Abstract
Carcinoma cells can lose their epithelial cell characteristics and dedifferentiate into a fibroblast-like cell during progression of a neoplasm. Aberrant expression of oligomeric transcriptional complexes contributes to progression of carcinomas. Although individual transcription factors initiating progression remain unknown, LIM-only protein (LMO) and LIM-domain binding protein (LDB) negatively regulate breast carcinoma cell differentiation. In this study, we investigated the expression of LMO4 and LDB in squamous cell carcinomas of the oral cavity. LMO4 mRNA was amplified in four of six carcinoma tissues and eight of 12 carcinoma cell lines, and LDB1 in three carcinoma tissues and 11 cell lines examined. Immunoprecipitation studies revealed that LMO4 and LDB1 interact with each other in the nuclear milieu of the carcinoma cells indicating the presence of an LMO4-LDB1-mediated transcription complex. Both LMO4 and LDB1 proteins were preferentially localised in the nuclei of carcinoma cells at the invasive front and the immunoreactivity was increased in less-differentiated carcinoma tissues (P<0.01). Carcinoma cells metastasised to the cervical lymph nodes with increased immunoreactivity compared to the primary site of neoplasm (P<0.05). These data suggest that the LMO4–LDB1 complexes may be involved in carcinoma progression possibly through dedifferentiation of squamous carcinoma cells of the oral cavity.
Keywords: LMO4, LDB1, LIM, carcinoma
Oral squamous cell carcinoma is the most common neoplasm of the head and neck. Worldwide, the annual incidence of new cases exceeds 300 000. The disease causes great morbidity, and the 5-year survival rate has not improved in more than two decades (Vokes et al, 1993; Lippman and Hong, 2001). With few exceptions, carcinomas are derived from single somatic cells and their progeny. Carcinoma cells in the emerging neoplastic clone accumulate within them a series of genetic and/or epigenetic changes that lead to changes in gene activity, and altered phenotypes which are subjected to selection of tumour progression (Ponder, 2001). The generation of cellular diversity in carcinomas frequently involves aberrant expression of transcriptional regulators acting in a combinational manner. Loss of epithelial morphology and acquisition of mesenchymal characteristics, often referred to as the epithelial–mesenchymal transition, are typical for carcinoma cells in dedifferentiation and correlate with tumour progression (Hay, 1995; Birchmeire et al, 1996; Thiery, 2002). However, the genetic basis of dedifferentiation and progression of carcinoma cells has not been determined.
The LIM-only (LMO) protein carries two tandemly repeat LIM zinc-binding domains and consists of four members (designated as LMO1–4). The LIM domain has been identified in a variety of nuclear proteins (Sanchez-Garcia and Rabbitts, 1994). It functions primarily as a module for the assembly of protein complexes through protein–protein interactions. The LIM domain does not directly interact with DNA, but acts as an adaptor molecule for transcription factors facilitating assembly of large transcriptional complexes (Breen et al, 1998; Jurata et al, 1998; Sugihara et al, 1998). LMO1 and LMO2 have been shown to specify neuronal and haematopoietic cell lineages in combination with their transcription partners (Hinks et al, 1997; Yamada et al, 1998; Herblot et al, 2000). Misexpression of these genes by chromosomal translocation abrogates proper differentiation of cells and is oncogenic within T cells (Boehm et al, 1991; Royer-Pokora et al, 1991). Little is known about LMO3, which was discovered on the basis of sequence homology. The most recently identified member, LMO4, shares only 50% homology with the LIM domains of other LMO proteins. The LMO4 gene is widely distributed in embryonic tissues (Kenny et al, 1998; Sugihara et al, 1998), and involved in negative regulation of breast carcinoma cell differentiation (Visvader et al, 2001). LMO4 binds with a high affinity to the LIM domain-binding proteins, LDB1 (CLIM2, NLI1) and LDB2 (CLIM1) (Agulnick et al, 1996; Jurata et al, 1996; Sugihara et al, 1998). LDB proteins bind to transcription factors directly or indirectly mediated through LMO proteins, and then bridge a unique bipartite DNA sequence separated by about one helix turn from each other (Agulnick et al, 1996; Jurata et al, 1996; Wadman et al, 1997; Rabbitts, 1998). Both LMOs and LDBs are widely expressed during development (Kenny et al, 1998; Millan et al, 1998; Toyama et al, 1998; Hermanson et al, 1999) and appear to have essential functions in cell proliferation and lineage determination, and oncogenesis (Jurata et al, 1998; Milán et al, 1998; Rabbitts, 1998; Thaler et al, 2002).
Prompted by the observation that LMO4 and LDB1 are expressed in embryonic epithelial tissues (Bach et al, 1997; Sugihara et al, 1998), we examined the expression pattern of LMO4 and LDBs in squamous carcinoma cells. In contrast to the negligible expression of LDB2, LMO4 and LDB1 were frequently detected in the less-differentiated carcinomas and carcinoma cells at the invasive front, and upregulated in metastasised lymph nodes, suggesting an involvement of the LMO4–LDB1 transcriptional complex in the pathology of carcinoma progression.
