Abstract
Although the size of an organism is a defining feature, little is known about the mechanisms that set the final size of organs and whole organisms. Here we describe Arabidopsis DA1, encoding a predicted ubiquitin receptor, which sets final seed and organ size by restricting the period of cell proliferation. The mutant protein encoded by the da1-1 allele has a negative activity toward DA1 and a DA1-related (DAR) protein, and overexpression of a da1-1 cDNA dramatically increases seed and organ size of wild-type plants, identifying this small gene family as important regulators of seed and organ size in plants.
Keywords: DA1; EOD1/BB; seed and organ size; cell proliferation, Arabidopsis
Many experiments suggest that organs possess intrinsic information about their final size and grow until they reach a final predetermined mass (Conlon and Raff 1999; Day and Lawrence 2000), but the mechanisms setting the limits of growth are not well characterized despite their central importance. Recently, a key pathway suppressing cell proliferation during organogenesis has been identified (Dong et al. 2007) that is conserved in insects and mammals. However, many of the factors regulating organ size in animals have no obvious counterparts in plants, suggesting that the control of plant organ size involves novel mechanisms. Although external cues such as light, day length, and temperature influence plant growth and adapt sessile plants to their prevailing environment, the final size of plant seeds and determinate organs is reasonably constant within a given species, whereas interspecific seed and organ size variation is remarkably large, demonstrating that developing seeds and organs also possess intrinsic information about their final size (Tsukaya 2006). The mechanisms that establish the final size of seeds and organs and mediate environmental inputs into growth are poorly understood, despite their fundamental importance and relevance to crop plant improvement.
Plant organ growth occurs by an initial proliferative phase in which cell numbers increase while their size remains fairly constant, followed by dramatic cell size increases that cease when the set size of the organ is reached. Increases in cell ploidy occur during later stages of organ growth that can be associated with the final size of cells (Sugimoto-Shirasu and Roberts 2003). In leaves, the transition from cell proliferation to cell expansion follows cell cycle arrest fronts that move from the tip to the base (Donnelly et al. 1999). Modulation of the time and location of cell proliferation arrest (Nath et al. 2003; Dinneny et al. 2004; Disch et al. 2006; White 2006) have been established as key regulatory points during leaf and petal formation that set final organ size and establish its shape. Interaction between organs also influences seed size. Reduced maternal integument size reduces final seed size (Garcia et al. 2005), and reduced endosperm proliferation arrests cell elongation in the integument (Garcia et al. 2003).
The growth regulator auxin promotes growth through ARGOS (Hu et al. 2003), which mediates expression of AINTEGUMENTA (ANT), a transcription factor that regulates expression of cell cycle genes (Mizukami and Fischer 2000). Elevated ANT levels increase cell numbers and increases the size of all shoot organs. Conversely, low ANT levels reduce the final size of organs (Krizek 1999; Mizukami and Fischer 2000). The transcription factors ANGUSTIFOLIA (AN3) and AtGRF5 function together to promote cell proliferation in leaves (Horiguchi et al. 2005), and their overexpression increases leaf growth. Several factors that limit cell proliferation during organ formation have been described. Mutations in ARF2, an auxin-responsive protein, cause extra cell divisions, resulting in larger organ and integument size, which leads to larger seeds (Schruff et al. 2006) as well as partial sterility and other phenotypes. ARF2 has also been shown to mediate auxin-responsive gene expression and negatively regulates cell expansion (Li et al. 2004). Several organ-specific growth regulators have been identified. The BIGPETALp gene limits petal size by restricting cell size (Szecsi et al. 2006). Deletion of the PEAPOD (PPD) genes increases leaf area and curvature due to prolonged division of dispersed meristematic cells in the leaf blade (White 2006). Loss-of-function mutations in AP2, encoding a transcription factor, lead to a range of floral defects that correlate with increased seed mass (Jofuku et al. 2005; Ohto et al. 2005).
