Abstract
The modular protein Alix is a central node in endosomal-lysosomal trafficking and the budding of HIV-1. The Gag p6 protein of HIV-1 contains a LYPXnLXXL motif that is required for Alix-mediated budding and binds to a region of Alix spanning residues 360-702. The structure of this fragment of Alix has the shape of the letter “V” and is termed the “V domain”. The V domain has a topologically complex arrangement of eleven α-helices, with connecting loops that cross three times between the two arms of the “V”. The conserved residue Phe676 is at the center of a large hydrophobic pocket and is critical for binding to a peptide model of HIV-1 p6. Overexpression of the V domain inhibits HIV-1 release from cells. This inhibition of release is reversed by mutations that block binding of the Alix V domain to p6.
The budding and release of nascent virus particles from the infected host cell is an essential phase of the retroviral replication cycle. The budding process requires the trafficking of the Gag polyprotein precursor from its site of synthesis in the cytosol to the appropriate cellular membrane at which virus assembly occurs. Concomitant with particle release from the host cell the viral protease (PR) cleaves the Gag precursor to generate the mature Gag proteins matrix (MA), capsid (CA), and nucleocapsid (NC). In the case of HIV-1 and equine infectious anemia virus (EIAV), additional domains known as p6 and p9, respectively, are located at the C-terminus of the Gag precursor 1. p6 and p9, and functionally analogous Gag proteins of other retroviruses, encode the so-called “late” domains that promote the release of virions from the infected cell 2-4. Late domains function by co-opting the host cell endosomal sorting machinery via direct interactions with defined components of the cellular protein trafficking machinery 2-4. Three retroviral late domains have been characterized to date, containing the short sequence motifs P(T/S)AP, PPPY, and LYPXnLXXL, respectively. P(T/S)AP, located within p6, promotes release by interacting with Tsg101, a component of the cellular ESCRT-I protein complex 5-10. The PPPY motif, found in the Gag proteins of a number of retroviruses including murine leukemia virus (MLV) 11, functions by interacting with Nedd4-like ubiquitin ligases 12-14. The human protein Alix was first discovered and named AIP1 or Alix for its association with the calcium binding protein ALG-2 (apoptosis-linked gene 2) 15,16; here we use the term “Alix” to differentiate it from other unrelated proteins named “AIP1”. EIAV and HIV-1 contain LYPXnLXXL motifs that promote release via a direct interaction with Alix 9,17-19.
Alix is at a nexus between endosomal-lysosomal trafficking, cell death pathways, and the cytoskeleton 20-22. Alix can function in multiple pathways because of its modular structure. Its first 359 amino acid residues comprise a Bro1 domain, so named for the Bro1 protein, which is the yeast ortholog of Alix. Bro1 domains are conserved boomerang-shaped structures that target to endosomal membranes via binding to the protein CHMP4, known in yeast as Snf7 23. Residues 360-716 of Alix comprise a second folded domain that is responsible for binding to viral LYPXnLXXL motifs 17,24, however the function of this region of Alix in normal physiology is still unclear. The C-terminal residues 717-868 comprise a proline-rich domain (PRD), which is predicted to be unstructured. This last region interacts with a wide array of proteins, many of which contain SH3 and other domains that bind to Pro-rich peptide sequences.
Much of the interest in Alix has focused on its role in endosomal membrane trafficking. The yeast homolog of Alix, Bro1, was discovered as the product of the BRO1/VPS31 gene, whose disruption led to a class E vacuolar protein sorting (VPS) defect. Class E VPS defects have a characteristic enlarged cargo-rich compartment adjacent to the vacuole 25, which is the yeast counterpart of the human lysosome. In addition to BRO1, yeast class E VPS genes encode the subunits of at least four hetero-oligomeric protein complexes: the Vps27/Hse1 complex and ESCRT-I, II, and III 25,26. The major normal function of the ESCRT network is to sort transmembrane proteins into multivesicular bodies en route to the lysosome 27-29. The ESCRT network is hijacked by certain viruses, such as HIV, that use this system to bud from cells by a process topologically equivalent to multivesicular body formation. Alix is a central node in the ESCRT network. The PRD of Alix directly interacts with the Tsg101 (human Vps23) subunit of ESCRT-I 18,19. The ESCRT-III subunit CHMP4 binds to the Alix Bro1 domain. Alix associates with endocytic proteins outside of the ESCRT network; for example, its PRD binds the endocytic proteins SETA 30 and endophilin 31. Alix also binds to lysobisphosphatidic acid (LBPA), which may contribute to its targeting to inward-budding vesicles in the late endosome 32.
The central domain (residues 360-716) is the least-studied region of Alix. Its best defined function is to bind viral LYPXnLXXL motifs 17,24. The characterization of this domain is of great interest given that its overexpression strongly inhibits HIV-1 budding in cell culture 24. To better understand Alix and the mechanism of its interaction with viral LYPXnLXXL motifs, we have determined the crystal structure of a C-terminally truncated variant of this domain from human Alix, Alix360-702. The structure forms the shape of the letter “V”. We go on to pinpoint the late domain binding site on one arm of the “V” and to define its role in viral budding.
