Abstract
The human β-globin genes constitute a large chromosomal domain that is developmentally regulated. In nonerythroid cells, these genes replicate late in S phase, while in erythroid cells, replication is early. The replication origin is packaged with acetylated histones in erythroid cells, yet is associated with deacetylated histones in nonerythroid cells. Recruitment of histone acetylases to this origin brings about a transcription-independent shift to early replication in lymphocytes. In contrast, tethering of a histone deacetylase in erythroblasts causes a shift to late replication. These results suggest that histone modification at the origin serves as a binary switch for controlling replication timing.
Keywords: Replication timing, chromatin structure, epigenetics, gene expression, development
Studies in both yeast and animal cells demonstrate that the time at which replication origins fire during S phase is established by a preset marking system that operates in early G1 at about the same point that nuclear structure is restored following cell division (Raghuraman et al. 1997; Dimitrova and Gilbert 1999). This epigenetic signal may involve histone modification, as suggested by genetic manipulation experiments in yeast showing that normally early replicating origins can actually be made to fire aberrantly in late S phase simply by causing forced local histone deacetylation (Vogelauer et al. 2002). Whereas in yeast cells each origin replicates at an invariable fixed time within S phase, replication timing in higher eukaryotes is often subject to developmental regulation, and this is accomplished by altering the firing schedule of fixed origins. The human β-globin domain represents a good example of this phenomenon. Genetic and biochemical studies have demonstrated that this entire ∼100-kb region is copied by employing a single bidirectional origin that fires early during S phase in erythroid cells, but is converted to a late replication mode in all nonerythroid cells (Kitsberg et al. 1993). Nothing, however, is known about how this timing profile is set up at the molecular level and, in particular, whether this is dependent on local transcription (Gilbert 2002).
Results and Discussion
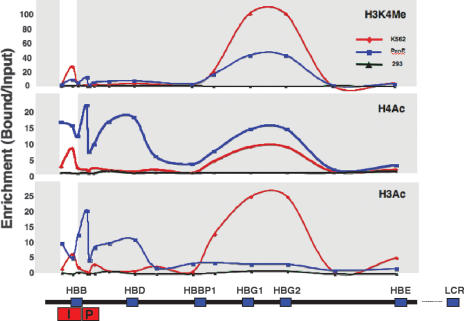
In order to assess whether this epigenetic switch may be mediated by changes in histone modification, we used chromatin immunoprecipitation (ChIP) analysis to map the mean histone acetylation pattern over the entire β-globin domain in different cell types (Fig. 1). In proerythroblast cells, we observed a large enrichment of histone H3 and H4 acetylation over the β-globin promoter and coding sequence in correlation with the transcription profile of this gene in these erythroid cells. Mapping studies indicate that the replication origin in this region is actually composed of two separate functional units (Wang et al. 2004), and it appears from our data that these sequences (see map) are also prepackaged with a combination of hyperacetylated histones and me-H3(K4) in the proerythroblast cells (Fig. 1). A similar pattern was also observed in other studies using this same culture system (Miles et al. 2007).
Figure 1.
Histone acetylation map of the human β-globin replication origin. Chromatin from human K562, 293, and proerythroblast (ProE) cells was subjected to ChIP analysis with antibodies against 2meH3K4, H4Ac, and H3Ac, and analyzed for relative enrichment by real-time PCR at 15 different sites along the β-globin domain on chromosome 11 (see map). The globin origin region, composed of two replicator subunits, RepI (I) and RepP (P) (Wang et al. 2004), is marked, as are the ε(HBE), γ(HBG1, HBG2), β-pseudo (HBBP1), δ(HBD), and β(HBB) gene sequences and the upstream LCR. Transcription is from right to left. Origin-specific histone acetylation and H3K4 methylation are characterized by a peak (highlighted by a white stripe). High-resolution ChIP-on-chip analysis for histone H3Ac in K562 cells over the origin region and the γ-globin genes can be seen in Supplemental Figure 1.