MATERIALS AND METHODS
Cell lines and tissue samples
Immortalised cell lines established from oral squamous carcinomas (Ca9.22, Ho1N1, HOC313, HSC2, HSC3, HSC4, KOSC2, KOSC3, OSC19, SCCKN, SCCTF, and TSU) were obtained from the Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer (Tohoku University, Sendai, Japan), Health Science Research Resources Bank (Osaka, Japan) or RIKEN Cell Bank (Tsukuba, Japan), and maintained in 10% fetal bovine serum (FBS)-containing DMEM or RPMI1640 medium (Invitrogen, Grand Island, NY, USA) in a 5% CO2 incubator. Normal gingival fibroblasts (GF12) were maintained in 10% FBS containing DMEM for 19 passages (Takahashi et al, 1997). HaCaT cells (Boukamp et al, 1988), immortalised normal keratinocytes, were grown in DMEM supplemented with 10% FBS.
A total of six oral carcinoma tissues (three well-differentiated carcinomas and three moderately differentiated carcinomas) and three normal oral tissues without a history of head and neck carcinoma were obtained from patients undergoing surgery for carcinoma resection or dental surgery at the Nippon Dental University Hospital, Meikai University Hospital, or Machida City Hospital under the informed consent of the patients.
Reverse transcription–PCR
Total RNA was isolated from cell lines at 70–85% confluency, oral squamous cell carcinomas, and normal oral tissues using TRIzol reagent (Invitrogen) followed by RNase-free DNase I treatment to eliminate DNA contamination in the sample. After reverse transcription (RT) to a single-stranded cDNA using SuperScript II (Invitrogen) and random hexamer (Invitrogen) at 42°C for 60 min, PCR reaction was performed with gene-specific primer sets for LMO4 (forward: exon 4; 5′-CGGGAGATCGGTTTCACTAC, reverse: exon 5; 5′-CCAGTGCCCTGCTAATTGTT), LDB1 (forward: exon 5; 5′-TGCCATGTTGACCATCACTT, reverse: exon 9; 5′-GGC-TGAGGCTGTAGGTCTTG), LDB2 (forward: exon 1; 5′-TTTCGA-AAAGCAGGCAAGAT, reverse: exon 6; 5′-TCGGGGACTGAGGTTGTAAG), or GAPDH (forward: 5′-GTCAGTGGTGGACCAGACCT, reverse: 5′-AGGGGAGATTCAGTGTGGTG) and Taq DNA polymerase (Invitrogen). PCR amplification was performed by running 30 cycles under the following conditions: denatured at 94°C for 40 s, annealed for 40 s at 60°C, and extended at 72°C for 1 min. PCR amplicons were analysed on 2% agarose gels.
Tissue specimen selection
Incisional or excisional biopsy specimens from 49 patients with oral squamous cell carcinomas were collected from the files of The Kanazawa University Hospital from 1991 to 2000 under informed consent of the patients. Clinical and pathological data were obtained from the patients' medical records and The Kanazawa University Hospital Surgical Pathology files. Clinical and pathologic variables included age, gender, tumour size, tumour location, grade of tumour differentiation, and presence or absence of cervical lymph node metastasis. Control normal tissues (n=5) were also obtained at surgery or autopsy from tongue and gingiva of patients without a history of head and neck cancer. These samples were immediately fixed in 10% neutral buffered formalin or periodate–lysine–paraformaldehyde (PLP) solution and embedded in paraffin wax.
Immunohistochemistry
The haematoxylin and eosin-stained slides from each of the cases were screened by light microscopy. Unstained serial sections (4 μm) were deparaffinised and rehydrated followed by the microwave treatment in 0.01 M sodium citrate buffer, pH 6.0 (500 W). After incubation with normal serum, sections were incubated with rabbit anti-LMO4 (5 μg ml−1, Chemicon, Temecula, CA, USA), goat anti-LDB1 (2 μg ml−1, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or goat anti-LDB2 (2 μg ml−1, Santa Cruz Biotechnology) antibodies for 12 h at 4°C. Biotinylated anti-rabbit or -goat IgG (DAKO, Glostrup, Denmark) was used for secondary antibody followed by incubation with avidin–biotin complexes (Vector Laboratories, Burlingame, CA, USA). The colour was developed with 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich, St Louis, MI, USA) under the microscope and counterstained with methyl green. To clarify the specificity of the antibody reactivity, primary antibodies were replaced with either nonimmune rabbit or goat IgG (DAKO) at matched protein concentrations. Carcinoma cells with a strong nuclear labelling were determined as positive reactions, but cells with a weak nuclear staining and/or diffuse cytoplasmic staining were not counted as positive. The percentage of positive nuclear staining of LMO4 or LDB1 was evaluated by counting at least 3000 carcinoma cells in randomly selected areas of each specimen at × 40. They were blinded as to the clinicopathological parameters.
Immunocytochemistry
Human oral squamous carcinoma cell lines (TSU, HOC313, HSC3, and OSC19) were cultured on glass slides (Lab-Tek Chamber II, NUNC, Naperville, IL, USA) in 10% FBS-containing medium and fixed in 1% paraformaldehyde for 3 min at 23°C. The cells were reacted with primary antibodies to LMO4 (10 μg ml−1), LDB1 (4 μg ml−1), or LDB2 (4 μg ml−1) for 16 h at 4°C. Alexa Fluor 546 anti-rabbit or -goat IgG (Molecular Probes, Eugene, OR, USA) was used for secondary antibody. To clarify the specificity of antibody reactivity, incubation with nonimmune rabbit IgG or goat IgG at matched protein concentrations instead of primary antibodies was performed.