One of the mechanisms setting final seed and organ size may potentially affect the time taken for an organ to reach its set final size, given that the period of proliferation is a key control point. The E3 ligase BIG BROTHER (BB) negatively regulates the duration of cell proliferation in leaves and petals, possibly by targeting growth stimulators for degradation, and is proposed as a bona fide organ size regulator (Disch et al. 2006). Despite being expressed in all plant tissues, loss-of-function bb mutations seem to influence primarily petal and stem growth. Here we describe the DA1 gene, which defines a novel growth control pathway that sets the final size of seeds and determinate organs in Arabidopsis by controlling the period of cell proliferation. A modifier screen identified an enhancer of da1-1 (EOD1), which is allelic to BB (Disch et al. 2006). Mutations in EOD1/BB synergistically enhance both seed and organ size phenotypes of da1-1, indicating that DA1 acts in parallel with EOD1/BB to control the final size of seeds and organs in Arabidopsis. DA1 expression is induced by the growth regulator abscisic acid (ABA) and organ growth has reduced sensitivity to ABA in da1-1, suggesting ABA may be an important factor in establishing final organ size.
Results and Discussion
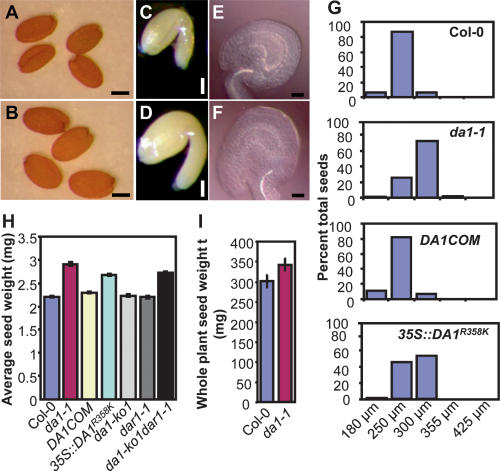
We identified the da1-1 (DA means “large” in Chinese) mutant in genetic screens in Arabidopsis thaliana that increased both seed and organ size (Figs. 1, 2A–L) and has normal fertility compared with wild type (data not shown). The seed mass of da1-1 was increased to 132% of wild type (Figs. 1H, 5D [below]); ovule and seed volume, embryo size, and cotyledon areas were substantially increased (Figs. 1A–G, 2A,B; Supplemental Fig. 1); and total seed yield per plant was increased (Fig. 1I). Seeds produced by a da1-1 mother, regardless of the genotype of the pollen donor, were consistently heavier and larger than those produced by maternal wild-type plants, and da1-1 mutant pollen in a wild-type mother produced seeds with wild-type mass (Supplemental Table 1), suggesting that DA1 can act maternally to control seed mass. The integuments surrounding the ovule are derived from maternal tissues and form the seed coat. Increased integument size such as that seen in da1-1 ovules (Fig. 1E,F) is known to contribute to increased seed size (Garcia et al. 2005; Schruff et al. 2006).
Figure 1.
Seed size in da1-1. (A,B) Dry seeds of Col-0 (A) and da1-1 (B). (C,D) Mature embryos of Col-0 (C) and da1-1(D). (E,F) Ovules of Col-0 (E) and da1-1 (F). (G) Preweighed batches of wild-type Col-0, da1-1, complementation line (DA1COM#2), and 35S∷DA1R358K #5 seeds from individual plants were passed through a series of wire sieves of decreasing mesh size (in microns) as described in the Supplemental Material. Seeds retained by each sieve were weighed, and the mass of each fraction was expressed as a percentage of the total mass of the seed sample analyzed. (H) Average seed weights of Col-0, da1-1, DA1COM#2, 35S∷DA1R358K #5, da1-ko1, dar1-1, and da1-ko1dar1-1 double mutants are shown as milligrams per 100 seeds. Standard deviations are shown (n = 5). (I) Average seed weight per plant. Standard deviation values were given (n = 5). Plants were grown under identical conditions. Bars: A,B, 200 μm; C,D, 100 μm; E,F, 50 μm.
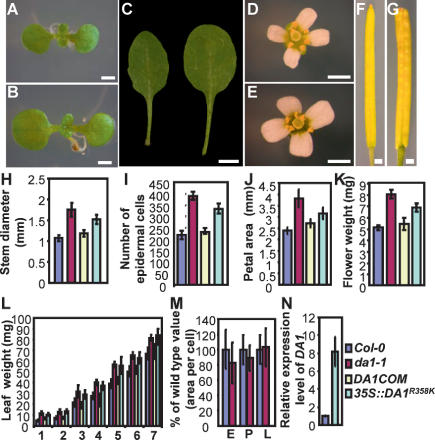
Figure 2.