To determine whether this acetylation pattern may be a common feature of other erythroid cell types as well, we next carried out ChIP analysis on the CML-derived (Konopka et al. 1985), γ-globin-expressing K562 cell line (Donovan-Peluso et al. 1987). Strikingly, the same replicator fragment still shows peaks of histone acetylation and me-H3(K4), even though the adult β-globin gene itself is inactive and underacetylated in these cells, and this has been confirmed at high resolution using ChIP-on-chip analysis, as well (Supplemental Fig. 1). In contrast to erythroid cells where the β-globin domain replicates early, this same origin region is completely deacetylated in the nonerythroid 293 cell line (Fig. 1) where this domain replicates late (data not shown). In keeping with these findings, the mouse β-globin origin (Forsberg et al. 2000) as well as the human lamin B2 origin (Abdurashidova et al. 2000) have also been shown to be packaged with acetylated histones in correlation with early replication. These studies clearly suggest that there is a straightforward transcription-independent correlation between early replication timing and histone acetylation at origin-associated DNA sequences.
In order to test whether histone acetylation at the origin may represent an epigenetic mark that actively controls replication timing, we devised a strategy for bringing about site-directed histone acetylation near the human β-globin origin in vivo. To this end, we utilized a 150-kb BAC (RP11622D14) containing the entire human globin domain to insert multiple Gal4-binding sites near the origin region (Materials and Methods) by recombineering (Zhang et al. 1998), and then generated transgenic mice. Five different founder mice were obtained and all were shown by multiplex PCR analyses to contain a single intact copy of the β-globin BAC construct. Two of these founder animals (B14 and B54) were then crossed with transgenic mice that express the histone acetylase genes VP16, CBP, or PCAF fused to a Gal4-binding domain (see Materials and Methods). In double-transgenic animals these HAT proteins should be tethered to the β-globin origin region, bringing about forced local histone acetylation in nonerythroid cell types.
To test whether this strategy actually brings about increased histone acetylation over the origin region, we isolated spleen lymphocytes from transgenic animals carrying the β-globin BAC, either alone or in combination with the HAT fusion constructs, and then performed ChIP analysis for histone H3 acetylation. As expected for nonerythroid cells, the origin located on the wild-type BAC is packaged with deacetylated histones. In contrast, the presence of a tethered HAT protein brings about a striking increase in histone acetylation over a 3.2-kb domain that includes the β-globin origin region (Supplemental Fig. 2A). This reacetylation was observed mainly for histone H3 with histone H4 showing only a modest increase (data not shown). The fact that very similar results were observed for two different founder animals (B14 and B54) targeted with a variety of HAT proteins (Supplemental Fig. 2B) strongly suggests that this tethering strategy is highly effective in vivo.
We next asked whether this imposed histone acetylation pattern over the origin region affects DNA replication timing of the entire locus. To this end, we isolated spleen cells from animals containing either the wild-type BAC or its artificially histone-acetylated counterpart and carried out interphase FISH analysis using a large probe specific for the human β-globin region (Materials and Methods). In this method, the appearance of a single hybridization dot in the nucleus represents the unreplicated form of this region, while a double dot indicates that replication has already occurred. In a nonsynchronized population of cells, the percentage of S-phase (BrdUpositive) nuclei with single as opposed to double dots provides a direct measurement of replication timing (Selig et al. 1988). Previous studies have demonstrated that human globin transgenes that contain an intact LCR replicate in a developmentally controlled manner (Simon et al. 2001). In keeping with this, FISH analysis of the wild-type human globin BAC in B14 nonerythroid tissues revealed a single hybridization signal in 56%–60% of cells, indicating that it undergoes replication a short time after mid-S, at a point similar to the endogenous mouse globin sequences (∼50% single dots). In contrast, BAC DNA that had undergone forced histone acetylation around the origin became earlier replicating (32%–38%) in both mouse embryonic fibroblasts (MEFs) and lymphoid cells, and similar results were also obtained for B54 transgenic founder animals (Fig. 2A,B). Control experiments demonstrated that this shift in replication timing does not occur when the Gal4-binding sequences are inserted at an alternate chromosomal site or in the presence of a mutant (CBPΔ) acetylase construct (Fig. 2), clearly indicating that this effect must be due to specific histone acetylation at the origin itself. It should be noted that the level of human β-globin transcription in these cells remained extremely low (see Materials and Methods) even following the shift to early replication.
Figure 2.
Histone acetylation causes a shift to early replication. (A) FISH analysis (percentage of single dots) of BrdU-positive nuclei from transiently cultured, BrdU-labeled spleen lymphocytes (SL) or MEFs using the human globin BAC (H) and a mouse globin construct (M) as probes. Results from HAT-tethered mice are marked in red. (B) Bar graph showing the normalized values (Supplemental Material) for replication time as a percentage of S phase as measured in spleen lymphocytes (B14-L and B54-L) or MEFs (B14-M). Standard replication time markers included mouse globin (mHbb), as well as both the early (S1) and late (S2) asynchronous Snrpn alleles. The ranges of wild-type (WT) (late) and HAT (early) replication times are indicated by dotted lines. (Inset) Transgenic mice carrying a randomly integrated nonorigin fragment (NOC2) were crossed with animals containing the Gal4-CBP transgene, and spleen nuclei then subjected to FISH analysis with the pLJ168 probes.
