Abstract
The crystal structure of an enzyme–substrate complex with histidyl-tRNA synthetase from Escherichia coli, ATP, and the amino acid analog histidinol is described and compared with the previously obtained enzyme–product complex with histidyl-adenylate. An active site arginine, Arg-259, unique to all histidyl-tRNA synthetases, plays the role of the catalytic magnesium ion seen in seryl-tRNA synthetase. When Arg-259 is substituted with histidine, the apparent second order rate constant (kcat/Km) for the pyrophosphate exchange reaction and the aminoacylation reaction decreases 1,000-fold and 500-fold, respectively. Crystals soaked with MnCl2 reveal the existence of two metal binding sites between β- and γ-phosphates; these sites appear to stabilize the conformation of the pyrophosphate. The use of both conserved metal ions and arginine in phosphoryl transfer provides evidence of significant early functional divergence of class II aminoacyl-tRNA synthetases.
Keywords: histidyl-adenylate, x-ray crystallography, class II aminoacyl-tRNA synthetase
Before their ligation to the 3′ ends of tRNAs, amino acids must be activated by condensation with ATP. This aminoacyl adenylylation reaction is catalyzed by the aminoacyl-tRNA synthetases (aaRS), which are divided into two classes of 10 members each that are based on two different catalytic folds (1, 2). Each class possesses diagnostic sequence motifs whose conserved residues are necessary for ATP binding (3, 4). Based on the crystallographic analyses of complexes of aaRS with ATP or their respective adenylates, ATP recognition is largely mediated through residues that are invariant among class members, whereas amino acid recognition occurs by using residues that are unique to the evolutionary family (5–14). For most systems, it has not yet been possible to obtain complexes containing both amino acid and unreacted ATP (or suitable analog), so significant mechanistic issues remain to be addressed, including the precise roles of specific catalytic residues, bound metal ions, and mobile loop elements. To provide further insight into these issues, we have characterized the structures of two different enzyme–substrate complexes of histidyl-tRNA synthetase (HisRS) from Escherichia coli, one with the histidyl-adenylate and the other with ATP and histidinol. In addition, data were collected from crystals into which MnCl2 had been diffused to provide information about divalent metal binding sites. Analysis of these complexes provides evidence for both general features of the catalytic mechanism of class II enzymes and features that are specific to the histidine system.
MATERIALS AND METHODS
Crystallization.
Crystals were grown as described (15), with the exception that histidinol (HisOH) was substituted for histidine in the case of the HisRS:HisOH:ATP complex. Both complexes crystallized in the triclinic space group P1 with unit cell dimensions a = 61.3 Å, b = 110.1 Å, c = 108.7 Å, α = 115.0°, β = 97.4°, and γ = 90.0° for the HisRS:HisAMP complex and a = 60.9 Å, b = 107.3 Å, c = 107.2 Å, α = 114.1°, β = 97.4°, and γ = 90.0° for the HisRS:HisOH:ATP complex. Crystals were stabilized in a solution containing 0.1 M Tris (pH 7.4), 15% polyethylene glycol (PEG) 8000, 0.2 M NaCl, 50 mM histidine or HisOH (depending on the complex), and 5 mM ATP as described (14).
Data Collection.
Diffraction intensities were measured on a MAR Research (Hamburg, Germany) phosphor image plate system using rotating anode or synchrotron x-rays [beamline W32, Laboratoire pour l’Utilisation du Rayonnement Electromagnétique, Orsay, France (LURE); ref. 16]. Data for the HisRS:HisOH:ATP complex were collected at LURE from one single crystal at −143°C. Before flash-cooling to this temperature, the crystal had been transferred gradually (in five steps) to a cryoprotecting solution containing 0.1 M Tris (pH 7.4), 5% PEG 8000, 30% PEG 400, 50 mM HisOH, and 5 mM ATP. Data for the HisRS:HisOH:ATP:Mn2+ complex were collected at −140°C by use of rotating anode-generated x-rays from one single crystal that had been soaked in solutions as indicated above but contained 10 mM MnCl2. Additional data were also collected at LURE for the HisRS:HisAMP complex from several crystals at 0°C. They were merged with the native data set used in the original structure determination (14), which improved its completeness. All data were processed with the marxds/marscale software (17). The statistics are summarized in Table 1.
Table 1.