Immunoprecipitation
For detection of endogenous protein binding, crude nuclear extracts (30 μg) of HSC3, TSU, OSC19, or HOC313 cells were prepared by the method described elsewhere (Digman et al, 1983; Lee et al, 1998), immunoprecipitated with goat anti-LDB1 antibody (2 μg) and protein G-Sepharose (Pharmacia, Wikströms, Sweden), and size fractionated by SDS–PAGE (10% total acrylamide) under reducing condition. After electrotransfer to nitrocellulose membranes, membranes were blocked by 3% bovine serum albumin and incubated with rabbit anti-LMO4 or goat anti-LDB1 antibodies, followed by incubation with biotinylated secondary antibodies. Avidin–biotin complex and 3,3′-diaminobenzidin tetrahydrochloride were used for the colour development.
Statistical analysis
One-way analysis of variance followed by contrast statements (Scheffe's F-test and Fisher's PLSD) was performed to compare LMO4 or LDB1 immunoreactivity with carcinoma differentiation and other clinical and pathological parameters recorded in the text. Statistical analysis of correlation between the immunoreactivity of LMO4 and LDB1 was conducted by simple linear regression and between primary site and metastasised lymph nodes by Wilcoxon's signed-rank test.
RESULTS
Expression of LMO4, LDB1, and LDB2 in oral carcinomas
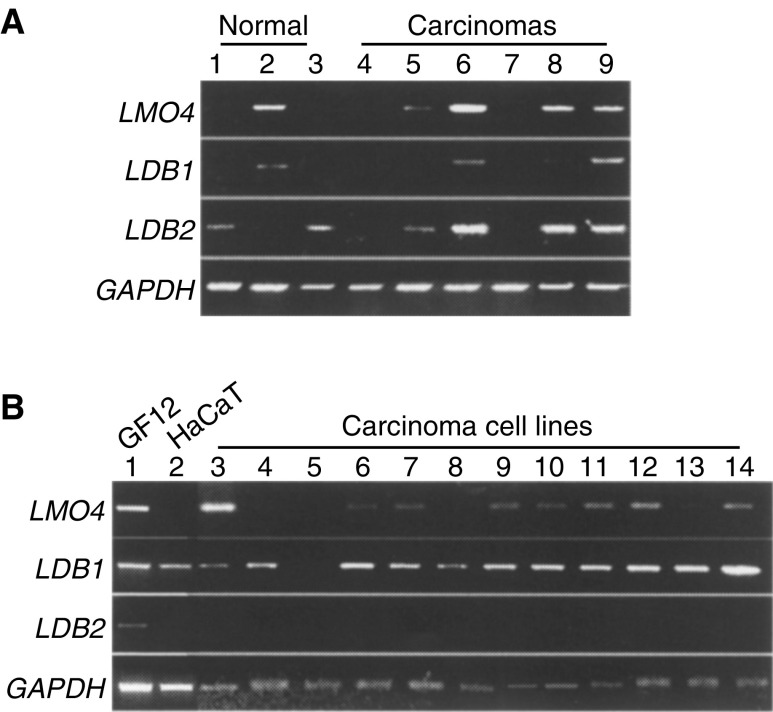
Expression of LMO4, LDB1, and LDB2 in squamous cell carcinomas has not been investigated. We therefore examined the expression pattern of these genes in carcinoma tissues of the oral cavity by RT–PCR (Figure 1A). Based on the human genome database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi), primer sets for LMO4, LDB1, or LDB2 were designed as spanning one or more introns to prevent amplification of genomic DNA. LMO4 and LDB1 exhibited an almost identical pattern of expression. Specific primer sets for each gene amplified a single product; four of six and three of six carcinomas for LMO4 (485 bp) and LDB1 (494 bp), respectively. Normal gingiva obtained from patients without a history of head and neck cancer also expressed the genes in one of the three samples. Four carcinomas and two normal samples augmented a single 702 bp fragment of the LDB2 transcript.
Figure 1.
Expression of LMO4 and LDBs mRNA in oral squamous cell carcinomas. (A) LMO4, LDB1, and LDB2 transcripts are amplified in normal gingiva (lanes 1–3) and carcinoma tissues (lanes 4–9) by RT-PCR. A single 485, 494, or 702 bp fragment was observed by a specific primer set for LMO4, LDB1, or LDB2, respectively. GAPDH (395 bp) was included as an internal control. (B) Oral squamous carcinoma cell lines were subjected to RT–PCR analysis (lane 3; HOC313, lane 4; TSU, lane 5; HSC3, lane 6; HSC4, lane 7; KOSC2, lane 8; Ho1N1, lane 9; Ca9.22, lane 10; SCCKN, lane 11; KOSC3, lane 12; SCCTF, lane 13; HSC2, lane 14; OSC19). RNA sample isolated from GF12 normal gingival fibroblasts (lane 1) and HaCaT cells (lane 2) were applied as controls.
We analysed RNA samples isolated from tissues containing epithelial and mesenchymal components. To remove mesenchymal cell contamination from carcinoma cells, we examined the expression of LMO4, LDB1, and LDB2 in 12 different squamous carcinoma cell lines (Figure 1B). LMO4 was amplified in eight of 12 cell lines and LDB1 in 11 of 12 carcinoma cell lines. LMO4 was amplified in GF12 normal fibroblasts and LDB1 in GF12 and HaCaT cell lines. LDB2 was only detected in GF12, but not in any of the cell lines of epithelial origin.