In order to confirm the findings obtained from FISH analysis, we also used an S-phase fractionation method to measure replication timing (Azuara et al. 2003). For this experiment, BrdU-labeled spleen-derived cells were sorted into six different fractions according to DNA content. The BrdU DNA was then purified by immunoprecipitation and real-time PCR was used to quantitate the amount of replicative DNA in each fraction. These data are then normalized relative to known early and late gene markers (Supplemental Material). According to this analysis, the wild-type human β-globin BAC replicates relatively late in S phase, but following targeted histone acetylation, replication of this entire region is markedly shifted to a much earlier time point (Fig. 3A). These experiments clearly show that histone acetylation at the origin has a dominant effect on replication timing in vivo.
Figure 3.

(A) Spleen lymphocytes from B14 wild-type (WT) and B14 VP16 (VP16) transgenic mice were transiently cultured and incubated with BrdU for 1 h and cell-cycle-sorted by FACS into G1, S1, S2, S3, S4, and G2 fractions. BrdU DNA was then purified from each sample and analyzed by real-time PCR using six different primer sets distributed over the human globin BAC. The results from both experiments were individually averaged over all probes (local abundance) and normalized to the peaks of standard early (Cd19 and Actb) and late (Hbb and Amy) replicating mouse genes and then mathematically adjusted to take into consideration variations between the separate FACS analyses (Supplemental Material). (B) Replicating fork directions were determined by isolating emetine-treated, BrdU-labeled cells and carrying out single-strand real-time PCR analysis on purified BrdU DNA at two loci (28K and 74K) positioned on opposite sides of the human β-globin origin (Supplemental Material). The ratio of normalized backward to forward synthesis is shown. Leading strand synthesis in the rightward direction shows a high ratio of backward/forward PCR synthesis, while left-hand leading strand synthesis gives an opposite result.
In normal human cells, replication timing of the globin locus is controlled by sequences in the LCR that operate by influencing the firing schedule of a single bidirectional replication origin (Simon et al. 2001). In order to show that histone acetylation actually works by altering the initiation of replication at this specific origin and not by some other mechanism, we carried out mapping studies to ascertain origin usage for both the normal and targeted BAC sequences. This was accomplished by detecting the leading strand of DNA synthesis at several positions along the globin locus (Supplemental Material; Handeli et al. 1989). In both erythroid and nonerythroid human cells containing the normal human β-globin locus, assays of probes located both upstream of and downstream from the β gene revealed bidirectional replication (Fig. 3B). Identical results were obtained for the human β-globin BAC in both wild-type and hyperacetylated transgenic animals, as well, clearly suggesting that this histone modification affects replication by altering the time of origin firing in the same way that replication timing is regulated during normal development (Kitsberg et al. 1993).
In light of our results demonstrating that forced acetylation at the origin causes an early replication time switch in nonerythroid cells, we next tested the effect of deacetylation on the early replication time pattern normally seen in erythroid cells. To this end, mice carrying the wild-type BAC were crossed with transgenic animals that express a gene chimera in which the Gal4-binding domain is fused to a histone deacetylase (HDAC2) (see Materials and Methods). We then isolated and purified adult proerythroblasts (Dickerman et al. 1976) from double-transgenic mice and carried out ChIP analysis over the human globin domain in these cells. The wild-type BAC appears to have a histone acetylation pattern similar to that of normal human proerythroblasts, with marked acetylation observed over the β-globin origin itself (data not shown), confirming previous results for human globin transgenes (Miles et al. 2007). Tethering of the HDAC protein brings about a relative decrease in histone acetylation over this region (Supplemental Fig. 2C).