Summary of crystallographic data collection
| HisRS complexed with
|
|||
|---|---|---|---|
| HisAMP | HisOH:ATP | HisOH:ATP:Mn2+ | |
| No. of crystals | 1, 7 | 1 | 1 |
| Temperature, °C | 0, 0 | −143 | −140 |
| X-ray source | Lab, LURE | LURE | Lab |
| Wavelength, Å | 1.542, 0.94 | 0.94 | 1.542 |
| Resolution, Å | 12.0–2.6 | 16.0–2.6 | 15.0–3.0 |
| No. of observations | 183,629 | 116,502 | 93,539 |
| Unique reflections | 66,471 | 63,562 | 42,518 |
| Redundancy | 2.7 | 1.8 | 2.2 |
| Rmerge, % | 9.6 | 3.6 | 7.5 |
| Completeness | 85.4 | 84.4 | 88.0 |
Rmerge = ΣhklΣi | 〈Ihkl〉 − Ihkl,i |/ΣhklΣi | Ihkl |, where i are the observations of reflection hkl.
Refinement.
The HisRS:HisAMP structure (14) was further refined with the program x-plor (18) by using merged data from eight crystals in the resolution range 12.0–2.6 Å and a bulk solvent mask (probe radius = 0.2 Å). The resulting model has good stereochemistry as determined with procheck (19). The structure of the HisRS:HisOH:ATP complex was determined by molecular replacement and rigid-body refinement with x-plor by using reflections in the 10.0–3.5 Å resolution range and the coordinates of HisRS without the adenylate or solvent molecules. It was then refined, and the resolution was extended to 2.8 Å, followed by the addition of the ligands. A bulk solvent mask (probe radius = 0.2 Å) was introduced, and reflections in the resolution range 12.0–2.8 Å were used for further refinement. To improve the agreement with the observed data, the torsion dynamics method (20) was used, and then the structure was adjusted by using the program o (21) and positionally refined with x-plor. The resulting model has good stereochemistry as determined with procheck. All refinement was done with protein parameters derived by Engh and Huber (22). The refinement statistics are summarized in Table 2.
Table 2.
Summary of crystallographic refinement
| HisRS:HisAMP | HisRS:HisOH:ATP | |
|---|---|---|
| Resolution | 12.0–2.6 | 12.0–2.8 |
| Reflections (F > 3σ) | 52,794 | 38,015 |
| Rcryst (%) | 22.1 | 24.9 |
| Rfree (%) | 29.6 | 34.8 |
| rms dev. bonds, Å | 0.012 | 0.015 |
| rms dev. angles, ° | 1.699 | 1.915 |
| rms dev. dihedrals | 24.492 | 24.799 |
| rms dev. impropers | 1.376 | 1.557 |
| Mean B-factor, Å2 | 47.0 | 44.0 |
| No. of protein atoms | 11,630 | 11,547 |
| No. of cofactor atoms | 132 | 164 |
| No. of solvent atoms | 197 | 39 |
dev., deviation. Rcryst = Σhkl | Fhklobs − Fhklcalc |/Σhkl | Fhklobs |, testhkl = 0. Rfree = Σhkl | Fhklobs − Fhklcalc |/Σhkl | Fhklobs |, testhkl = 1 (2.5% of reflections were used for this calculation; ref. 18, no. 1836).
Mutagenesis and Kinetics.
The R259H mutant HisRS protein was constructed by oligonucleotide-directed mutagenesis by using the method of Kunkel et al. (23). The DNA fragment for the resulting construct was then inserted into pQE-30 (Qiagen, Chatsworth, CA), which encodes a His6 affinity tag that allows expression and purification of the mutant protein, as described previously. The tag has previously been shown to have no significant effect on the kinetic parameters of the enzyme (24). The rate of adenylate formation was measured by using the pyrosphosphate exchange assay (25). The assay was performed at 37°C with use of an enzyme concentration of 500 nM and the following substrate concentration ranges: ATP, 10–2,000 μM and histidine, 10–500 μM. Saturating concentrations of histidine and ATP were 2 mM and 5 mM, respectively. MgCl2 was present at a 2.5-fold excess over the ATP concentration. The aminoacylation assays were conducted as described (24) with 25 nM enzyme and 0.2–5.0 μM tRNA. The initial rates were measured over time intervals that were determined empirically. The apparent kinetic parameters were determined from at least two data sets by using the program enzfitter (Elsevier), which calculates curve fits to the Michaelis–Menten equation.