RESULTS
Structure of the Alix V domain
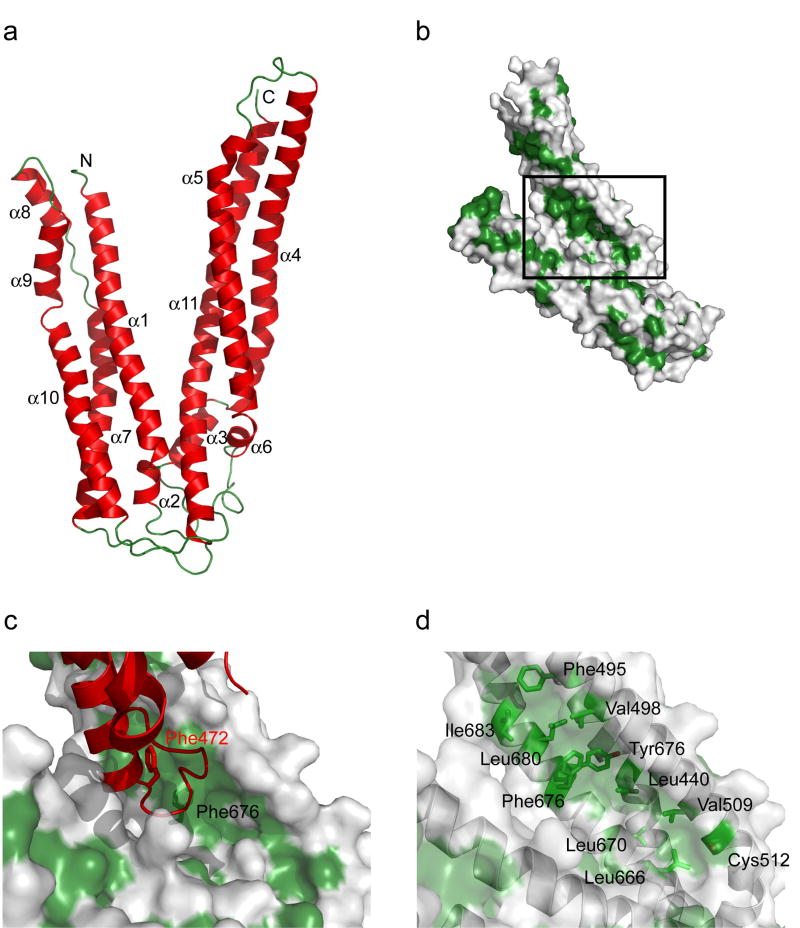
The crystal structure of the Alix V domain was determined by multiwavelength anomalous dispersion (MAD) from selenomethionyl protein and refined at 2.9Å resolution (Fig. 1a, Supplementary Fig. 1). The structure is almost completely α-helical and contains two arms of unequal length arranged at ~40° to each other in the shape of a letter “V”. The shorter of the two arms is 75 Å long, contains the N-terminus, and comprises six α-helices (α1, α2, α7, α8, α9, α10) and 150 residues. The long arm is 100 Å in extent, contains the C-terminus, five α-helices (α3, α4, α5, α6, α11), and 194 residues. The topological arrangement of helices in the two arms is complex. The polypeptide chain crosses between the arms three times, between α2 and α3; α6 and α7; and α10 and α11. The overall architecture of each arm is centered around three long helices. The long arm is built around a three-stranded coiled coil comprising α4, α5, and α11. Helices α7 and α10 of the short arm have some supercoiling, reflecting coiled-coil character, but the other long helix with which they are partnered, α1, is straight. The region of contact between the two arms is modest. Much of the interaction is formed between the C-terminal three turns of α1 from the short arm, and three turns of long arm helix α11 near its N-terminus. Each arm buries an average of 760 Å2 of solvent-accessible surface area in the overall structure.
Figure 1. Structure and LYPXnLXXL motif binding site of the Alix V domain.
(a) Overall fold of the Alix V domain. (b) Location of the hydrophobic pocket with respect to the overall structure. (b) Lattice-related molecule shown in red highlights the potential of this pocket for hydrophobic interactions. (c) Stick model of hydrophobic residues (green) shown under a translucent surface in the vicinity of the main hydrophobic pocket.
