Abstract
Polycomb group (PcG) proteins regulate important cellular processes such as embryogenesis, cell proliferation, and stem cell self-renewal through the transcriptional repression of genes determining cell fate decisions. The Polycomb-Repressive Complex 2 (PRC2) is highly conserved during evolution, and its intrinsic histone H3 Lys 27 (K27) trimethylation (me3) activity is essential for PcG-mediated transcriptional repression. Here, we show a functional interplay between the PRC2 complex and the H3K4me3 demethylase Rbp2 (Jarid1a) in mouse embryonic stem (ES) cells. By genome-wide location analysis we found that Rbp2 is associated with a large number of PcG target genes in mouse ES cells. We show that the PRC2 complex recruits Rbp2 to its target genes, and that this interaction is required for PRC2-mediated repressive activity during ES cell differentiation. Taken together, these results demonstrate an elegant mechanism for repression of developmental genes by the coordinated regulation of epigenetic marks involved in repression and activation of transcription.
Keywords: EZH2, Polycomb, histone methyl transferase, histone demethylase, epigenetics, embryonic stem cell
PcG proteins are essential regulators of development, and their function is highly conserved during evolution (Ringrose 2007; Schuettengruber et al. 2007; Schwartz and Pirrotta 2007). The PcG proteins form mainly two different Polycomb-repressive multiprotein complexes, named PRC1 and PRC2 (Francis and Kingston 2001; Simon and Tamkun 2002). PRC1 is a large-sized complex that includes a variety of different PcG proteins, and its composition can vary in a cell type-specific manner. The PRC1 complex mediates the ubiquitylation of histone H2A through the ubiquitin E3 ligase activity of the Ring1A and Ring1B subunits and is required for transcriptional silencing of target genes, possibly by mediating chromatin compaction (Shao et al. 1999; Saurin et al. 2001; de Napoles et al. 2004; Francis et al. 2004; Wang et al. 2004). The PRC2 complex is smaller, and the three PcG proteins EZH2, EED, and SUZ12 form the core complex. The catalytic subunit EZH2 of the PRC2 complex can modify histone H3 N-terminal tails by specifically trimethylating (me3) Lys 27 (K27). This modification correlates with transcriptional repression, and it has been proposed to serve as a docking site for PRC1 recruitment (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002).
The three PcG proteins forming the core of the PRC2 complex are all required for EZH2 lysine methyl transferase (KMT) activity in vitro and in vivo (Cao and Zhang 2004; Pasini et al. 2004b). Moreover, consistent with a potential essential role of H3K27me3 in regulating transcriptional silencing during development, the three PcG proteins are essential for embryonic development during the gastrulation stage (Faust et al. 1995; O’Carroll et al. 2001; Pasini et al. 2004b; Montgomery et al. 2005).
The identification of PRC2 and PRC1 downstream regulatory pathways by genome-wide location analysis has demonstrated that the two complexes share a highly significant number of target genes in different cell types (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006). The analysis of PcG target genes in embryonic stem (ES) cells revealed that PcG proteins directly control the expression of a large number of genes that are essential for determining cell fate decisions during embryonic development (Boyer et al. 2006; Lee et al. 2006). Despite this key role of the PcG protein in developmental control, the mechanisms by which epigenetic silencing through H3K27me3 occurs are poorly understood, and the pathways regulating PcG recruitment to target genes, the factors, and enzymatic activities involved in PcG-mediated silencing are still unknown.
The recent isolation and characterization of enzymes with histone lysine demethylase activity (KDM) have shown that histone methylation is more dynamic than previously anticipated and have introduced an additional layer of complexity in the regulation of transcription (Shi 2007; Agger et al. 2008). Among the histone demethylases, we and others have shown that members of the Jumonji protein family, JARID1, specifically catalyze the demethylation of H3K4me3 and H3K4me2, regulate the expression of HOX genes, and are required for normal development (Christensen et al. 2007; Eissenberg et al. 2007; Iwase et al. 2007; Klose et al. 2007; N. Lee et al. 2007; Secombe et al. 2007; Tahiliani et al. 2007; Yamane et al. 2007). However, our knowledge regarding the mechanisms by which the JARID1 proteins contribute to developmental control is limited, and the identification of the downstream pathways regulated by the JARID proteins together with the mechanisms by which these factors regulate transcription are important questions to address.