Organ size in da1-1. (A,B) (A) Nine-day-old seedlings of Col-0 (A) and da1-1 (B). da1-1 has larger cotyledons than wild type. (C) The fifth leaves of Col-0 (left) and da1-1 (right). da1-1 has larger and rounder leaves compared with wild-type Col-0. (D,E) Flowers of Col-0 (D) and da1-1 (E). (F,G) Siliques of Col-0 (F) and da1-1 (G). (H–J) Stem diameter (H), epidermal cell number in stem cross-sections (I), and petal area (J) of Col-0, da1-1, DA1COM#2, and 35S∷DA1R358K#5. (K,L) Mass of five fresh flowers (stage 14) (K) and leaves (first through seventh) of 35-d-old plants (L). (M) Cell area of embryos (E), petals (P), and leaves (L) in Col-0 and da1-1. Values are given as mean ± SD relative to the respective wild-type value, set at 100%. (N) Relative expression levels of DA1 in Col-0 and 35S∷DA1R358K#5 seedlings were measured by quantitative real-time RT–PCR. Bars: A,B, 1 mm; C, 0.5 cm; D–G, 1 mm.
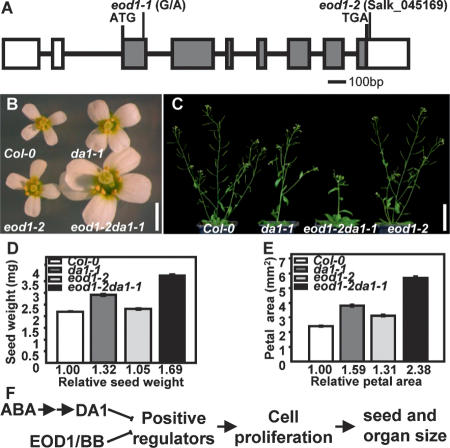
Figure 5.
Mutations in EOD1/BB synergistically enhance the phenotypes of da1-1. (A) EOD1/BB gene structure, showing the mutated sites of the two eod1 alleles. The start codon (ATG) and the stop codon (TGA) are indicated. Closed boxes indicate the coding sequence, and lines between boxes indicate introns. The mutated site in eod1-1 and T-DNA insertion site in eod1-2 also are shown. (B) Flowers of Col-0, da1-1, eod1-2, and eod1-2da1-1 double mutants. (C) Soil-grown plants of Col-0, da1-1, eod1-2da1-1 double mutant, and eod1-2 are shown. (D) Average seed weights of Col-0, da1-1, eod1-2, and eod1-2da1-1 double mutants are shown as milligrams per 100 seeds. Standard deviations are shown (n = 5). Plants were grown under identical conditions. (E) Petal areas of Col-0, da1-1, eod1-2, and eod1-2da1-1 double mutant. Standard deviation values are shown (n > 50). (F) A model of DA1 and EOD1/BB controlling seed and organ size. Bars: B, 2 mm; C, 50 mm.