RESULTS AND DISCUSSION
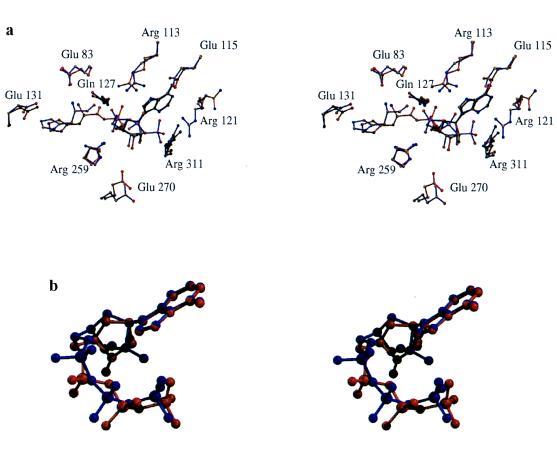
Although informative with respect to interactions made to the histidyl-adenylate, the HisRS complex described previously (14) provided no information about the positions of the β- and γ-phosphates of ATP. To obtain information about earlier steps in the reaction, HisRS was crystallized in the presence of ATP and the competitive inhibitor histidinol. This analog lacks the carboxyl group characteristic of the amino acid and serves as a competitive inhibitor of the PPi exchange reaction with a Ki of 3.5 × 10−5 M (26). When the histidinol:ATP and histidyl-adenylate complexes are compared (Fig. 1a), no large-scale rigid body motions or conformational changes of the motif 2 loop or the HisA loop (L257VRGLDYY264) are observed. Many enzyme-ligand contacts are unchanged, including contacts between ATP and class II invariant residues, such as Phe-125, semiconserved Gln-127, and various main chain contacts. The conformation of ATP in the histidinol complex agrees with that observed previously in the yeast aspartyl-tRNA synthetase (AspRS; ref. 8; Fig. 1b) and Thermus thermophilus seryl-tRNA synthetase (SerRS; ref. 11) complexes. Unlike the adenosyl moiety, interactions with the phosphate groups undergo significant change during the reaction. These are treated in detail below.
Figure 1.
Stereoviews of superpositions of the histidinol:ATP (blue bonds) and histidyl-adenylate (red bonds) complexes (a), and the ATPs from the HisRS:histidinol:ATP complex (red) and yeast AspRS:tRNA:ATP complex (blue) (b). The figure was prepared by using molscript (27) and raster3d (28, 29).
Although there is a shift of nearly 1 Å in the position of histidinol and histidine (Fig. 1a), interactions with the imidazole ring are virtually unchanged in the two complexes. Specificity for the imidazole group is provided by contacts with conserved Glu-131 in motif 2 and Tyr-264 in the HisA loop. The orientations of these residues are maintained by additional hydrogen bonds to Thr-85, Tyr-288, and Arg-259. Multiple interactions with the imidazole group may account for the ability of the enzyme to discriminate between histidine and related analogs (26). In most class II aaRS, there is a direct interaction between the α-amino group and a highly conserved carboxylate in motif 2 (8–10, 12), which is also conserved in eukaryotic HisRS. However, this residue is a glycine (Gly-129) in most prokaryotic HisRS, and the interaction is replaced by a salt bridge to Glu-83, conserved in prokaryotic HisRS (14). In addition, the α-amino group makes a hydrogen bond to an ordered water molecule coordinated to Gln-127 and Tyr-107, along with a hydrogen bond to Gln-127.
Phosphate Interactions.