Protein expression of LMO4 and LDB1, and interaction in carcinoma cells
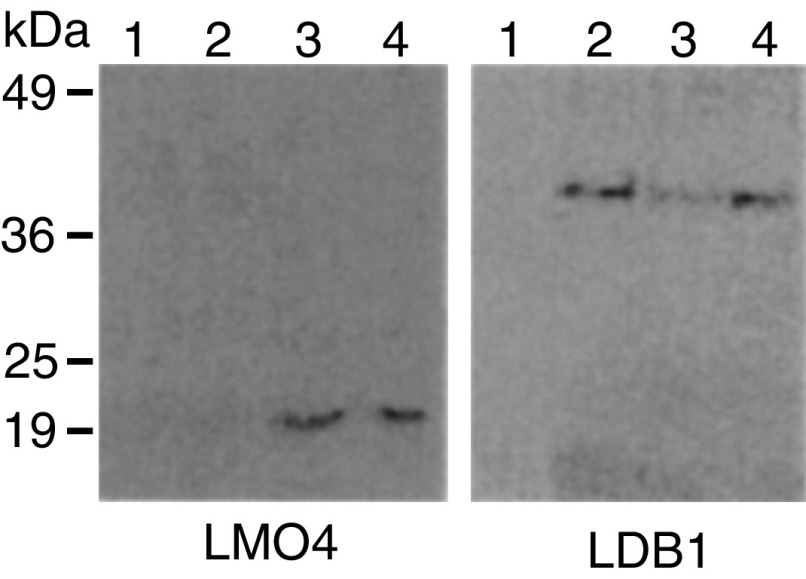
Since LMO4 and LDB1 were expressed in oral carcinomas, we examined the binding between LMO4 and LDB1 proteins by immunoprecipitation. LMO4 was coimmunoprecipitated with an anti-LDB1 antibody in OSC19 and HOC313 cells, which expressed LMO4 and LDB1 genes (Figure 2). The molecular size of LDB1 has not been determined, but is expected to be around Mr 42 809 from the deduced amino-acid sequence. LDB1 exhibits a single reactive protein band at Mr 40 000. TSU cells, which amplified LDB1, but not LMO4 genes, were immunoprecipitated only by LDB1 protein. As expected, LMO4- and LDB1-negative HSC3 cells did not react with the antibodies.
Figure 2.
Analysis of protein interaction in squamous carcinoma cell lines. Nuclear extracts from HSC3 (lane 1), TSU (lane 2), OSC19 (lane 3), or HOC313 cells (lane 4) were immunoprecipitated using anti-LDB1 antibody. After SDS–PAGE, immunoblotting was performed using antibodies specific to LMO4 or LDB1.
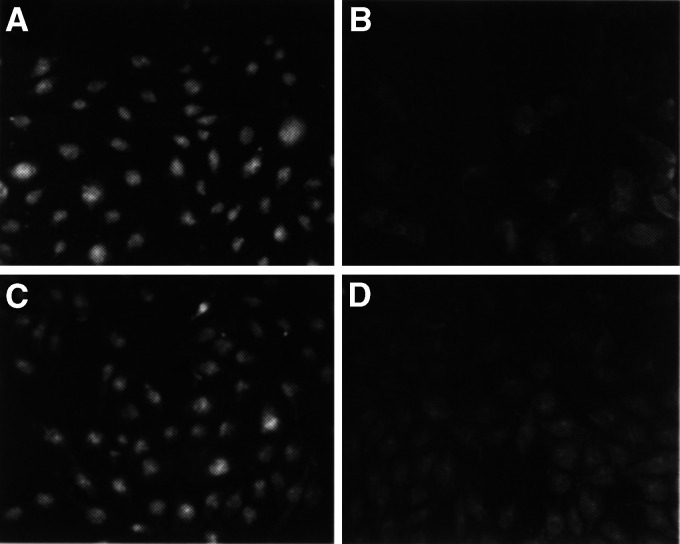
Immunocytochemistry stained the nuclei of HOC313 and OSC19 cells for LMO4, OSC19, HOC313, and TSU for LDB1, and none for LDB2 (Figure 3A and C). HSC3 cells were not stained by antibodies against LMO4 or LDB1 (Figure 3B and D). No staining was observed with nonimmune IgG (data not shown). Specificity of the antibodies used in this study was ascertained by nuclear immunostainings and by the appropriate size of a single-reactive band to the predicted molecular weight. Thus, these data demonstrate that LMO4 and LDB1 established a protein complex in oral squamous carcinoma cells.
Figure 3.
Immunocytochemisty of LMO4 or LDB1 in carcinoma cells. Squamous carcinoma cells cultured on the slide glasses were applied for immunostaining to LMO4 (A, B) or LDB1 (C, D). (A, C) HOC313 cells (B, D) HSC3 cells. Bar=50 μm.
Tissue localisation of LMO4 and LDB1 in oral carcinoma tissues
Clinical and pathological characteristics of the 49 patients with oral squamous cell carcinoma were collected and analysed. The samples were obtained from the tongue (20 cases), gingiva (15 cases), oral floor (six cases), buccal mucosa (five cases), lip (two cases), or maxillal sinus (one case). The patients (21 males and 28 females) ranged from 37 to 92 years of age (mean, 66 years old). Histopathological typing of carcinomas was classified into three groups; well (21 cases), moderately (23 cases), and poorly (five cases) differentiated carcinoma groups. None of the cases represented were classified as squamous carcinomas with a spindle-cell morphology. Lymph node tissues with metastasis of carcinomas were also subjected to the immunohistochemistry procedure (n=7).