In order to test the effect of this change in histone status on replication timing, we carried out FISH analysis on transient cultures of dividing proerythroblasts (Fig. 4A,B). In keeping with previous findings, the wild-type BAC was found to replicate early in S phase (35%–38% single dots) at a point similar to that of the endogenous mouse globin domain. Strikingly, however, even partial deacetylation of the origin region causes a marked shift in domain-wide replication timing to late S phase (56%–62% single dots), thus mimicking what occurs in nonerythroid cells (see Fig. 2). This change appears to occur even though β-globin transcription remains relatively high in these cells (see Materials and Methods). Forced histone deacetylation (Coffee et al. 1999) may also be responsible for the dramatic shift to late replication seen at the FMR1 locus in Fragile X syndrome as a result of de novo DNA methylation on origin sequences (Gray et al. 2007) located in the gene promoter.
Figure 4.
Histone deacetylation causes a shift to late replication. (A) FISH analysis (percentage of single dots) of BrdU-positive nuclei from transiently cultured, BrdU-labeled erythroid cells using the human globin BAC (H) and a mouse globin construct (M) as probes. Results from HDAC2 mice are marked in red. (B) Bar graph showing the normalized values (Supplemental Material) for replication time as a percentage of S phase in erythroblasts (B14-E and B54-E). Standard replication time markers include mouse globin (mHbb), as well as both the early (S1) and late (S2) asynchronous Snrpn alleles. The ranges of wild-type (WT) (early) and HDAC2 (late) replication times are indicated by dotted lines.
Previous studies have shown that histone acetylation affects replication timing of origins in yeast (Vogelauer et al. 2002). Using a similar experimental approach, we now demonstrate that this same mechanism may play a role in the developmental regulation of replication timing in animal cells. Proper firing of each origin takes place as a two-step process involving the initial assembly of a multicomponent origin recognition complex (ORC) at the end of mitosis, followed by actual activation of the origin during S phase (Goren and Cedar 2003; DePamphilis 2005). The timing of activation at each origin appears to be determined by structural cues that are set up during early G1 phase at about the time when nuclear structure is being reconstituted following the previous round of mitosis (Raghuraman et al. 1997; Dimitrova and Gilbert 1999).
Our studies indicate for the first time that there are indeed origin-associated structural differences that definitively control early versus late firing. While this appears to be accomplished through a straightforward binary histone modification code, it should be noted that the relationship between histone acetylation levels and replication timing may actually be quite complex (Figs. 2, 3), and other epigenetic signals, such as DNA methylation, could also play a role in this program (Jablonka et al. 1985; Hansen et al. 2000). It has been suggested that replication timing may merely reflect local transcription, with active regions being early replicating (Gilbert 2002). Our data clearly show that transcription levels of β-globin do not correlate with the replication timing profile of this locus in human erythroid cells (Fig. 1). Furthermore, a forced shift to either early or late replication is not accompanied by changes in local transcription. Taken together, these data indicate that switches in replication timing are controlled independently of transcription. Whatever the details of this binary switch on the β-globin origin, it appears to be set up for both the early and late replication modes by long-range cis-acting sequences located in the upstream LCR (Simon et al. 2001).
In the next step of replication timing control, the signals that mark replication origins as being early or late must be read and interpreted during S phase. Judging by other cell-cycle-controlled processes, this might require the generation of a gradient over time, and protein factors that can recognize the predetermined histone acetylation pattern that is probably set up during G1. It has been shown that cyclins themselves are indirectly implicated in the control of replication timing. In yeast, for example, the β-cyclin Clb5 is actually required specifically for the replication of late origins, and in its absence the entire genome is copied by early-firing origins exclusively (Donaldson et al. 1998). This discrimination could work either because these cyclins recognize the modification pattern at the origin as a histone code, or because the increased cyclin concentration during late S phase eventually overcomes the relative lack of accessibility caused by underacetylation at the origin. In either case, the histone acetylation state appears to play a critical role as a molecular switch for controlling replication timing. The Rad53 and Mec1 checkpoint genes also seem to be involved in the regulation of replication timing, but these proteins apparently work by delaying late origin firing until the early replicons have completed elongation in a process that operates independently of the histone acetylation marking system (Early et al. 2004).
It has already been shown that replication timing has a decisive effect on the initial acetylation state of newly added histones H3 and H4 during nucleosome reassembly in the wake of DNA synthesis (Zhang et al. 2002). According to this model, the entire globin domain is repackaged in a relatively closed structure following each round of replication in nonerythroid cells, and this maintenance process may constitute one of the mechanisms for preventing activation of this entire region. In contrast, the switch to early replication timing in erythroid cells immediately brings about a major change in the initial reassembly process, thus facilitating the creation of an open chromatin structure by other, more stable mechanisms. This may help explain how local changes at key targets on the DNA (e.g., origins) can exert major structural and functional effects over large genomic domains.