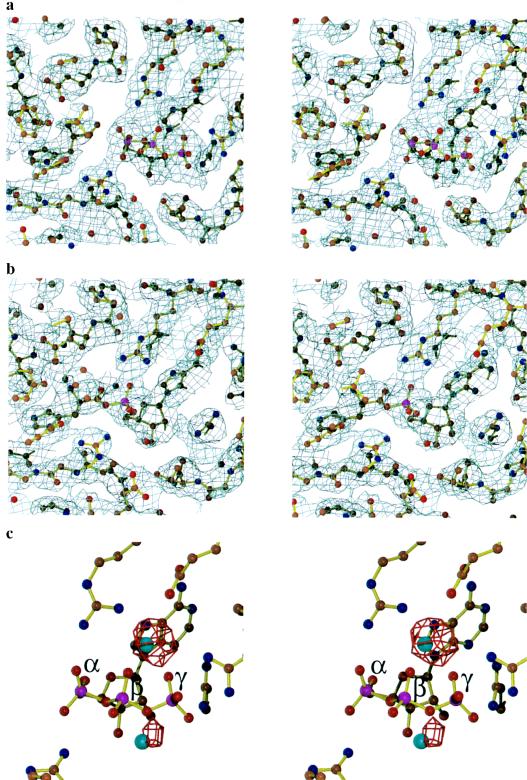
The most significant difference between the two complexes concerns interactions made with the triphosphate group of ATP. In the histidinol:ATP complex (Fig. 2a), density for all three phosphates is observed, whereas only the α-phosphate is seen in the adenylate complex (Fig. 2b). Contacts to the triphosphate group are made by four arginines located in motif 2, motif 3, and the HisA loop (Fig. 1a and 3). Arg-113, a motif 2 invariant residue, makes a bridging interaction with the α- and β-phosphate in the histidinol complex but shifts slightly to make a bifurcated interaction with the pro-R oxygen of the adenylate α-phosphate. The salt bridges Arg-121 and Arg-311 make to the γ-phosphate in the initial complex are absent in the adenylate, and Arg-121 adopts different conformations in the different monomers. Inversion of the α-phosphate is evident in a comparison of the two structures (Figs. 1a and 3), as predicted from the in-line mechanism (31).
Figure 2.
Stereoviews of electron density maps of ligands in the active site of HisRS. Final 2Fo − Fc map contoured at 1σ of the active site of the HisRS:HisOH:ATP complex (a) and HisRS:HisAMP complex (b), superposed on the final refined structures. (c) HisRS:HisOH:ATP:Mn2+ complex. An Fo − Fc map contoured at 4.5 σ (red); the map was computed with structure factors and phases calculated from the model in a. The figure was prepared by using minimage (30), molscript (27), and raster3d (28, 29).
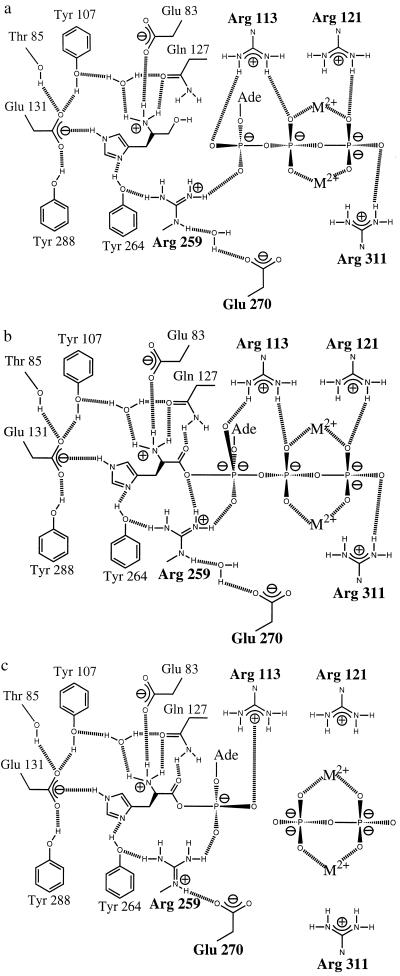
Figure 3.
Schematic diagram showing steps in the activation of histidine by HisRS: (a) binding of histidinol and ATP, (b) proposed transition state with pentavalent α-phosphate, and (c) formation of histidyl-adenylate (14). a and c are based on crystallographic data, whereas b is a proposed model for the transition state.
Arg-259 maintains a close interaction with the α-phosphate in both complexes. This active site arginine is conserved in all HisRS, but has not been observed in the active sites of other class II enzymes. Unlike Arg-113, whose aliphatic side chain is relatively unconstrained in both structures, the side chain of Arg-259 is maintained both by hydrophobic interactions with conserved Leu-257 and Val-268 and by a hydrogen bond between one ηN of the guanidinium side chain and the phenolic oxygen of Tyr-264. The other ηN is within 3 Å of the pro-R oxygen of the α-phosphate. Arg-259 makes a third interaction that is different in the two complexes and that may be of particular functional significance. In the histidinol complex (Fig. 3a), there is a water-mediated interaction between the ɛN and one of the ɛ-carboxylate oxygens of the class II conserved Glu-270. In the adenylate complex (Fig. 3c), Glu-270 moves to make a direct contact with the guanidinium group of the arginine. This movement excludes the water molecule and creates a salt bridge that may act to weaken the ionic interaction between Arg-259 and the α-phosphate. The resulting decrease in the strength of electrostatic interaction is likely to stabilize the adenylate in the active site.