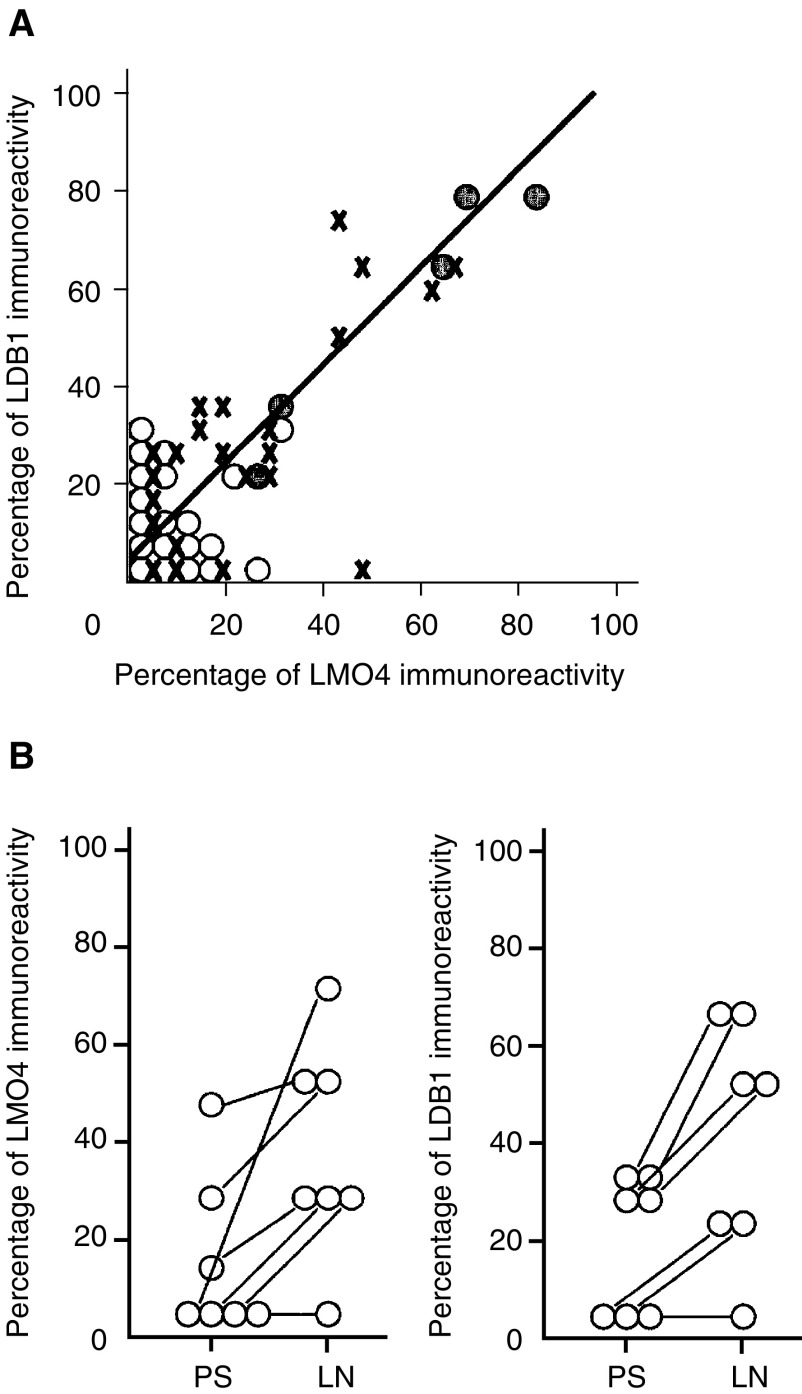
We localised LMO4 and LDBs in tissue sections by immunostaining using specific antibodies. Nuclear staining of LMO4 and LDB1 was observed in 30 (61%) and 37 (76%) of 49 carcinomas, respectively. Although minimal reaction of LDB2 immunostaining was noted in inflammatory cells that infiltrated into the carcinoma tissues, carcinoma cells did not react (data not shown). Normal epithelium from gingiva or tongue did not stain with each antibody, while an occasional reaction was observed in lymphocytes that infiltrated into the submucosal layer (Figure 4C). LMO4- and/or LDB1-immunoreactive carcinoma cells were usually found at the periphery of the carcinoma cell nests. In addition, the reaction became more prominent in carcinoma cells at the invasive front (Figure 4A and B). Fibroblast-like cells and infiltrating lymphocytes surrounding carcinoma cells at the invasive front also exhibited nuclear staining. To quantify the immunoreactivity, we calculated the percentage of nuclear staining in carcinoma cells and compared this to the clinicopathological parameters of the samples. Immunoreactive carcinoma cells were observed in 18.69±22.89% (mean±1 s.d.) of LMO4 staining and 23.41±22.44 of LDB1 staining. However, the percentage of immunoreactive carcinoma cells was significantly increased in less-differentiated carcinoma tissues (P<0.01) (Table 1 ) and their immunoreactivity of LMO4 and LDB1 showed a positive linear correlation (Figure 5A). We also compared the immuno-reactivity between carcinoma cells at the primary site and metastasised lymph nodes. Carcinoma cells in the lymph nodes significantly increased the immunoreactivity of LMO4 (37.67±22.60) and LDB1 (38.07±23.84) compared to the corresponding primary sites (LMO4; 13.46±18.29, LDB1; 17.63±14.99) (Figure 5B). There was no statistical difference in the immunoreactivity with patient age, gender, and tumour size and grade (data not shown).