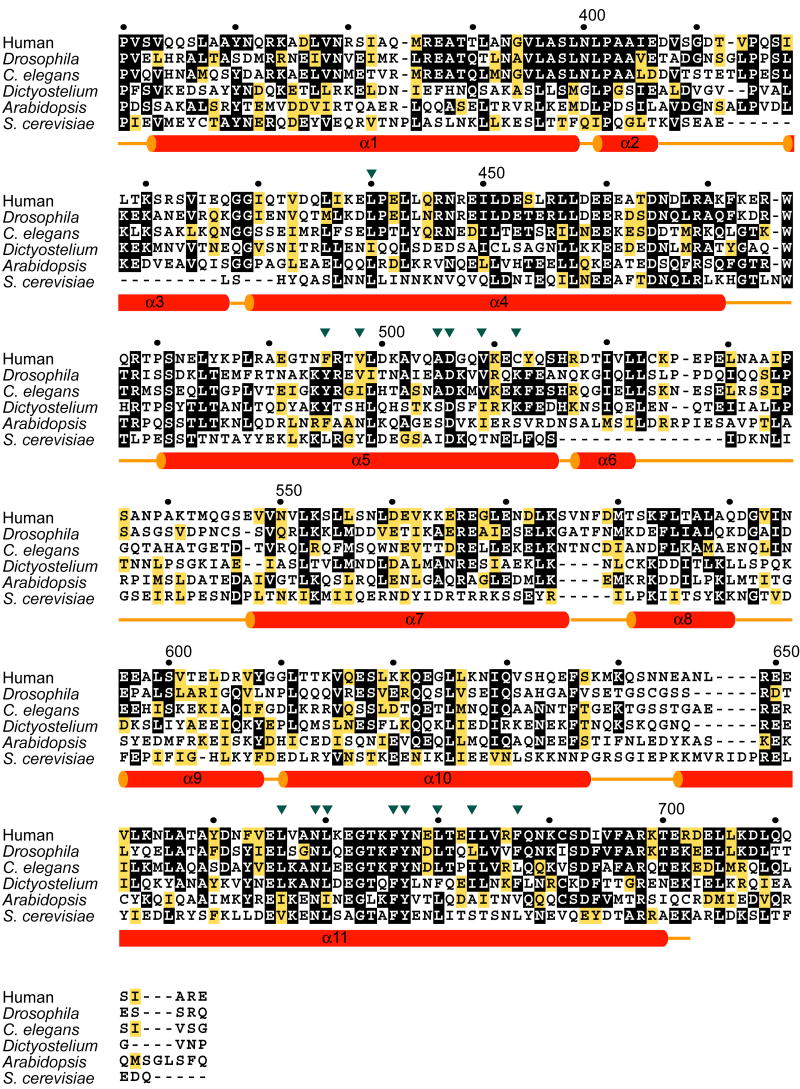
The most dramatic surface feature of the structure is a deep and extensive hydrophobic pocket on the long arm (Fig. 1b). The hydrophobic surface in these pockets stabilizes one of the major symmetry contacts in the crystal lattice. The C-terminus of helix α4 and the α4-α5 loop of a lattice-related molecule protrude deep into the gap between the two arms of the V to contact this site. Major lattice contacts are sometimes formed by “frustrated” interaction sites when crystals are grown in the absence of the normal interaction partner. As such, these contacts can sometimes be useful guides to the identification of functional interaction sites. The exposed side-chains of Phe472 and Trp476 of the lattice-related molecule make extensive contacts with the pocket (Fig. 1c). Both the guanidine and aliphatic portions of the side-chain of Arg475 of the lattice-related molecule interact with the pocket as well. The pocket faces into the gap between the arms of the “V”. The side-chain of Leu440 of α4 is at the very bottom of the pocket. The surrounding walls are formed by Phe495, Val498, Ala505, Asp506, Val509, and Cys512 of α5; and Leu666, Asn669, Leu670, Phe676, Tyr677, Leu680, Ile683, and Phe687 of α11 (Fig. 1d). The side-chains of Asp506 and Tyr677 form a hydrogen bond near the bottom to the pocket, one of very few polar features in the pocket. Val498, Phe676, and Tyr677 form a ridge that divides the pocket. Many of these residues are conserved in Alix orthologs (Fig. 2).
Figure 2. Secondary structures and sequence alignment of Alix and Alix orthologs.
Strictly and highly conserved residues are shown by black and yellow boxes, respectively. Putative LYPXnLXXL motif binding site residues are indicated with inverted triangles.