The H3K4me3 mark is restricted to transcription start sites (TSS) and most often found associated with active promoters (Mikkelsen et al. 2007). However, recent studies have demonstrated that H3K27me3 and H3K4me3 can coexist on transcriptional silent promoters in ES cells and have suggested that this “bivalent mark” maintains a promoter in a “poised” state accessible for activation during cellular commitment (Bernstein et al. 2006; Mikkelsen et al. 2007). This intriguing observation suggests a mechanism for how transcriptional programs are achieved during development and the existence of a cross-talk between these two antagonistic modifications that could require a tight regulation mediated by both KMTs and KDMs. Consistent with this, recent studies found that a specific H3K27me3 demethylase, UTX, physically interacts with the MLL complex (a histone H3K4-specific methyltransferase), and that the UTX activity is involved in the transcriptional activation of HOX genes in NT2 cells (Agger et al. 2007; M.G. Lee et al. 2007b).
We investigated the functional role of the H3K4 demethylase RBP2 (JARID1A) and its interplay with the PRC2 complex. We demonstrate that RBP2, in association with the PRC2 complex, is critical for the transcriptional repression of a large number of Polycomb target genes involved in developmental control. Based on our results, we propose a mechanism for transcriptional control as cells undergo differentiation, which involves the coordinated regulation of H3K4 demethylation and H3K27 trimethylation.
Results
Identification of Rbp2 target genes in mouse ES cells
To understand how RBP2 can contribute to development and differentiation, we identified Rbp2 target genes in mouse ES cells by genome-wide location analysis. To this extent, we used an Rbp2-specific antibody that in agreement with previously published results (Benevolenskaya et al. 2005) specifically enriched for the BRD8 promoter in human cells (Fig. 1A) and that recognized mouse Rbp2 (Fig. 4A, below). Hence, we performed chromatin immunopreciptation (ChIP) experiments in mouse ES cells and hybridized the precipitated material to an oligonucleotide array covering ∼2.5 kb (−2 kb to +0.5 kb with respect to the TSS) of 19,489 annotated transcripts in the NCBI database. The analysis identified 606 promoters that were significantly bound by Rbp2 in ES cells (Supplemental Table 1). Functional clustering of the Rbp2-bound promoters revealed that Rbp2 is present on the promoters of a considerable number of genes that are involved in the regulation of development and differentiation (Fig. 1B), suggesting that Rbp2 could play a direct role in regulating the transcription of these genes during development.
Figure 1.
Identification of Rbp2 target genes in mouse ES cells by genome-wide location analysis. (A) ChIP assays in control and RBP2 shRNA-treated 293T cells on the BRD8 promoter. ChIP with antibody against the hemagglutinin (HA) tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal. (B) Clustering of Rbp2-bound promoters in mouse ES cells into functional groups. (C) Comparison of PcG target genes and their regulatory pathways published elsewhere (Bracken et al. 2006) with those regulated by Rbp2. (D) Statistically significant overlap between promoters bound by Rbp2 and promoters bound by PcGs published elsewhere (Boyer et al. 2006) in ES cells. The P-value was calculated by hypergeometric probability test using Genespring software (Agilent).
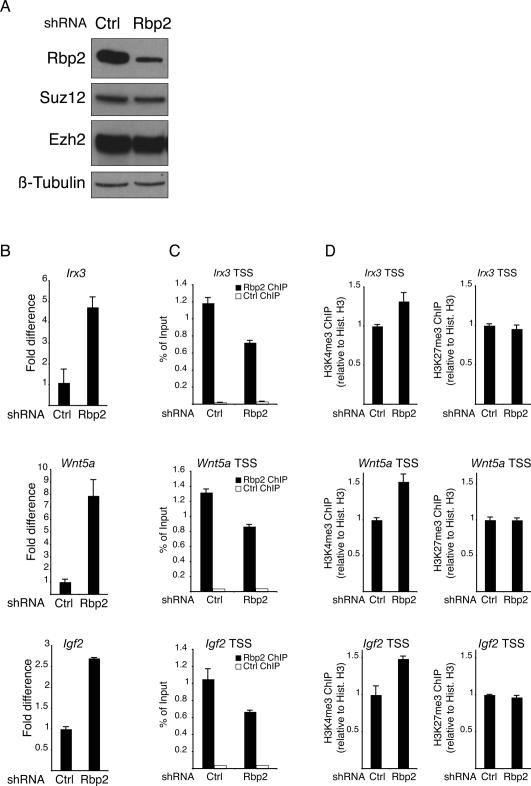
Figure 4.