Figure 4.
Immunolocalisation of LMO4, LDB1, or LDB2 in carcinoma or normal tissues. LMO4 (A) and LDB1 (B) were localised to carcinoma cells and stromal cells at the invasive front. Lymphocytes infiltrated into the submucosal layer of normal gingival were immunoreactive to LDB2 staining (C). Broken line depicted the borderline between epithelium and mesenchyme. Negative control staining with nonimmune goat IgG was performed instead of primary antibody (D). Bar=250 μm (A, B, and D) and 125 μm (C).
Table 1. Association between LMO4 or LDB1 immunoreactivity and histological tumour differentiation.
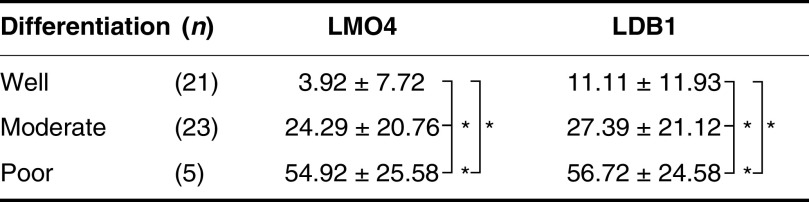 |
Percentage of immunoreactive carcinoma cells were counted and compared to histological differentiation grade by one-way analysis of variance and contrast statements (mean±1 s.d.). *P<0.01.
Figure 5.
Immunoreactivity of LMO4 and LDB1 at the primary site of carcinomas (A) and in metastasised lymph nodes (B). (A) A positive direct correlation between LMO4 (horizontal line) and LDB1 (vertical line) immunoreactivity was found by simple linear regression (r2=0.669, P<0.01, n=49). Open circles, crossed, and shaded circles indicated well, moderately, and poorly differentiated squamous cell carcinomas, respectively. (B) The percentage of immunoreactive carcinoma cells in primary sites and metastasised cervical lymph nodes (LMO4; left, LDB1; right) was analysed by Wilcoxon's signed-rank test (P<0.05).
DISCUSSION
Metastasis of squamous carcinoma cells to distant organs or lymph nodes requires several steps, including detachment from the primary site, dedifferentiation, invasion of the surrounding stroma and vessel walls, embolism, and stromal invasion and proliferation. Dedifferentiation of carcinoma cells initiates carcinoma progression toward a metastatic neoplasm (Vokes et al, 1993; Birchmeire et al, 1996; Lippman and Hong, 2001). Investigation of the molecular mechanism of carcinoma cell differentiation/dedifferentiation is a momentous subject in cancer cell biology and may contribute to the development of a novel strategy to relieve patient suffering from the disease. Although investigators reported that various factors are involved in carcinoma cell differentiation and dedifferentiation, accumulating evidence implies that transcriptional misregulation takes a critical part in the events (Angel et al, 1996; Hunter, 1997; Visvader et al, 1997; Battle et al, 2000; Cano et al, 2000). In the present study, our data demonstrate for the first time that LMO4 and LDB1 form protein complexes in the nucleus and are expressed in carcinoma cells at the invasive front, and their immunoreactivity is increased in less-differentiated carcinomas and in metastasised lymph nodes.
LMO4 and LDB1 are ubiquitously expressed in the mouse embryos, including epithelial and mesenchymal areas, when compared to the exclusive expression of other LMO family members and LDB2 in neuronal and haematopoietic cells (Visvader et al, 1997; Kenny et al, 1998; Sugihara et al, 1998; Toyama et al, 1998; Hermanson et al, 1999; Thaler et al, 2002). The LIM domain of LDBs contributes to the binding of transcription factors, including LIM-homeodomain, zinc-finger and basic helix–loop–helix (bHLH) proteins (Agulnick et al, 1996; Jurata et al, 1996; Bach et al, 1997; Morcillo et al, 1997). Formation of protein complexes synergistically activates the expression of target genes (Jurata et al, 1998; Milán et al, 1998). In the presence of LMO protein, it mediates the transcription factor binding to the LDB protein or competes with the direct binding between LDB and the transcription factor (Rabbitts, 1998; Thaler et al, 2002). These combinational actions are involved in the diversity of transcriptional regulation and specification of cell types (Visvader et al, 1997; Thaler et al, 2002). Misexpression of LMO1 and LMO2 by the chromosomal translocation is observed in T-cell leukaemia and facilitates the formation of an aberrant multimeric complex (Grütz et al, 1998; Rabbitts, 1998). Although the role in the pathology of carcinomas of epithelial origin is not clear, Visvader et al 2001 recently indicated that LMO4 and LDB1 are required to maintain the undifferentiation state of invasive breast carcinoma cells, and the forced expression of LMO4 inhibits differentiation of mammary epithelial cells. Increased expression of LMO4 and LDB1 in less-differentiated oral carcinomas represented in this study suggests an involvement in cellular dedifferentiation. Carcinoma cells located at the invasive front also exhibited an increase in immunoreactivity. Carcinoma cells located at the invasive front enhance the characteristics of epithelial–mesenchymal transition, which initiates invasion into the collagen matrices (Gobbert et al, 1985; Behrens et al, 1989; Frixen et al, 1991; Imai et al, 1995). Although the molecular mechanism of LMO4–LDB1 complex formation in carcinoma dedifferentiation is not clearly defined yet, LMO4 acts as a dominant negative by interacting with LDB1, thereby competing for binding between LDB1 and transcription factors. This dominant-negative effect of LMO4 inhibits differentiation of neuronal cells (Thaler et al, 2002). It is possible to speculate that increased expression of LMO4 may inhibit differentiation and accelerate invasion of oral carcinoma cells.