Arg-259 Required for Efficient Catalysis.
To provide additional evidence for the importance of Arg-259, an arginine-to-histidine substitution was introduced by oligonucleotide-directed mutagenesis. The mutant protein was expressed and purified, and kinetic parameters for the pyrophosphate exchange and aminoacylation were determined (Table 3). These measurements showed that replacement of the arginine side chain reduced the rates of exchange and transfer by factors of 1,000 and 500, respectively. By contrast, the minimal effect of the substitution on the Michaelis constants suggests that the local folding has not been perturbed substantially. The slight decrease in Km (2- to 3-fold) observed for ATP is consistent with a differential requirement for Arg-259 in the transition state relative to ground state binding, but this will have to be confirmed through the measurement of elementary rate constants that make up the Michaelis constant. These results confirm the importance of Arg-259 for the exchange reaction, but due to the requirement for adenylylation to precede aminoacylation, further studies will be needed to explicitly define the role of Arg-259 in the aminoacylation reaction.
Table 3.
Apparent steady state kinetic parameters for pyrophosphate exchange and aminoacylation catalyzed by R259H HisRS at 37°C
|
Km, μM
|
kcat, sec−1
|
||||
|---|---|---|---|---|---|
| His | ATP | tRNA | PPi exchange | aminoacylation | |
| wt HisRS | 30 (±5) | 890 (±64) | 1.4 (±0.6) | 130 (±5) | 2.6 (±0.4) |
| R259H | 25 (±1) | 307 (±12) | 3.3 (±0.4) | 0.103 (±0.010) | 0.006 (±0.001) |
Values shown represent the mean of at least two independent determinations. Standard errors, shown in parentheses, were obtained in the curve fits to the Michaelis–Menten equation calculated by the program enzfitter. wt, wild type.
The Position of Coordinated Metal Ions.
The comparison of the two complexes and the results of the mutagenesis experiment provided strong evidence for the catalytic role of Arg-259 but did not rule out a potential role for one or more bound Mg2+ ions. In the electron density map of the HisRS:HisOH:ATP complex, weak density consistent with a bound Mg2+ ion could be identified between the β- and γ-phosphates. To provide more definitive evidence for this location of magnesium, diffraction data were collected from a crystal of the HisRS:HisOH:ATP complex that had been soaked in 10 mM MnCl2 (Table 1). The omit maps calculated from these data showed two Mn2+ ions coordinated to the β- and γ-phosphates, one at 6 σ above background and a second at 4 σ above background (Fig. 2c). The remaining coordination sites of both metal ions are filled with water molecules, as all potential carboxylated ligands are more than 3.0 Å away from the ions.
Measurement of the interatomic distances between the principal Mn2+ ion and the phosphate oxygens revealed that the distance to the β-phosphate oxygen is approximately 0.5 Å shorter than the distance to the γ-phosphate oxygen. Positioning the metal closer to the β-phosphate would be expected to contribute to catalysis by weakening the bond between the α- and β-phosphates, as opposed to merely neutralizing the negative charge (−4) on the leaving group. However, at least part of this asymmetry in the location of the principal Mn2+ ion may be the result of chemical differences between Mn2+ and Mg2+, and a more symmetrical coordination in the Mg2+ complex cannot be ruled out.
This relatively secondary role of Mg2+ in the adenylylation reaction catalyzed by HisRS relative to the catalytic Arg-113 and Arg-259 is in marked contrast to other class IIa aaRS, most notably SerRS. In the SerRS system, the locations of the divalent cations have been determined previously by manganese soaking experiments (11). This work showed that the principal divalent cation in the active site is coordinated to the α- and β-phosphates, in a position occupied by Arg-259 in HisRS. This principal metal ion has also been proposed to fulfill the role of electrophilic catalyst by virtue of transition state stabilization. The generality of this metal-catalyzed variation of the adenylylation reaction is suggested by the observation of a similarly located Mn2+ ion in an adenylate complex of glycyl-tRNA synthetase from T. thermophilus (J. G. Arnez, A. C. Dock-Bregeon, and D.M., unpublished work), and by the presence of two class II invariant carboxylates that are involved in the coordination of the catalytic Mg2+. Significantly, the residues in HisRS that correspond to these two highly conserved carboxylates (Glu-270 and Thr-281) participate in the arginine salt bridge switch and have poor geometry for metal coordination.