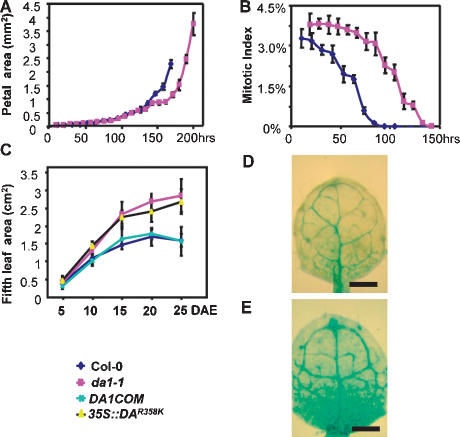
da1-1 plants have large flowers with extra petals and carpels (Fig. 2D,E; Supplemental Fig. 2), enlarged and somewhat flattened siliques (Fig. 2F,G), rounder and large leaves (Figs. 2C, 3C), and thick stems as well as increased biomass compared with wild type (Fig. 2H,I,K,L). In all organs of da1-1 plants, the size of cells was comparable with that measured in wild type (Fig. 2M), whereas the number of cells was increased—for example, in the stem epidermal layer, up to 180% of wild-type numbers (Fig. 2I). Kinematic analysis of petal and leaf growth (Fig. 3A,C) illustrated that da1-1 plants undergo a longer phase of proliferative growth and have a longer life span than wild-type plants (Fig. 5C [below]; Supplemental Fig. 9D,E). Measurement of the mitotic index using a pCYCB1:1∷GUS reporter fusion that is specifically expressed in mitosis during petal and leaf growth demonstrated that cell cycle activity was arrested later in growth (Fig. 3B,D,E). These analyses show that DA1 functions to limit the period of proliferative growth during early stages of organ development and controls the time taken for most organs to reach their final size.
Figure 3.
Kinematic analysis of petal and leaf growth. (A) Growth of Col-0 and da1-1 mutant petals. The largest petals of each series are from opened flowers (stage 14). (B) Mitotic index in wild-type and da1-1 mutant petals. The time axis in B corresponds to that in A. (C) Fifth leaf area of Col-0, da1-1, DA1COM#2, and 35S∷DA1R358K#5 measured over 25 d. (DAE) Days after emergence. (D,E) pCyclinB1;1∷GUS activity in the first leaves (9 d after germination) of wild-type (D) and da1-1 (E) seedlings grown on MS medium containing 1% glucose. Bar, 1 mm.
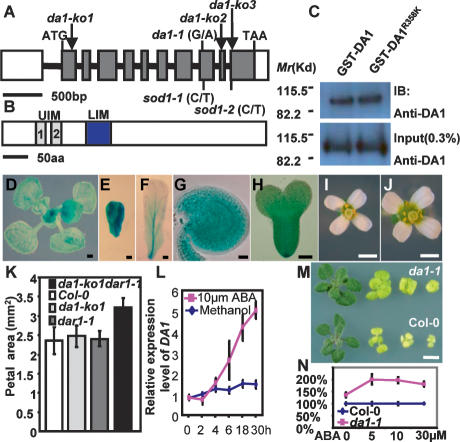
The DA1 gene was identified by map-based cloning (Supplemental Fig. 3). DNA sequencing revealed that the da1-1 allele has a single-nucleotide G-to-A transition in gene At1g19270 predicted to cause an arginine-to-lysine change in a conserved amino acid at position 358 (Fig. 4A; Supplemental Figs. 3, 4A, 5). All da1-1 phenotypes were complemented by the wild-type DA1 gene and by wild-type DA1 full-length cDNA expressed from a 35S promoter, indicating that At1g19270 is indeed the DA1 gene (Figs. 1G,H, 2H–L, 3C). The DA1 gene is predicted to encode a 532-amino-acid protein containing two ubiquitin interaction motifs (UIM) typical of ubiquitin receptors (Hicke et al. 2005) and a single zinc-binding LIM domain defined by its conservation with the canonical Lin-11, Isl-1, and Mec-3 domains (Fig. 4B; Supplemental Fig. 4A–C; Freyd et al. 1990; Hiyama et al. 1999). To test whether mutation in da1-1 affects the predicted ubiquitin-binding activity of DA1, we expressed full-length GST-DA1 and GST-DA1R358K fusion proteins in Escherichia coli, incubated protein extracts with ubiquitin–agarose beads, and identified bound proteins using DA1 antibody. No significant difference in ubiquitin-binding activity between GST-DA1 and GST-DA1R358K was detected (Fig. 4C), demonstrating that the da1-1 mutation does not affect DA1 ubiquitin binding. The ubiquitin receptor function of the UIMs was demonstrated using a GST fusion protein in E. coli to bind ubiquitin in pull-down assays (Supplemental Fig. 6). Seven other predicted proteins in Arabidopsis share extensive amino acid similarity with DA1 and have been named DA1-related (DAR) proteins (Supplemental Fig. 7A), of which four are found in a tandem cluster on chromosome 5. Proteins that share significant homology with DA1 outside the UIM and LIM domains are found in crop plants including rice and maize (Supplemental Fig. 7B), but not animals, suggesting the family has evolved to control growth in plants.