Rbp2 is required for the repression of PRC2 target genes. (A) Western blot analysis of protein lysates prepared from mouse ES cells transduced with either control or Rbp2 shRNA lentiviruses. (B–D) Expression levels of the Irx3, Wnt5a, and Igf2 (B) and ChIP assays of the Irx3, Wnt5a, and Igf2 promoters using antibodies specific for Rbp2, histone H3, H3K4me3, and H3K27me3 (C,D) in control (Ctrl) or Rbp2 shRNA-treated mouse ES cells. ChIPs with antibody against HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal. ChIPs for H3K4me3 and H3K27me3 are further normalized to histone H3 density. Gene expression is normalized to Gapdh levels.
Previously, we and others have identified the downstream regulatory pathways of PcG proteins, showing that PcGs repress the expression of a large number of genes essential for cell fate decisions during development (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006). Interestingly, we observed a substantial overlap between the genes and the pathways regulated by the PcG proteins (Bracken et al. 2006) and Rbp2 (Fig. 1C). To obtain statistical data for this observation, we compared the PcG target genes identified in mouse ES cells (Boyer et al. 2006) with our list of Rbp2 target genes. Despite the Rbp2 and PRC2 ChIP–chip analysis were performed in different laboratories using different platforms, this analysis revealed that Rbp2 and the PcG proteins share a statistically highly significant number of overlapping target genes in ES cells (32% of targets, 195 out of 606, Fig. 1D), suggesting a functional interplay between the PcG proteins and Rbp2.
To validate the Rbp2 location analysis, we performed independent ChIP experiments for 31 Rbp2 target genes that were or were not identified previously as PcG targets with a high, medium, and low false discovery rate (FDR) (Supplemental Table 1). Rbp2 binding was confirmed for most of the target genes (29/31), showing the high accuracy of the location analysis also for target genes with an FDR close to the cutoff (0.2) (Fig. 2A, top panel).
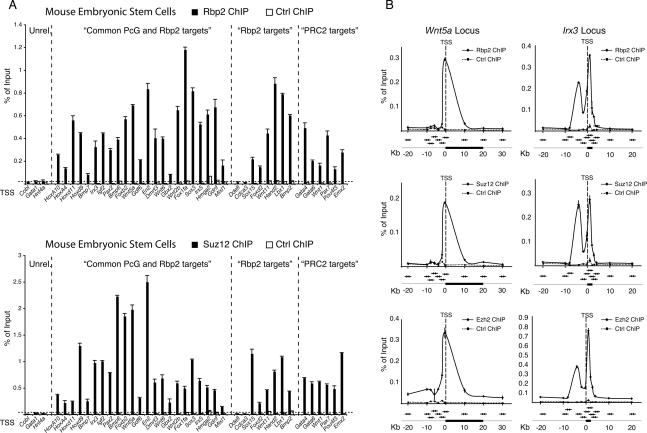
Figure 2.
Rbp2 and PRC2 occupy overlapping target genes. (A) ChIP analysis in ES cells using antibodies against Rbp2 and Suz12. The first group includes unrelated control genes, the second group includes genes found in both Rbp2 and Suz12 genome-wide location analysis, the third group provides genes found bound by Rbp2 only, and the fourth group of genes are bound by PRC2 only. Antibody against the HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal. (B) ChIP analysis in mouse ES cells of Rbp2, Suz12, and Ezh2 along the Wnt5a and Irx3 gene loci. ChIPs with antibody against the HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal. The position of the qPCR amplicons (arrows) relative to the genes (black boxes). TSS (dashed line) are also presented in the figure.
To confirm that the PcG proteins share common target genes with Rbp2, we performed ChIP analysis for the PRC2 protein, Suz12. The results from this analysis confirmed that Rbp2 and Suz12 are associated with the same promoters (Fig. 2A) and, surprisingly, that Rbp2 targets that were not identified previously as PcG target genes (Fig. 2A, Rbp2 targets) also showed a significant binding of Suz12. These data suggest that the binding overlap between Rbp2 and the PcG proteins is more extensive than suggested by the location analysis. To confirm this possibility, we tested and found that Rbp2 is associated with PcG targets that were not identified in the Rbp2 location analysis (Fig. 2A, PRC2 targets). These results further support the fact that PcGs and Rbp2 share a larger number of target genes than were identified by the location analysis, and this is most likely due to the technical differences between the compared experiments.