A General Mechanism for the Adenylylation Reaction Catalyzed by Class II aaRS.
These observations and reports describing other complexes between class II aaRS and their substrates (8–11) provide evidence for a common class II mechanism for the adenylylation reaction involving several main features. First, the class II active site is configured to bind both the amino acid and the ATP in fixed geometries that reduce rotational and translational entropy (Fig. 3a). Second, neutralization of the negative charge of the pyrophosphate is mediated by metal ions coordinated to the β- and γ-phosphate oxygens and by the invariant arginine from motif 3. Additional neutralization of the pyrophosphate leaving group is provided by an additional basic residue in the motif 2 loop, typically arginine or histidine. Structural analyses of the AspRS and SerRS systems suggest that this basic residue has an important role in the transfer reaction by virtue of direct contact with cytosine 74 of the tRNA substrate (8, 11). Lastly, a major feature of class II aaRS contributing to rate enhancement of the adenylylation reaction is the electrophilic catalysis provided by two electron-deficient moieties in the active site that are directly coordinated to the α-phosphate. The first of these is the class II invariant arginine of motif 2, which contacts the α-phosphate directly in all ATP and adenylate complexes described so far (8–11). Mutagenesis of this residue in AspRS showed that the motif 2 invariant arginine is absolutely required for activity (8). The second contributor to electrophilic catalysis varies between different class II aaRS, such that a divalent cation strongly coordinated to the α-phosphate provides one mode, and a basic residue such as arginine provides a second. Either coordinated metal ions or Arg-259 would be expected to increase the electropositivity of the α-phosphate while also providing additional transition state stabilization by helping to bring O5′ and the nonbridging oxygens into the pentacoordinate geometry required by the associative mechanism (ref. 32 and Fig. 3b).
In the phosphoryl transfer reactions catalyzed by polymerases and RNAs, coordinated metal ions are essential catalytic moieties. For RNAs such as the Tetrahymena group I intron (33), RNase P (34), and the hammerhead ribozyme (35–37), metal replacement experiments have demonstrated the requirement of coordinated Mn2+ ions for catalytic activity, which is in addition to the more general requirements for divalent and monovalent ions in RNA folding and substrate binding. These requirements may reflect fundamental similarities with the 3′–5′ exonuclease domain of DNA polymerase I, where two Mn2+ ions coordinated to invariant carboxylates activate the nucleophile and stabilize the transition state (38–40). By contrast, the involvement of catalytic arginines in other enzymes that carry out phosphoryl transfer, such as alkaline phosphatase (41), staphylococcal nuclease (42), and fructose 1,6-bisphosphatase (43) has been well established. The presence of two distinct catalytic strategies in the class II aaRS raises the question of whether or not this role by arginine is unique to HisRS or is instead found in other aaRS whose structures are currently unknown. Given the ability of RNA to self-aminoacylate in a metal-dependent reaction and bind arginine specifically (44), this mechanistic diversity could reflect fundamental properties of catalytic RNAs that were conserved in the transition from RNA-based to protein-based enzymes.
Acknowledgments
We thank A. Mitschler for his invaluable assistance with data collection, J.-C. Thierry for advice on data processing, R. Fourme and staff at LURE for beamline support. This work was supported by National Institutes of Health Grant GM 48146 and a North Atlantic Treaty Organization Collaborative Research Award (C.S.F.), by the Centre National de la Recherche Scientifique and Université Louis Pasteur (D.M.), and by a joint National Science Foundation/Centre National de la Recherche Scientifique exchange program (C.S.F. and D.M.). J.G.A. was supported by a fellowship from the Fondation pour la Recherche Médicale.
ABBREVIATIONS
- aaRS
aminoacyl-tRNA synthetase(s)
- HisRS
histidyl-tRNA synthetase(s)
- AspRS
aspartyl-tRNA synthetase(s)
- SerRS
seryl-tRNA synthetase(s)
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11974 (accession codes 1KMM and 1KMN).