The LYPXnLXXL motif binding site
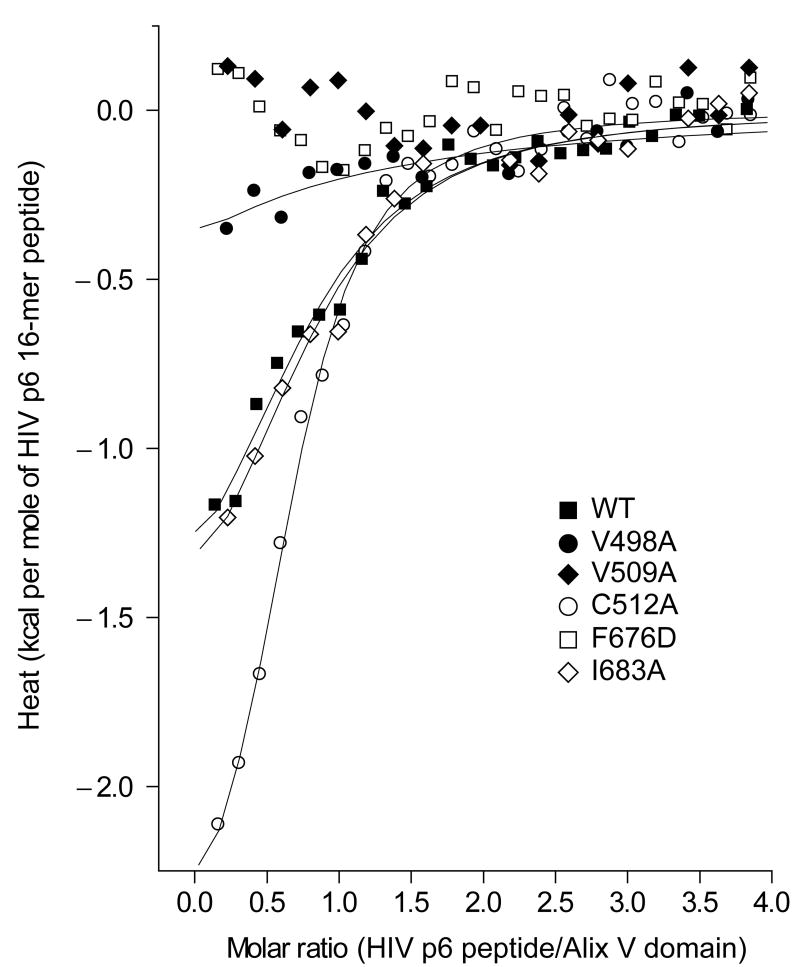
To test whether the Phe-676 pocket is important for LYPXnLXXL motif binding and function, Phe-676 and several adjoining residues within the pocket were mutagenized and tested for their ability to bind to a p6-derived peptide in vitro. We have established that the 16-residue synthetic peptide p6-16 (DKELYPLTSLRSLFGN), which encompasses the LYPXnLXXL motif, binds to the Alix V domain with Kd = 5 - 6 μM. This is nearly identical to the 8 μM affinity measured for full-length HIV-1 p6 24, validating it as an in vitro model system for quantitating the interaction. The most prominent exposed hydrophobic residue, Phe676, was mutated to Asp to determine whether these residues are involved in essential hydrophobic interactions with HIV-1 p6. The Asp substitution was selected because the insertion of a charged residue into a hydrophobic interface is highly destabilizing. The mutant F676D expressed at normal levels, but shows no binding to the p6-16 peptide as measured by isothermal titration calorimetry (ITC) at concentrations of up to 100 μM (Fig. 3, Table 1).
Figure 3. In vitro binding of a HIV-1 p6 LYPXnLXXL-based 16 mer peptide to Alix.
ITC analysis of the binding of the Alix V-domain mutants to the HIV-1 p6 16-mer peptide (32DKELYPLTSLRSLFGN) in solution. Representative one-site model fitted curves of the wild-type and the mutants along with the buffer-subtracted data points are shown.
Table 1.
Binding affinities of Alix mutants to HIV p6 16-mer peptidea
| Alix mutant | Kd (μM)b |
|---|---|
| WT | 5.8 ± 2.4 |
| V498A | 30 ± 14 |
| V509A | N.D.c |
| C512A | 3.0 ± 0.3 |
| F676D | N.D.c |
| I683A | 4.6 ± 1.0 |
Sequence of the HIV p6 16-mer peptide is 32DKELYPLTSLRSLFGN.
Apparent dissociation constant (Kd) calculated from fitting the data to an one-site model. For each mutant, Kd value from three independent experiments is shown with standard deviation.
Not determinable due to no binding or very weak binding preventing reliable quantification.
Having confirmed a role for the Phe676 pocket in LYPXnLXXL motif binding, we explored the surrounding surface with less drastic perturbations. A spectrum of effects was observed. The V509A mutant exhibited no detectable binding by ITC. Val-509 is 12 Å from Phe676, on the opposite side of the groove between helices α5 and α11, and proximal to the crotch of the V. Val-498 is 5 Å from Phe-676, directly across the α5 and α11 groove, and V498A reduces binding by 5 to 6-fold. Ile683 is 7Å from Phe676 and more distal to the crotch of the V; its mutation I683A has no effect on binding. Cys512 is located at the end of the hydrophobic pocket closest to the crotch of the V, and is highly solvent exposed. C512A has marginally higher affinity than wild-type. All of these Ala mutant proteins had essentially normal stability on the basis of their equal levels of expression as recombinant proteins. They collectively span most of the hydrophobic pocket and provide strong confirmation of a direct role in LYPXnLXXL motif binding.