To further investigate the potential functional interplay between Rbp2 and the PcG proteins, we speculated that, if it exists, they should show a similar binding distribution on their target genes. Therefore, we analyzed the binding patterns of Rbp2, Suz12, and Ezh2 at the Irx3, Igf2, Wnt5a, and Brachyury loci by ChIP. The distribution of the real-time quantitative PCR (qPCR) amplicons covered a region of ∼20 kb upstream of and downstream from the TSS in the four loci with a higher resolution (∼2 kb) within the first 10 kb upstream of the TSS, as indicated in Figure 2B and in Supplemental Figure 1A. This analysis revealed a perfect overlap between the binding profiles of Suz12, Ezh2, and Rbp2 (Fig. 2B; Supplemental Fig. 1A) and confirmed that Rbp2 and the PRC2 complex bind to the same promoters in the proximity of the TSS. In conclusion, these results suggest that the overlap between PcG and Rbp2 target genes is even more extensive than the data presented in Figure 1D suggest.
Rbp2 binds directly to the PRC2 complex
The significant overlap between PcG and Rbp2 target promoters indicates a physical interaction between the proteins. To investigate this, we tested and showed that ectopically expressed RBP2 associates with endogenous components of the PRC2 complex (Fig. 3A). This association was confirmed also with the endogenous proteins: Immunoprecipitation (IP) experiments using antibodies specific to different PRC2 subunits (Ezh2 and Suz12) and to Rbp2 showed coimmunoprecipitation of endogenous Rbp2 with the members of the PRC2 complex in both mouse ES cells and in human 293T cells (Fig. 3A,B; Supplemental Fig. 1B,C). Moreover, the association between the proteins is independent of DNA, since Rbp2 associates with the PRC2 in the presence of ethidium bromide (Supplemental Fig. 1D).
Figure 3.
RBP2 interacts with the PRC2 complex. (A) IP assays using 293T cell lysates expressing HA-tagged wild-type (WT) or enzymatically inactive RBP2 mutant showing specific coimmunoprecipitation of RBP2 with EZH2 and EED. (B) IPs of mouse ES cell lysates using specific antibodies to Ezh2 and Suz12 showing specific coimmunoprecipitation of Rbp2. IP with anti-HA-tag antibody served as negative control. (C, top panels) Size exclusion chromatography (Superose-6, GE Healthcare) of mouse ES cell protein lysate showing coelution of Rbp2, Ezh2, Suz12, and Eed in high-molecular-weight fractions. (Bottom panels) The indicated fractions were immunoprecipitated with a Suz12-specific antibody. IP with anti-HA-tag antibody served as negative control (Ctrl). (D) Coomassie-stained gel and Western blot of protein-immunoprecipitated material from Sf9 cells expressing recombinant RBP2, EZH2, SUZ12, and Eed with anti-SUZ12 and anti-HA-tag (as negative control) antibodies showing the binding of RBP2 to the PRC2 complex.
To further support these findings, we tested if Rbp2 and the components of the PRC2 complex are present in the same molecular weight fractions. For this, we fractionated ES cells nuclear extracts on a Superose-6 gel filtration column and analyzed the different fractions by Western blot. Importantly Ezh2, Eed, Suz12, and Rbp2 coelute in fractions corresponding to a molecular weight ≥1 MDa (Fig. 3C, top panels). This result is consistent with previous findings showing the existence of a large-sized complex containing PRC2 subunits and suggests that this includes the H3K4 demethylase, Rbp2. To test if Rbp2 and the components of the PRC2 complex physically interact in the high molecular weight fractions, we incubated the different gel filtration fractions with a specific antibody to Suz12 and demonstrated that Rbp2 and the PRC2 complex interact in fractions with complexes larger than ∼1 MDa (Fig. 3C, bottom panels).
Finally, coexpression of recombinant RBP2, EZH2, EED, and SUZ12 in insect cells confirmed the efficient binding of RBP2 with the PRC2 complex (Fig. 3D). Taken together, these results provide evidence for a strong direct binding between PRC2 and RBP2 and suggest the existence of high molecular weight biochemical structures that contain RBP2 and the PRC2 complex.
Rbp2 is required for maintaining repression of PRC2 target genes
To understand if Rbp2 plays a role in maintaining repression of PcG target genes, we tested the effects of inhibiting Rbp2 expression (Fig. 4A) on genes that were reported previously to be activated by loss of PRC2 (Boyer et al. 2006; Lee et al. 2006). For this, we infected ES cells with lentiviral particles expressing specific shRNA against Rbp2 (Fig. 4A) and isolated RNA from these cells for qPCR expression analysis. As shown in Figure 4B, down-regulation of Rbp2 leads to increased expression of Irx3, Wnt5a, and Igf2. This demonstrates that Rbp2 activity is required for the maintenance of the repression of PRC2 target genes in ES cells. Importantly, ChIP experiments performed in the same cells showed that inhibition of Rbp2 expression also correlates with decreased binding of Rbp2 to its target promoters (Fig. 4C) and, consistent with Rbp2 demethylase activity, to increased levels of H3K4me3 (Fig. 4D, left panels). However, no changes in H3K27me3 levels (Fig. 4D, right panels; Supplemental Fig. 2B) and PRC2 binding (Supplemental Fig. 2A) were observed upon inhibition of Rbp2 expression. Taken together, these results show that Rbp2 contributes to maintenance of PcG–Rbp2 target gene repression, and that Rbp2 is not required for the binding of PRC2 to its target genes.
