MJ. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am J Hum Genet. 2006;79:935–941. doi: 10.1086/508433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, McMahon AP. Expression pattern of the FGF-related proto-oncogene int-2 suggests multiple roles in fetal development. Development. 1989;105:131–136. doi: 10.1242/dev.105.1.131. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228:267–272. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003a;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. FGF signaling in ear development and innervation. Curr Top Dev Biol. 2003b;57:225–259. doi: 10.1016/s0070-2153(03)57008-9. [DOI] [PubMed] [Google Scholar]
- Wu DK, Nunes FD, Choo D. Axial specification for sensory organs versus non-sensory structures of the chicken inner ear. Development. 1998;125:11–20. doi: 10.1242/dev.125.1.11. [DOI] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–391. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dong C. Regulatory mechanisms of mitogen-activated kinase signaling. Cell Mol Life Sci. 2007:7012–3. doi: 10.1007/s00018-007. [DOI] [PMC free article] [PubMed] [Google Scholar]