Function of the Phe676 site in HIV-1 budding
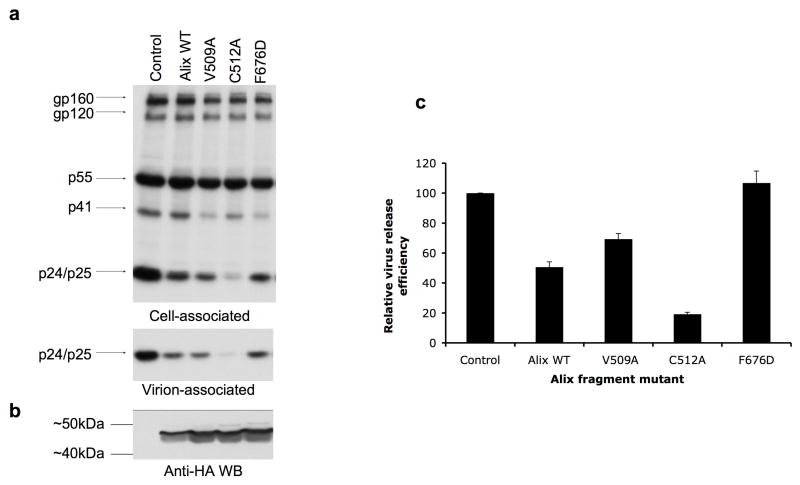
We previously demonstrated that overexpression of the V domain of Alix resulted in an inhibition of HIV-1 release. This inhibition was abrogated by mutations in p6 (e.g., L41A and L41R) that blocked the p6/Alix interaction24. To validate the importance of Alix Phe-676 in p6 binding in vivo, we measured the ability of the F676D-mutant Alix V domain to disrupt HIV-1 particle release. HeLa cells were cotransfected with the full-length infectious pNL4-3 molecular clone and control plasmid or were cotransfected with pNL4-3 and vectors expressing wild-type or mutant Alix V domains. The efficiency of virus release was determined by radioimmunoprecipitation analyses (Fig. 4a) and levels of Alix V domain expression were determined by immunoblotting (Fig. 4b). As previously observed24, overexpression of the wild-type V domain significantly (p<2 × 10-5) inhibited particle release. In contrast, the F676D mutation completely abrogated the ability of the V domain to disrupt budding (Fig 4a, c). The loss of inhibition was not due to lower expression of the F676D-mutant Alix V domain relative to wild-type (Fig. 4b). The V509A mutant, which displayed reduced binding activity, inhibited particle release to a lesser extent than wild-type. Intriguingly, the C512A mutant, which has slightly enhanced binding in vitro, exhibited a significant (p<1.5 × 10-4) increase relative to the wild-type V domain in its ability to inhibit HIV-1 budding (Fig. 4a and 4c).
Figure 4. A functional hydrophobic pocket is required for inhibition of HIV-1 release.
(a) Hela cells were cotransfected with pNL4-3 and control plasmid (control) or were cotransfected with pNL4-3 plus vector expressing wild-type (Alix364-716) or the indicated mutant Alix V domain expression vectors. Transfected cells were metabolically labeled with [35S]Cys/Met. Cell- and virion-associated proteins were immunoprecipitated with HIV-Ig (see Experimental Procedures). Positions of the envelope glycoprotein species gp160 and gp120, the Pr55Gag precursor protein (p55), the Gag processing intermediates p41 and p25, and the mature CA protein p24 are indicated. (b) Detection of the Alix V domains by anti-HA Western blotting. The positions of molecular mass markers are indicated. Relative levels of fragment expression for the different mutants was highly consistent between experiments. (c) Virus release efficiency, calculated as the amount of virion-associated Gag as a fraction of total (cell plus virion) Gag synthesized during the 2-h labeling period. The effect of the Alix V domain fragments on virus release was normalized to the amount of Alix V domain expressed. Data are shown +/- SD, n = 3.
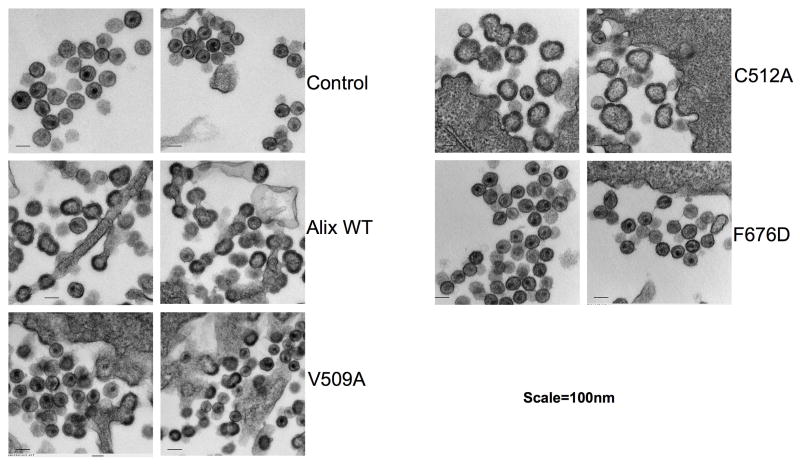
To corroborate the biochemical virus release data, we performed thin section transmission electron microscopy (EM) with cells expressing wild-type or mutant Alix V domains. Consistent with the radioimmunoprecipitation data, the wild-type V domain and the C512A mutant potently inhibited budding, as visualized by the accumulation of tethered, immature particles at the plasma membrane and the presence of numerous chains of tethered virions and multi-budded particles (Fig. 5). We also observed that Alix V domain expression induced the release of non-spherical particles with irregular electron density in the Gag shell; this phenotype was particularly striking for the C512A mutant. Cultures expressing the V509A mutant V domain showed a mix of normal and tethered particles. Importantly, the F676D mutation, which blocked Alix/p6 peptide binding in vitro and abrogated the ability of the Alix V domain to disrupt virus particle production, had no effect on virion release or morphogenesis (Fig. 5). These results confirm the importance of Alix Phe-676 in p6 binding in vivo.
Figure 5. Virus budding defects visualized by EM.