Figure 4.
Identification, expression, function, and genetic interactions of the DA1 gene. (A) DA1 gene structure showing the mutated sites of da1-1, sod1-1, and sod1-2 alleles. The start codon (ATG) and the stop codon (TAA) are indicated. Closed boxes indicate the coding sequence, and lines between boxes indicate introns. T-DNA insertion sites (da1-ko1, da1-ko2, and da1-ko3) in DA1 gene are shown. (B) The predicted DA1 protein contains two UIM and one LIM domains. (C) In vitro ubiquitin-binding assay. GST-DA1 and GST-DA1R358K fusion proteins expressed in E. coli were incubated with ubiquitin–agarose beads. Bound GST-DA1 and GST-DA1R358K fusion proteins (top panel) and 0.3% input proteins (bottom panel) were detected by immunoblotting (IB) with an antibody against DA1 (anti-DA1). (D–H) DA1 expression activity was monitored by pDA1∷GUS transgene expression. GUS staining during early leaf development (D), developing (E) and mature (F) petals, a developing ovule (G), and early-stage embryo (H). (I,J) Flowers of Col-0 (I) and da1-ko1dar1-1 double mutant (J). (K) Petal area of Col-0, da1-ko1, dar1-1, and da1-ko1dar1-1 double mutants. The da1-ko1dar1-1 double mutant displays a da1-1 phenotype including large flowers and petals. (L) Quantitative RT–PCR analysis revealed that expression of DA1 is slowly induced by ABA. Seven-day-old wild-type seedlings were treated with 10 μM ABA for 2, 4, 6, 18, and 30 h. (M) Seedlings of Col-0 (bottom) and da1-1 (top) were established on MS medium for 4 d and transferred to MS + 1% glucose medium without ABA and with 5 μm ABA, 10 μM ABA, and 30 μM ABA (from left to right) for 2 wk, respectively. The da1-1 mutant seedlings exhibit ABA-insensitive organ growth, compared with Col-0. (N) The shoot fresh weight of Col-0 and da1-1 in M. Values are given as mean ± SE relative to the respective wild-type value, set at 100%. Bars: D,M, 0.5 mm; E,F, 0.1 mm; G,H, 50 μm; I,J, 1 mm; (O, 2 mm.
A screen for modifiers of da1-1 function was conducted to identify other components in the DA1 pathway. We identified two suppressors of large seed and organ phenotypes of da1-1 (sod) alleles that mapped to the original DA1 locus. Sequencing revealed that these sod1 alleles harbored second site mutations in DA1 that were predicted to reduce gene function (Fig. 4A; Supplemental Fig. 4A), indicating that the R358K mutation in DA1 is responsible for increased seed and organ size. Consistent with this, disruption of the DA1 gene with T-DNA insertions in the predicted coding region (da1-ko1, da1-ko2, and da1-ko3) (Fig. 4A) did not cause an obvious growth phenotype, nor did T-DNA insertion alleles in DAR1, the most closely related family member (Figs. 1H, 4K). Lines heterozygous for the original da1-1 mutation and wild-type Col-0 had a seed and organ size nearly similar to the wild type, whereas all the F1 plants from crosses of da1-1 with da1-ko lines displayed a similar phenotype to da1-1 (Supplemental Fig. 8A,B). The da1-1 phenotype was also observed in wild-type Col-0 plants expressing a 35S∷DA1R358K transgene (Figs. 1G,H, 2H–L,N, 3C; Supplemental Fig. 8C). These data suggested that the protein encoded by the da1-1 allele exerts a negative activity toward DA1 in a dose-dependent manner. This result also indicated that this negative activity of the da1-1 allele is weak because da1-1/Col-0 F1 plants display very similar phenotypes to Col-0 (Supplemental Fig. 8A,B). Because the loss-of-function alleles of DA1 display no obvious phenotype, we postulated that DA1 may act redundantly with DAR proteins and that the da1-1 allele may also exert a negative activity on the function of DAR proteins. Consistent with this hypothesis, double da1-ko1dar1-1 knockout mutants exhibited the original da1-1 phenotype (Figs. 1G,H, 4I–K). These analyses demonstrate that the R358K mutation in DA1 is necessary and sufficient to cause the da1-1 phenotype and the DA1R358K mutant protein has a negative effect on the function of DA1 and DAR1 proteins.