Another intriguing possibility comes from a study that GATA zinc-finger proteins interact with the LMO2–LDB1 complex and specifies a haematopoietic lineage differentiation (Wadman et al, 1997). However, overexpression of LMO2 in T-cell leukaemia results in the formation of a novel aberrant complex that substitutes a GATA protein to the E-box (CANNTG) binding bHLH and inhibits T-cell differentiation (Grütz et al, 1998). The C. elegans homologue of GATA, Elt, is a prerequisite for ectodermal cell differentiation (Gilleard and McGhee, 2001). GATA3 is expressed in the normal cervical squamous epithelial cells, but downregulated in progressive carcinoma cells (Steenbergen et al, 2002), suggesting a role of GATA in epithelial cell differentiation. The cis-acting E-box element is found in the E-cadherin promoter region and binding of the zinc-finger protein, SNAIL or SIP1, represses gene expression. E-cadherin has a central role in maintenance of the epithelial cell-type characteristic and the SNAIL/SIP1 inhibition of E-cadherin expression in squamous carcinoma cells accelerates dedifferentiation and invasion (Battle et al, 2000; Cano et al, 2000; Carver et al, 2001; Comijn et al, 2001). Our preliminary study showed significant expression of the SNAIL and SIP1 genes in oral carcinomas (T Chiba and K Imai, manuscript in preparation). Although GATA expression in oral carcinomas is not yet known, it could be intriguing if misexpression of LMO4 results in substitution of GATA to SNAIL/SIP1 and inhibits carcinoma cell differentiation.
It is interesting to note that the carcinoma cells that metastasised to the cervical lymph nodes exhibited increased immunoreactivity to both LMO4 and LDB1 when compared to the corresponding primary sites of tumour. An increased reaction in the metastasised carcinoma cells suggests that LMO4- and LDB1-expressing carcinoma cells at the primary sites may ease the progress toward metastasis. It might also be plausible that the metastasised carcinoma cells upregulate LMO4 and LDB1 in the milieu of the lymph node. It is known that the local microenvironment modifies carcinoma cell differentiation (Aboseif et al, 1999; Liotta and Kohn, 2001). Further studies should be addressed to demonstrate a direct role for LMO4 and LDB1 in carcinoma metastasis.
The present study demonstrated that LMO4 and LDB1 form a protein complex and are overexpressed at the carcinoma invasive front, and in less-differentiated and metastasised squamous carcinoma cells. It suggests that misexpression of LMO4 and LDB1 expression may play a role in progression of neoplasm. Future avenues of research will clarify transcriptional partners and target genes of LMO4–LDB1 complexes, and elucidate the role of this pathway in the pathology of squamous cell carcinomas.
Acknowledgments
We thank Drs Shirakawa (Nippon Dental University), Sakashita (Meikai University), and Ioroi (Machida City Hospital) for providing us oral carcinoma and normal tissue samples, and Dr Satoh (Nippon Dental University) and Dr Fusenig (German Cancer Research Center) for a generous gift of GF12 gingival fibroblasts and HaCaT cells. We are also grateful to Dr D'Armiento (Columbia University) for critical reading of the manuscript, and Drs Yamamoto and Kawashiri (Kanazawa University) for a gift of oral carcinoma tissue specimens. A portion of this study was supported by a grant from the Uehara Memorial Foundation to K Imai.