HeLa cells were transiently cotransfected with pNL4-3 plus control plasmid (control) or pNL4-3 plus vectors expressing wild-type (Alix364-716) or the indicated mutant Alix V domain protein. Transfected cells were processed for EM as described in the Experimental Procedures. Two representative micrographs for each sample are shown. Bar = 100nm.
DISCUSSION
Here we have determined the overall fold of the central domain of Alix, from the end of the Bro1 domain to the start of the presumably unstructured proline-rich domain. This region of Alix has a surprisingly intricate topology. Taken together with the structure of the Bro1 domain from the yeast Alix ortholog Bro1 23, we now have a structural picture of essentially all of the ordered portion of Alix. The major goal of this project was to determine how viral LYPXnLXXL motifs bind to Alix. While we have been unable thus far to obtain a structure of an Alix/ LYPXnLXXL motif peptide complex, we have been able to infer the location of the LYPXnLXXL motif binding site on Alix. The Phe676 pocket is the largest contiguous region of exposed hydrophobic surface area on the structure. Another exposed hydrophobic region on a lattice-related molecule interacts with the Phe676 site, highlighting its potential for interactions. Phe676 and surrounding residues are highly conserved. We were able to confirm the functional requirement for this site by mutating Phe676 and surrounding residues. F676D abrogates binding in vitro to the p6-16 peptide model of the LYPXnLXXL motif. While overexpression of the wild-type Alix V domain inhibits viral budding and release, and leads to the production of virions with a defective morphology, the F676D mutation completely reverses these phenotypes. Taken together with observations that mutations of other residues surrounding Phe-676 modulate function both positively and negatively, this establishes with high confidence the location of the LYPXnLXXL motif binding site on Alix. Other mutations tested corroborate these findings. It is especially satisfying that one of the mutations, C512A, which has increased affinity, also increased the potency of budding inhibition. It is unclear whether the modest enhancement of binding alone is responsible for increasing the effectiveness of the V domain as an HIV-1 release inhibitor. Cysteine is subject to a wide range of post-translational modifications, including palmitoylation, S-glutathionylation, S-nitrosylation, and oxidation to sulfenic acid. Palmitoylation seems unlikely in this context, but the possibility of other Cys modifications of Alix may warrant investigation. Cys512 is solvent exposed and could be a candidate for a post-translational modification that could interfere with binding.
It is interesting to note that the effect of the Alix fragment on virion morphogenesis differs from that typically observed with late domain mutations. Not only is a marked increase in the number of particles tethered to the plasma membrane observed24 (Fig. 5) but also many of the virions released in the presence of the Alix V domain fragment are non-spherical and display an irregular pattern of electron density in their Gag shell. This phenotype is particularly striking with the Alix C512A mutant (Fig. 5), which has higher affinity for Gag than wild-type. We speculate that binding of the Alix fragment to p6 inhibits proper particle assembly in addition to disrupting virion budding. This hypothesis is consistent with our recent observation24 that the impact of the Alix V domain fragment on Gag processing and particle release is more severe than, and distinct from, the defect induced by mutation of the Alix binding site in p6.
The secondary structure of HIV-1 p6 has been analyzed in solution by NMR 33. HIV-1 p6 is largely unstructured, but contains two helical regions, one (α1) from residues 14-18, and the other (α2) from residues 33-43. These studies were carried out in part in an aqueous trifluoroethanol solution, which enhances α-helix formation, and has been postulated to mimic the hydrophobic helix stabilization provided by membranes or hydrophobic protein surfaces. These NMR results are consistent with secondary structure predictions. Helix α2 of p6 corresponds closely to the central ten residues, 35-44, of the LYPXnLXXL motif. In one attractive model for the p6/Alix interaction, p6 α2 might be aligned roughly coaxial with the Alix V domain long arm. The hydrophobic pocket is ~30 Å long, which is more than sufficient to accommodate a three-turn, ten residue helix. In this model, helix α2 of p6 would form a small four helix bundle together with Alix α4, α5, and α11. An example of such a recognition mechanism is provided by the helical LD motifs of paxillin, which bind co-axially to the four-helix bundle FAT domain 34,35.
Portions of the LYPXnLXXL motif binding site are conserved in organisms as diverse as yeast (Fig. 2), while other portions of the site are not well conserved. Phe676 and adjacent residues on α11 are among the most highly conserved in the Alix family, suggesting a conserved physiological function. On the other hand, residues on α5 are less well conserved. Val-498, which we find has a role in LYPXnLXXL motif binding, is replaced by hydrophilic residues in Dictyostelium and Arabidopsis, for example. The partial conservation of the site suggests that it possesses physiological ligands. Motifs of this form are abundant in proteomes. For example, a search of the yeast proteome where Leu or Ile were allowed at the “L” position, and Phe or Tyr at the “F” position, identified 150 occurrences in 143 different yeast proteins. Most of these proteins had no obvious connection to pathways in which Alix is known to participate. The identification of host V domain ligands remains a significant challenge for the field. Other challenges for the Alix field include understanding the physiological significance of its interactions with lipids and with endosomal, cytoskeletal, and apoptotic proteins. Together with previous work on the Bro1 domain of the yeast homolog of Alix 23, these findings for the V domain lay the structural foundation for advancing understanding of Alix and its homologs.