We also identified an enhancer of da1-1 (eod1-1) in the screen for modifiers, and an eod1-1da1-1 double mutant exhibited substantially larger seed and organ size and longer life span than da1-1 (Fig. 5C; Supplemental Fig. 9). The EOD1 gene was mapped to the Big Brother (BB) locus (At3g63530), encoding an E3 ubiquitin ligase that also represses organ (but not seed) growth in Arabidopsis (Disch et al. 2006). Sequencing revealed that the eod1-1 allele is a single-nucleotide G-to-A transition resulting in an alanine-to-threonine change in BB protein (Fig. 5A). Both a T-DNA insertion in the 3′ untranslated region (UTR) (eod1-2) and the original bb-1 mutation also enhanced the da1-1 phenotypes (Fig. 5B–E; Supplemental Fig. 9C–E), and transformation of eod1-1 with a binary vector expressing the At3g63530 cDNA from a 35S promoter rescued the eod1 phenotype (data not shown), demonstrating that EOD1 is the BB gene. Seed and petal measurements in eod1-2da1-1 and bb-1da1-1Ler double mutants demonstrated a synergistic enhancement of seed and organ size of da1-1 by mutations in EOD1/BB (Fig. 5B–E; Supplemental Fig. 9C–E), suggesting that the two genes act in parallel pathways to control the final size of seeds and organs in Arabidopsis. Double mutants of da1-1 with ant-5, axr1-12, arf2-7, and ap2-7 mutants that also exhibit altered seed or organ size were assessed (Lincoln et al. 1990; Krizek 1999; Mizukami and Fischer 2000; Jofuku et al. 2005; Ohto et al. 2005; Schruff et al. 2006). The genetic interactions between da1-1 and these mutants were essentially additive for petal or seed size, compared with their parental lines (Supplemental Fig. 10), suggesting that da1-1 acts independently of ANT, AXR1, ARF2, and AP2.
A DA1 promoter∷GUS fusion (pDA1∷GUS) was expressed most strongly during the early stages of leaf, petal, integument, and embryo formation (Fig. 4D–H) and at much reduced levels later in petal and leaf development, consistent with its role in controlling the period of cell proliferation. Microarray data showed that DA1 is expressed at higher levels in ABA-treated seedlings (Li et al. 2006), and further experiments established that DA1 is slowly induced by ABA (Fig. 4L) and not by other growth regulators such as jasmonic acid, auxin, cytokinin, brassinosteroids, gibberellins, or glucose (data not shown). Interestingly, seedling and root growth, seed germination, and seedling establishment in the da1-1 mutant are relatively insensitive to inhibition by ABA (Fig. 4M–N; Supplemental Fig. 11), suggesting ABA may be an endogenous or environmental cue that limits the period of proliferative growth during organogenesis through regulation of DA1 expression. To gain a better understanding of ABA function in size control, we generated double mutants of da1-1 with abi4-1 and abi5-1 mutants that control ABA growth and developmental responses (Soderman et al. 2000; Lopez-Molina et al. 2001). Genetic analysis revealed that the seed and petal size phenotypes of abi4-1da1-1 and abi5-1da1-1 double mutants were essentially additive (data not shown), compared with their parental lines, suggesting that DA1 is independent of ABI4 and ABI5, two key regulators of ABA responses in Arabidopsis.
Our genetic, physiological, biochemical, and kinematic data established that the novel growth regulator gene DA1 plays a major role in setting final sizes of both seeds and organs by restricting the duration of proliferative growth. The mutant DA1R358K protein has a negative activity toward DA1 function in a dose-dependent manner, and it also has a negative activity toward the activity of the closely related DAR1 protein. This type of effect has also been reported for the wol and sleepy1 mutants (Mahonen et al. 2000, 2006; Strader et al. 2004). The R358K change affects an invariant amino acid in an extensive region conserved in all members of the DA1 family, two of which can function redundantly. It is possible that the R358K mutation in an invariant amino acid in this conserved region interferes with the interaction of DA1 family members with a common target—for example, by forming an unproductive complex that normally restricts growth.