References
- Aboseif S, El-Sakka A, Young P, Cunha G (1999) Mesenchymal programming of adult human epithelial differentiation. Differentiation 65: 113–118 [DOI] [PubMed] [Google Scholar]
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H (1996) Functional and physical interaction of Ldb1, a novel LIM domain binding factor, with the LIM homeodomain protein Lhx1/Xlim-1. Nature 384: 270–272 [DOI] [PubMed] [Google Scholar]
- Angel P, Szabowski A, Schorpp-Kistner M (1996) Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 20: 2413–2423 [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Anderson B, Rosenfeld MG (1997) A family of LIM domain-associated cofactors confer transcriptional synergism between LIM, and Otx homeodomain proteins. Genes Dev 11: 1370–1380 [DOI] [PubMed] [Google Scholar]
- Battle E, Sancho E, Franci C, Dominguez D, Monfan M, Baulida J, Garcia de Herreros A (2000) The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2: 84–89 [DOI] [PubMed] [Google Scholar]
- Behrens J, Mareel M, van Roy F, Birchmeire W (1989) Dissecting tumor cell invasion: epithelial cells acquire invasive properties after the loss of uvomorulin-mediated cell–cell adhesion. J Cell Biol 108: 2435–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeire C, Birchmeire W, Brand-Saberi B (1996) Epithelial–mesenchymal transition in cancer progression. Acta Anat 156: 217–226 [DOI] [PubMed] [Google Scholar]
- Boehm T, Foroni L, Kaneko Y, Perutz MP, Rabbitts TH (1991) The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci USA 88: 4367–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106: 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen JJ, Agulnick AD, Wistphal H, David IB (1998) Interaction between LIM domains and the LIM domain-binding protein Ldb1. J Biol Chem 273: 4712–4717 [DOI] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Neito MA (2000) The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2: 76–83 [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T (2001) The mouse snail gene encodes a key regulator of the epithelial–mesenchymal transition. Mol Cell Biol 21: 8184–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 7: 1267–1278 [DOI] [PubMed] [Google Scholar]
- Digman JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frixen U, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Loechner D, Birchmeire W (1991) E-cadherin mediated cell–cell adhesion prevents invasiveness of human carcinoma cell lines. J Cell Biol 111: 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD (2001) Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol 21: 2533–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbert HR, Wagner R, Moll R, Gerharz CD (1985) Tumor dedifferentiation: an important step in tumor invasion. Clin Exp Metast 3: 257–279 [DOI] [PubMed] [Google Scholar]
- Grütz G, Forster A, Rabbitts TH (1998) Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLK1: a candidate for displacement by LMO proteins in T-cell acute leukemia. Oncogene 17: 2799–2803 [DOI] [PubMed] [Google Scholar]
- Hay ED (1995) A review of epithelo-mesenchymal transformation. Acta Anat 154: 8–20 [DOI] [PubMed] [Google Scholar]
- Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T (2000) SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol 1: 138–144 [DOI] [PubMed] [Google Scholar]
- Hermanson O, Sugihara TM, Anderson B (1999) Expression of the LMO4 in the central nervous system of the embryonic and adult mouse. Cell Mol Biol 45: 677–686 [PubMed] [Google Scholar]
- Hinks GL, Shah B, French SJ, Campos LS, Staley K, Hughes J, Sofronine MV (1997) Expression of LIM protein genes Lmo1, Lmo2, and Lmo3 in adult mouse hippocampus and other forebrain regions: differential regulation by seizure activity. J Neurosci 17: 5549–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T (1997) Oncoprotein networks. Cell 88: 333–346 [DOI] [PubMed] [Google Scholar]
- Imai K, Kumagai S, Nakagawa K, Yamamoto E, Nakanishi I, Okada Y (1995) Immunolocalization of desmoglein and intermediate filaments in human oral squamous cell carcinomas. Head Neck 17: 204–212 [DOI] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN (1996) Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA 93: 11693–11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Phaff SL, Gill GN (1998) The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem 273: 3152–3157 [DOI] [PubMed] [Google Scholar]
- Kenny DA, Jurata LW, Saga Y, Gill GN (1998) Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc Natl Acad Sci USA 95: 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Bindereif A, Green MR (1998) A small scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene 5: 22–32 [DOI] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC (2001) The microenvironment of the tumour–host interface. Nature 411: 375–379 [DOI] [PubMed] [Google Scholar]
- Lippman SM, Hong WK (2001) Molecular marker of the risk of oral cancer. N Engl J Med 344: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Milán M, Diaz-Benjumea FJ, Cohen SM (1998) Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev 12: 2912–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D (1997) Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev 11: 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder BA (2001) Cancer genetics. Nature 411: 336–341 [DOI] [PubMed] [Google Scholar]
- Rabbitts TH (1998) LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev 12: 2651–2657 [DOI] [PubMed] [Google Scholar]
- Royer-Pokora B, Loos U, Ludwig WD (1991) TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukemia with the t(11;14)(p13;q11). Oncogene 6: 1887–1893 [PubMed] [Google Scholar]
- Sanchez-Garcia I, Rabbitts TH (1994) The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet 10: 315–320 [DOI] [PubMed] [Google Scholar]
- Steenbergen RDM, OudeEngberink VE, Kramer D, Schrijnemakers HFJ, Verheijen RHM, Meijer CJLM, Snijders PJF (2002) Downregulation of GATA-3 expression during human papillomavirus-mediated immortalization and cervical carcinogenesis. Am J Pathol 160: 1945–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara TD, Bach I, Kioussi C, Rosenfeld MG, Anderson B (1998) Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO4, and CLIM coregulators. Proc Natl Acad Sci USA 95: 15418–15423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Sato T, Niwa M (1997) Cytotoxicity and mutagenicity to human adult gingival cells of sodium fluoride releasing devices. Biomed Res Trace Elements 8: 107–118 [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL (2002) LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein–protein interactions. Cell 110: 237–249 [DOI] [PubMed] [Google Scholar]
- Thiery JP (2002) Epithelial–mesenchymal transitions in tumor progression. Nat Rev Cancer 2: 442–454 [DOI] [PubMed] [Google Scholar]
- Toyama R, Kobayashi M, Tomita T, Dawid IB (1998) Expression of LIM-domain binding protein (ldb) genes during zebrafish embryogenesis. Mech Dev 71: 197–200 [DOI] [PubMed] [Google Scholar]
- Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH (1997) The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulator of erythroid differentiation. Proc Natl Acad Sci USA 94: 13707–13712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Ventor D, Hahm K, Santamaria M, Sum EYM, O'Reilly L, White D, Williams R, Armes J, Linderman GJ (2001) The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA 98: 14452–14457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes EE, Weichselbaum RR, Lippman SM, Hong WK (1993) Head and neck cancer. N Engl J Med 328: 184–194 [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grüz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH (1997) The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J 16: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH (1998) The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci USA 91: 3890–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]