Materials and methods
Transgenic mice
BAC RP11622D14, containing the entire human β-globin domain (Chr11: 5,175,878–5,363,071; University of California at Santa Cruz Genome Browser) was subject to recombineering (Zhang et al. 1998) in order to inject multiple (6×) Gal4-binding sites within the replication origin region (position 5,205,520) that overlaps the second exon of the β-globin gene itself (see map in Supplemental Fig. 2A). Five transgenic mice containing this modified BAC were generated and were found to contain intact copies of the globin domain as determined by PCR analysis at multiple points as well as FISH. Two single-copy transgenic founders (B14 and B54) were then used for subsequent experiments. As a non-origin-containing control, we prepared multiple-copy, single random integration site transgenic mice carrying the pLJ-168 plasmid (A. Klar) that contains eight tandem Gal4-binding site repeats (NOC2). Restriction fragments containing chimeric gene constructs made up of the Gal4-binding domain bound to histone acetylases or HDAC were excised from the plasmids pCMV × Gal4CBP, pCMV × Gal4CBPΔ, pCMV × Gal4P/CAF (T. Kouzarides), pBEF1α-Gal4VPI6 (G. Levkowitz), and pBind-HDAC2 (L. Vardimon) and were used to make transgenic mice. A single expressing founder of each type was used in crosses with transgenic mice containing the β-globin domain BAC (B14 and B54). MEFs were cultured from 13.5-d embryos. Lymphocytes from adult spleens of either wild-type BAC or double-transgenic mice were grown in DMEM supplemented with concanavalin-A for 3–4 d and erythroblasts were isolated from Phenylhydrazin-treated animals (Dickerman et al. 1976) by magnetic-activated cell sorting using biotinilated α-ter119 antibodies (PharMingen) (Goren et al. 2006). Blood-derived human proerythroblasts were enriched by culturing in specific media (Fibach et al. 1989). The levels of human β-globin transcription in mouse transgenic embryonic fibroblasts and spleen erythrocytes were determined by quantitative real-time PCR analysis as described previously (Goren et al. 2006). In MEFs from B14, both in control and early replicating cells, β-globin RNA was found to be present at levels of 10−6 as compared with Aprt (set at 1). The level of β-globin RNA in spleen erythrocytes was found to be 180 (in comparison with Aprt) in wild-type animals and 107 for HDAC-expressing mice.
Replication time assays
FISH (see the Supplemental Material) was performed as described previously (Lichter et al. 1988; Lichter et al. 1990) by culturing cells for 1 h with BrdU (3 × 10−5 M) and isolating nuclei using hypotonic KCl (0.5%) treatment and fixation with methanol:acetic acid (3:1) (Selig et al. 1992), and S-phase PCR analysis of BrdU-labeled replicating DNA was carried out as described (Azuara et al. 2003). These techniques are detailed in the Supplemental Material.
ChIP
Cells were cross-linked and chromatin was extracted and sonicated to an average size of 300–3000 base pairs (Schlesinger et al. 2007). Immunoprecipitation with anti-H4Ac, anti-H3Ac, or anti-H3K4me antibodies (Upstate Biotechnologies) was carried out (∼5 μg per 10–30 μg of DNA) using a ChIP assay kit. Incubation with the various antibodies was followed by precipitation with salmon sperm DNA and protein A Agarose (60 μL per 10 μg of DNA; Sigma) to isolate the bound fraction. Amplification was carried out by quantitative real-time PCR and the bound/input values were then normalized by setting the negative control (Cryaa or Pck1) to 1. Multiple assays of the same sample or the same gene sequence in separate immunoprecipitations from a given chromatin preparation showed an average coefficient of variance of ∼17%. Primer sequences are displayed in Supplemental Table 1.
Acknowledgments
We thank Yoav Smith, Roy Navon, and Zohar Yachini for assistance in the ChIP-on-chip analysis, and Yoav Smith for his help in normalizing the replication timing data. Proerythroblast cultures were obtained from Eitan Fibach. Recombineering was accomplished with the kind assistance of Francis Stewart and Susan Schneider. This work was supported by grants from the NIH, the Israel Science Foundation, and the Israel Cancer Research Fund.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.468308.