Analysis of double eod1-2da1-1 mutants shows that DA1 and EOD1/BB, another negative regulator of the proliferative phase of organogenesis (Disch et al. 2006), function in parallel pathways and may modulate the activity of a common target (Fig. 5F). However, the DA1R358K mutant protein appears to have a stronger negative influence on growth inhibition than EOD1/BB loss-of-function mutants, and affects the size of most determinate organs, including seeds (Fig. 5D). The ubiquitin-binding activity of DA1 is consistent with a potential signaling function. UIMs bind molecules modified by monoubiquitin or ubiquitin chains (Hicke et al. 2005) and promote coupled monoubiquitination, leading to conformational changes that regulate diverse cellular processes, including signaling and endocytosis (Hurley et al. 2006; Penengo et al. 2006). LIM domains, which serve as protein interaction interfaces, are involved in a variety of fundamental biological processes (Dawid et al. 1998; Kadrmas and Beckerle 2004). The close involvement of E3 ligase activity from EOD1/BB, the ubiquitin receptor activity, and potential coupled monoubiquitination activities from DA1 suggests direct involvement of ubiquitin-based cell signaling mechanisms in plant organ size control.
The induction of DA1 expression in response to ABA and the ABA-insensitive growth seen in the da1-1 mutant suggests a mechanism for limiting proliferative growth during organogenesis by ABA. ABA signaling has the potential to link environmental inputs, such as stress, to final organ size (Finkelstein et al. 2002), and ABA is required for normal plant development (Phillips et al. 1997; Cheng et al. 2002). The large effects of DA1 on the duration of cell proliferation during seed and organ formation, the absence of pleiotropic effects, and DA1’s genetic interactions with EOD1/BB, emphasize the central role of the period of cell proliferation in setting final seed and organ size in plants and define a major role for DA1 in coordinating size control at the whole-plant level. Our current knowledge of DA1 function and the identification of conserved DA1-like genes suggest that the da1-1 allele can be used to engineer large seed size and increased biomass in crop plants.
Materials and methods
Plant materials and growth conditions
A. thaliana Columbia (Col-0) was the wild-type line used. All mutants were in the Col-0 background, except for da1-1Ler, bb-1, and ant-5, which were in Landsberg erecta (Ler). Plant materials and growth conditions are available in the Supplemental Material.
Genetic screening
da1-1 was identified as a novel seed and organ size mutant from an ethyl-methanesulphonate (EMS)-treated M2 population of Col-0 accession. sod1-1, sod1-2, and eod1-1 were identified as suppressors and enhancers, respectively, of da1-1 from EMS-treated M2 populations of da1-1.
Morphological and cellular analysis
Detailed protocols, sample preparation, measurement, microscopy, and histochemical staining for β-glucuronidase activity are described in the Supplemental Material.
Map-based cloning and plant transformation
F2 mapping populations were generated from a single cross of Ler/da1-1, Ler/sod1-3da1-1, and da1-1Ler/eod1-1da1-1 plants. The DA1, SOD1, and EOD1 genes were isolated by using a map-based cloning method. Detailed information for map-based cloning, constructs, and plant transformation is described in the Supplemental Material.
Protein expression and ubiquitin-binding assays
GST-DA1 and GST-DA1R358K fusion proteins were cloned in pGEX-4T-2 (Amersham-Pharmacia), expressed in E. coli BL21(DE3), and incubated with ubiquitin–agarose beads (Santa Cruz Biotechnologies). Bound GST-DA1 and GST-DA1R358K fusion proteins were detected by immunoblotting (IB) with an antibody against DA1 (anti-DA1). The UIM1 + 2 domain from DA1 (residues 63–127) was cloned into pGEX-4T-2 and expressed in E. coli BL21 as a fusion with N-terminal GST tag. Detailed methods for protein expression and ubiquitin-binding assays are described in the Supplemental Material.