METHODS
Cloning and protein preparation
Human Alix V domain constructs consisting of residues 360-702 and 364-702 were cloned into parallel GST2 vector 36 using a standard PCR-based cloning strategy from cDNA encoding Alix. Site-directed single and double mutants were generated using the QuikChange Mutagenesis Kit (Stratagene). Construction of the mammalian cell expression vector encoding the HA-tagged Alix fragment has been described previously 24. All the constructs were verified by DNA sequencing. Alix V domains were overexpressed as an N-terminal glutathione S-transferase (GST)-fusion protein in Escherichia coli strain BL21(DE3). Cells were grown at 37 °C, induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside at optical density of 1.0 and further incubated at 20 °C overnight. Harvested cells were resuspended in buffer A (50 mM TrisHCl, pH 7.4, 150 mM NaCl, 7 mM β-mercaptoethanol), lysed and centrifuged. The supernatant was applied to a glutathione-Sepharose column (Amersham) in buffer A and washed with buffer B (50 mM TrisHCl, pH 7.4, 500 mM NaCl, 7 mM β-mercaptoethanol). His6-tagged Tobacco etch virus protease was added to cleave the Alix V domains from the GST fusion partner, and then removed by passing through TALON resin (BD Bioscience) in the buffer A. The cleaved protein was concentrated and applied to a Superdex 200 column (Amersham) in the buffer C (50 mM TrisHCl, pH 7.4, 150 mM NaCl, 5 mM dithiothreitol). Fractions containing Alix V domains were pooled and concentrated using VivaSpin concentrator (Viva Science). Selenomethione derivative was expressed in B834(DE3)pLysS (Calbiochem) and purified in the same way as the native protein. All the mutants were purified as described above.
Crystallization, data collection and structure determination
Crystals of Alix(360-702) were grown by hanging drop method at 22 °C. The reservoir solution contained 0.1 M TrisHCl pH 8.5, 13-15 % (w/v) polyethylene glycol 6000, and 6-10% (w/v) ethylene glycol. Crystals appeared overnight and were flash frozen under liquid nitrogen. Data were collected at 100° K at beamline X25 in the National Synchrotron Light Source and processed with HKL2000 (HKL Research). A three-wavelength MAD dataset from a selenomethionyl crystal was used for phasing, with the peak, inflection, and remote wavelength data sets collected at 0.9795, 0.9799, and 0.9600 Å, respectively. Three selenium sites were located out of five expected in the sequence using automated Patterson searching 37. Subsequence solvent flattening by RESOLVE 38 produced an interpretable map. Iterative manual model building and initial refinement were performed using O 39 and CNS 40, respectively. Final refinement was done using REFMAC with TLS parameters incorporated 41. Statistics for data processing and refinement are listed in Table 2. All the structural figures were generated using PyMol 42.
Table 2.
Data collection, phasing and refinement statistics
| Seleniuma | ||||||
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P21 | |||||
| Cell dimensions | ||||||
| a, b, c (Å) | 37.6, 55.4, 113.0 | |||||
| α, β, γ (°) | 90, 92.4, 90 | |||||
| Peak | Inflection | Remote | ||||
|
|
||||||
| Resolution (Å) | 2.9 (3.0 – 2.9)b | 2.9 (3.0 – 2.9) | 2.9 (3.0 – 2.9) | |||
| Rsym or Rmerge | 0.067 (0.317) | 0.062 (0.338) | 0.074 (0.394) | |||
| I / σI | 17.1 (3.1) | 23.2 (2.9) | 16.4 (2.5) | |||
| Completeness (%) | 97.1 (86.6) | 96.7 (81.6) | 95.5 (76.4) | |||
| Redundancy | 4.0 (3.6) | 4.0 (3.6) | 4.0 (3.5) | |||
| Refinement | ||||||
| Resolution (Å) | 2.9 | |||||
| No. reflections | 10,483 | |||||
| Rwork / Rfreec | 0.237/0.319 | |||||
| No. atoms | ||||||
| Protein | 2705 | |||||
| Ligand/ion | 0 | |||||
| Water | 0 | |||||
| B-factors | ||||||
| Protein | 49.3 | |||||
| Ligand/ion | None | |||||
| Water | None | |||||
| R.m.s deviations | ||||||
| Bond lengths (Å) | 0.009 | |||||
| Bond angles (°) | 1.16 | |||||
One crystal was used to determine the structure.
Values in parentheses are for highest-resolution shell.
Rfree is calculated for a randomly chosen 10% of relections.
Isothermal titration calorimetry
The HIV-1 p6-16 peptide was described previously 24. The peptide and the Alix(364-702) protein samples were in 10 mM Hepes pH 7.4, 150 mM NaCl, 2 mM DTT. The concentrations were 400 μM for the peptide and 15 -20 μM for the proteins. ITC measurements were performed on a MicroCal VP-ITC at 25 °C. The peptide was injected to the Alix(364-702) samples in 29 aliquots of 10 μl each at 240 sec intervals. Data obtained from the peptide injections to the buffer blanks were subtracted from the experimental data for analysis using MicroCal Origin software.