References
- 1.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature (London) 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 2.Moras D. Trends Biochem Sci. 1992;17:159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- 3.Carter C W., Jr Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 4.Cusack S. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 5.Brick P, Bhat T N, Blow D M. J Mol Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- 6.Rould M A, Perona J J, Söll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 7.Brunie S, Zelwer C, Risler J L. J Mol Biol. 1990;216:411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- 8.Cavarelli J, Eriani G, Rees B, Ruff M, Boeglin M, Mitschler A, Martin F, Gangloff J, Thierry J C, Moras D. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poterszman A, Delarue M, Thierry J-C, Moras D. J Mol Biol. 1994;244:158–167. doi: 10.1006/jmbi.1994.1716. [DOI] [PubMed] [Google Scholar]
- 10.Belrhali H, Yaremchuk A, Tukalo M, Larsen K, Berthet C C, Leberman R, Beijer B, Sproat B, Als N J, Grübel G, Legrand J F, Lehmann M, Cusack S. Science. 1994;263:1432–1436. doi: 10.1126/science.8128224. [DOI] [PubMed] [Google Scholar]
- 11.Belrhali H, Yaremchuk A, Tukalo M, Berthet-Colominas C, Rasmussen B, Bösecke P, Diat O, Cusack S. Structure (London) 1995;3:341–352. doi: 10.1016/s0969-2126(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 12.Onesti S, Miller A D, Brick P. Structure (London) 1995;3:163–176. doi: 10.1016/s0969-2126(01)00147-2. [DOI] [PubMed] [Google Scholar]
- 13.Doublié S, Bricogne G, Gilmore C, Carter C W. Structure (London) 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 14.Arnez J G, Harris D C, Mitschler A, Rees B, Francklyn C S, Moras D. EMBO J. 1995;14:4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francklyn C, Harris D, Moras D. J Mol Biol. 1994;241:275–277. doi: 10.1006/jmbi.1994.1498. [DOI] [PubMed] [Google Scholar]
- 16.Fourme R, Dhez P, Benoit J P, Kahn R, Dubisson J M, Besson P, Frouin J. Rev Sci Instrum. 1992;63:982–987. [Google Scholar]
- 17.Kabsch W. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 18.Brünger A T. x-plor: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 19.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Rice L M, Brünger A T. Proteins Struct Funct Genet. 1994;19:277–290. doi: 10.1002/prot.340190403. [DOI] [PubMed] [Google Scholar]
- 21.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Yan W, Augustine J, Francklyn C. Biochemistry. 1996;35:6559–6568. doi: 10.1021/bi952889f. [DOI] [PubMed] [Google Scholar]
- 25.Calender R, Berg P. Biochemistry. 1966;5:1690–1695. doi: 10.1021/bi00869a034. [DOI] [PubMed] [Google Scholar]
- 26.Lepore G C, Di Natale P, Guarini L, Di Lorenzo F. Eur J Biochem. 1975;56:369–374. doi: 10.1111/j.1432-1033.1975.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 27.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 28.Bacon D J, Anderson W F. J Mol Graphics. 1988;6:219–220. [Google Scholar]
- 29.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 30.Arnez J G. J Appl Crystallogr. 1994;27:649–653. [Google Scholar]
- 31.Langdon S P, Lowe G. Nature (London) 1979;281:320–329. doi: 10.1038/281320a0. [DOI] [PubMed] [Google Scholar]
- 32.Knowles J R. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 33.Piccirilli J A, Vyle J S, Caruthers M H, Cech T R. Nature (London) 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 34.Smith D, Pace N R. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 35.Dahm S, Uhlenbeck O C. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 36.Koizumi M, Ohtsuka E. Biochemistry. 1991;30:5145–5150. doi: 10.1021/bi00235a005. [DOI] [PubMed] [Google Scholar]
- 37.Slim G, Gait M J. Nucleic Acids Res. 1991;19:1183–1188. doi: 10.1093/nar/19.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freemont P S, Friedman J M, Beese L S, Sanderson M R, Steitz T A. Proc Natl Acad Sci USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beese L S, Steitz T A. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steitz T A, Steitz J A. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim E E, Wyckoff H W. J Mol Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 42.Cotton F A, Hazen E E, Legg M J. Proc Natl Acad Sci USA. 1979;76:2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y-H, Ogata C, Pflugrath J W, Levitt D G, Sarma R, Banaszak L J, Pilkis S J. Biochemistry. 1996;35:6010–6019. doi: 10.1021/bi9600613. [DOI] [PubMed] [Google Scholar]
- 44.Illangasekare M, Sanchez G, Nickles T, Yarus M. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]