References
- Abdurashidova G., Deganuto M., Klima R., Riva S., Biamonti G., Giacca M., Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- Azuara V., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 2003;5:668–674. doi: 10.1038/ncb1006. [DOI] [PubMed] [Google Scholar]
- Coffee B., Zhang F., Warren S.T., Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- DePamphilis M.L. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- Dickerman H.W., Cheng T.C., Kazazian H.H., Spivak J.L. The erythropoietic mouse spleen-a model system of development. Arch. Biochem. Biophys. 1976;177:1–9. doi: 10.1016/0003-9861(76)90408-2. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Gilbert D.M. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D., Raghuraman M.K., Friedman K.L., Cross F.R., Brewer B.J., Fangman W.L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Donovan-Peluso M., Acuto S., Swanson M., Dobkin C., Bank A. Expression of human γ-globin genes in human erythroleukemia (K562) cells. J. Biol. Chem. 1987;262:17051–17057. [PubMed] [Google Scholar]
- Early A., Drury L.S., Diffley J.F. Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:31–38. doi: 10.1098/rstb.2003.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibach E., Manor D., Oppenhein A., Rachmilewitz E.A. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 1989;73:100–103. [PubMed] [Google Scholar]
- Forsberg E.C., Downs K.M., Christensen H.M., Im H., Nuzzi P.A., Bresnick E.H. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.M. Replication timing and transcriptional control: Beyond cause and effect. Curr. Opin. Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Goren A., Cedar H. Replicating by the clock. Nat. Rev. Mol. Cell Biol. 2003;4:25–32. doi: 10.1038/nrm1008. [DOI] [PubMed] [Google Scholar]
- Goren A., Simchen G., Fibach E., Szabo P.E., Tanimoto K., Chakalova L., Pfeifer G.P., Fraser P.J., Engel J.D., Cedar H. Fine tuning of globin gene expression by DNA methylation. PLoS ONE. 2006;1:e46. doi: 10.1371/journal.pone.0000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.J., Gerhardt J., Doerfler W., Small L.E., Fanning E. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Mol. Cell. Biol. 2007;27:426–437. doi: 10.1128/MCB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Stoger R., Wijmenga C., Stanek A.M., Canfield T.K., Luo P., Matarazzo M.R., D’Esposito M., Feil R., Gimelli G., et al. Escape from gene silencing in ICF syndrome: Evidence for advanced replication time as a major determinant. Hum. Mol. Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- Jablonka E., Goitein R., Marcus M., Cedar H. DNA hypomethylation causes an increase in DNase-I sensitivity and an advance in the time of replication of the entire inactive X chromosome. Chromosoma. 1985;93:152–156. doi: 10.1007/BF00293162. [DOI] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Keshet I., Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- Konopka J.B., Watanabe S.M., Singer J.W., Collins S.J., Witte O.N. Cell lines and clinical isolates derived from Ph1-positive chronic myelogenous leukemia patients express c-abl proteins with a common structural alteration. Proc. Natl. Acad. Sci. 1985;82:1810–1814. doi: 10.1073/pnas.82.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Lichter P., Tang C.C., Call K., Hermanson G., Evans G.A., Housman D., Ward D.C. High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Miles J., Mitchell J.A., Chakalova L., Goyenechea B., Osborne C.S., O’Neill L., Tanimoto K., Engel J.D., Fraser P. Intergenic transcription, cell-cycle and the developmentally regulated epigenetic profile of the human β-globin locus. PLoS ONE. 2007;2:e630. doi: 10.1371/journal.pone.0000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M.K., Brewer B.J., Fangman W.L. Cell cycle-dependent establishment of a late replication program. Science. 1997;276:806–809. doi: 10.1126/science.276.5313.806. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., et al. Polycomb mediated histone H3(K27) methylation pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Selig S., Ariel M., Goitein R., Marcus M., Cedar H. Regulation of mouse satellite DNA replication time. EMBO J. 1988;7:419–426. doi: 10.1002/j.1460-2075.1988.tb02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig S., Okumura K., Ward D.C., Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Tenzen T., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H. Developmental regulation of DNA replication timing at the human β globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Rubbi L., Lucas I., Brewere B.J., Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Wang L., Lin C.M., Brooks S., Cimbora D., Groudine M., Aladjem M.I. The human β-globin replication initiation region consists of two modular independent replicators. Mol. Cell. Biol. 2004;24:3373–3386. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Buchholz F., Muyrers J.P., Stewart A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- Zhang J., Feng X., Hashimshony T., Keshet I., Cedar H. The establishment of transcriptional competence in early and late S-phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]