Cell culture, metabolic labeling, virus release assays, and Western blotting
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum, L-glutamine (2mM), penicillin and streptomycin. Transfections were performed with lipofectamine 2000 (Invitrogen) as recommended by the supplier. Methods used for metabolic radiolabeling, preparation of cell and virus lysates, and immunoprecipitation have been previously described in detail 43,44. Briefly, transfected HeLa cells were starved in Cys/Met-free medium for 30 min and then metabolically radiolabeled for 2 h with [35S]Cys/Met Pro-mix (Amersham). Virions were pelleted by ultracentrifugation. Cell and virus lysates were immunoprecipitated with pooled immunoglobulin from HIV-1-infected patients (HIV-Ig) obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. The radioimmunoprecipitated proteins were separated by SDS-PAGE, followed by fluorography and quantification by phosphorimager analysis. Virus release efficiency was calculated as the amount of released virion-associated Gag as a fraction of total (cell plus virion) Gag synthesized during the 2-h labeling period, normalized for the level of Alix fragment expression. Alix V domain expression was monitored by Western blotting with an anti-HA antibody (HA-7, Sigma) and quantified using an Alpha Innotec FluorTec SP chemiluminescence imager. Statistical significance of the virus release data was determined with the Student’s t-test.
Electron microscopy
Cells were cotransfected with pNL4-3 plus control plasmid or were cotransfected with pNL4-3 and Alix V domain expression vectors. Cells were washed in phosphate-buffered saline and fixed in glutaraldehyde for EM analysis as described previously 45. Cells were examined by a Hitachi H7000 electron microscope.
Accession codes
Coordinates have been deposited in the protein data bank with accession code xxxx.
Supplementary Material
Acknowledgments
We thank J. Kim, G. Miller, and U. Munshi for many contributions to the early stages of this project, F. Soheilian for EM support, D. Eric Anderson for mass spectrometry support, B. Beach and T. Leonard for advice and discussions, and J. Bonifacino for the use of his calorimeter. C. Hill and W. Sundquist independently determined this structure, and suggested the term “V domain”, which we have adopted. Use of the NSLS, BNL was supported by the U. S. DOE, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-98CH10886. The HIV-Ig was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. This research was supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (JHH) and National Cancer Institute, Center for Cancer Research (EOF). Funds were also obtained from the Intramural AIDS Targeted Antiviral Program (JHH and EOF) and from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400 (KN).
Footnotes
COMPETING INTERESTS STATEMENT The authors declare they have no competing financial interests.
References
- 1.Freed EO. HIV-1 Gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 3.Demirov DG, Freed EO. Retrovirus budding. Virus Research. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 5.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 7.Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J Virol. 2003;77:6507–6519. doi: 10.1128/JVI.77.11.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VerPlank L, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan B, Li XQ, Goff SP. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. Embo J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kikonyogo A, et al. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci U S A. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vito P, Pellegrini L, Guiet C, D’Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- 16.Missotten M, Nichols A, Rieger K, Sadoul R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death and Differentiation. 1999;6:124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- 17.Chen CP, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) Endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280:40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- 18.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 19.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 20.Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- 21.Sadoul R. Do Alix and ALg-2 really control endosomes for better or for worse? Biol Cell. 2006;98:69–77. doi: 10.1042/BC20050007. [DOI] [PubMed] [Google Scholar]
- 22.Pan S, et al. Involvement of the adaptor protein alix in actin cytoskeleton assembly. J Biol Chem. 2006 doi: 10.1074/jbc.M602263200. in press. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, et al. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munshi U, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2006 doi: 10.1074/jbc M607489200. [DOI] [PubMed] [Google Scholar]
- 25.Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochimica Et Biophysica Acta-Molecular Cell Research. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomolec Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babst M. A Protein’s Final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 28.van der Goot FG, Gruenberg J. Intra-endosomal membrane traffic. Trends Cell Biol. 2006;16:514–521. doi: 10.1016/j.tcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends In Cell Biology. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, Borinstein SC, Gillis J, Sykes VW, Bogler O. The glioma-associated protein SETA interacts with AIP1/Alix and AZIG-2 and modulates apoptosis in astrocytes. J Biol Chem. 2000;275:19275–19281. doi: 10.1074/jbc.M908994199. [DOI] [PubMed] [Google Scholar]
- 31.Chatellard-Causse C, et al. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo H, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 33.Fossen T, et al. Solution structure of the human immunodeficiency virus type 1 p6 protein. J Biol Chem. 2005;280:42515–42527. doi: 10.1074/jbc.M507375200. [DOI] [PubMed] [Google Scholar]
- 34.Hurley JH. Leucine in the sky with diamonds. Structure. 2003;11:1192–1193. doi: 10.1016/j.str.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Hoellerer MK, et al. Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure. 2003;11:1207–1217. doi: 10.1016/j.str.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr Sect D-Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr Sect D-Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron- Density Maps and the Location of Errors in These Models. Acta Crystallogr Sect. 1991;A 47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D-Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr Sect D-Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- 43.Freed EO, Martin MA. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci U S A. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.