RT–PCR, quantitative real-time RT–PCR, and Northern blot analysis
Total RNA was extracted from Arabidopsis seedlings using an RNeasy Plant Mini kit (Qiagen). Detailed protocols for RT–PCR, quantitative real-time RT–PCR, and Northern blot analysis are described in the Supplemental Material.
Acknowledgments
We thank Kim Findlay for assistance with scanning electron microscopy, John Doonan for pCyclinB1:1∷GUS seeds, Michael Lenhard for bb-1 seeds, NASC for T-DNA insertion lines, and colleagues for their comments on the manuscript. This work was supported by BBSRC Grant BB/C515620/1, EC Contract 037704 (AGRO-nomics), and the core strategic grant to the John Innes Centre.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.463608.
References
- Cheng W.H., Endo A., Zhou L., Penney J., Chen H.C., Arroyo A., Leon P., Nambara E., Asami T., Seo M., et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon I., Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Dawid I.B., Breen J.J., Toyama R. LIM domains: Multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Day S.J., Lawrence P.A. Measuring dimensions: The regulation of size and shape. Development. 2000;127:2977–2987. doi: 10.1242/dev.127.14.2977. [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Yadegari R., Fischer R.L., Yanofsky M.F., Weigel D. The role of JAGGED in shaping lateral organs. Development. 2004;131:1101–1110. doi: 10.1242/dev.00949. [DOI] [PubMed] [Google Scholar]
- Disch S., Anastasiou E., Sharma V.K., Laux T., Fletcher J.C., Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 2006;16:272–279. doi: 10.1016/j.cub.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14 (Suppl.):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G., Kim S.K., Horvitz H.R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Garcia D., Saingery V., Chambrier P., Mayer U., Jurgens G., Berger F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003;131:1661–1670. doi: 10.1104/pp.102.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Fitz Gerald J.N., Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Schubert H.L., Hill C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hiyama H., Yokoi M., Masutani C., Sugasawa K., Maekawa T., Tanaka K., Hoeijmakers J.H., Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.T., Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- Hu Y., Xie Q., Chua N.H. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell. 2003;15:1951–1961. doi: 10.1105/tpc.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Lee S., Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K.D., Omidyar P.K., Gee Z., Okamuro J.K. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. 2005;102:3117–3122. doi: 10.1073/pnas.0409893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J.L., Beckerle M.C. The LIM domain: From the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Krizek B.A. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 1999;25:224–236. doi: 10.1002/(SICI)1520-6408(1999)25:3<224::AID-DVG5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J.M., Ecker J.R. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell. 2004;7:193–204. doi: 10.1016/j.devcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Li Y., Lee K.K., Walsh S., Smith C., Hadingham S., Sorefan K., Cawley G., Bevan M.W. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Res. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes & Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen A.P., Higuchi M., Tormakangas K., Miyawaki K., Pischke M.S., Sussman M.R., Helariutta Y., Kakimoto T. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 2006;16:1116–1122. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. 2005;102:3123–3128. doi: 10.1073/pnas.0409858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L., Mapelli M., Murachelli A.G., Confalonieri S., Magri L., Musacchio A., Di Fiore P.P., Polo S., Schneider T.R. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Phillips J., Artsaenko O., Fiedler U., Horstmann C., Mock H.P., Muntz K., Conrad U. Seed-specific immunomodulation of abscisic acid activity induces a developmental switch. EMBO J. 1997;16:4489–4496. doi: 10.1093/emboj/16.15.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff M.C., Spielman M., Tiwari S., Adams S., Fenby N., Scott R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Soderman E.M., Brocard I.M., Lynch T.J., Finkelstein R.R. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Ritchie S., Soule J.D., McGinnis K.M., Steber C.M. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc. Natl. Acad. Sci. 2004;101:12771–12776. doi: 10.1073/pnas.0404287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. ‘Big it up’: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Szecsi J., Joly C., Bordji K., Varaud E., Cock J.M., Dumas C., Bendahmane M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- White D.W. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. 2006;103:13238–13243. doi: 10.1073/pnas.0604349103. [DOI] [PMC free article] [PubMed] [Google Scholar]