The PRC2 complex recruits RBP2 to target genes
To test if the PRC2 complex is required for Rbp2 binding to its target genes, we performed ChIP experiments in wild-type and Suz12 knockout mouse ES cells (Fig. 5A) as well as in mouse ES cells expressing an shRNA to Suz12 (Supplemental Fig. 3A). As shown in Figure 5B and Supplemental Figure 3B, Rbp2 binding to its target promoters was dramatically decreased in the absence of the PRC2 complex. Consistent with this, H3K27me levels are reduced from the Irx3, Wnt5a, and Igf2 promoters, while H3K4me3 are increased at the Irx3 and Wnt5a promoters in Suz12 knockout ES cells (Supplemental Fig. 2C). These experiments strongly suggest that the PRC2 complex contributes to the recruitment of Rbp2 to its target genes.
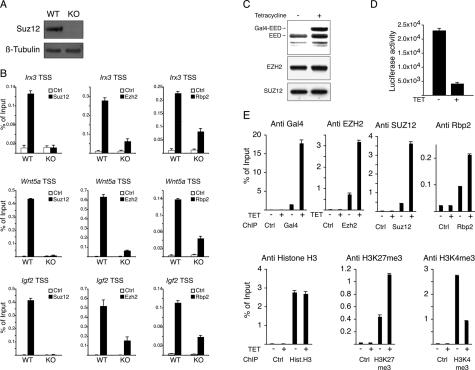
Figure 5.
The PRC2 complex is required for Rbp2 recruitment to target genes. (A) Western blots from Suz12 wild-type (WT) and knockout (KO) mouse ES cells using the indicated antibodies. β-Tubulin served as a loading control. (B) ChIP assays for Suz12, Ezh2, and Rbp2 on the Irx3, Wnt5a, and Igf2 promoters in wild-type (WT) and knockout (KO) ES cells. ChIPs with antibody against HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal. (C) Western blot analysis using antibodies specific for EED, SUZ12, and EZH2 of lysates prepared from a Gal4-EED-inducible 293T cell line grown with or without tetracycline. (D) Luciferase activity on cell lysates prepared from GAL4-EED 293T cells grown with or without tetracycline. (E) ChIP assays of lysates prepared from GAL4-EED 293T cells grown with or without tetracycline using the indicated antibodies. ChIP with antibody against HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal.
To obtain evidence that the PRC2 complex could mediate the recruitment of Rbp2 to target genes, we used a heterologous reporter system in which the expression of GAL4-EED from a tetracycline-inducible promoter represses the expression of an integrated Luciferase reporter plasmid containing GAL4-binding sites (K.H. Hansen, A.P. Bracken, D. Pasini, N. Dietrich, A. Monrad, and K. Helin, in prep.). As expected, expression of GAL4-EED, upon addition of tetracycline (Fig. 5C), led to ∼80% repression of the Luciferase activity (Fig. 5D). ChIP analysis performed in the same cells showed that GAL4-EED was recruited to the Gal4 DNA-binding site present at the promoter of the reporter system, and that this led to the recruitment of the endogenous PRC2 components EZH2 and SUZ12. Importantly, GAL4-EED expression also led to the recruitment of RBP2 to the same promoter, demonstrating that the PRC2 complex has the ability to recruit RBP2 to target genes. Consistent with this, PRC2–RBP2 recruitment to the reporter promoter led to increased H3K27me3 levels and to reduced H3K4me3 levels (Fig. 5E). Furthermore, RBP2 knockdown by siRNA led to reduced GAL4-EED repression of Luciferase activity, suggesting that RBP2 plays a role in this repressive mechanism (Supplemental Fig. 3C). Taken together, these results show that the PRC2 complex directly recruits RBP2 activity to target genes.
Rbp2 and PRC2 activities cooperate during ES cell differentiation
The PRC2 complex is required to maintain the repressive state of genes involved in development, and its displacement during ES cell differentiation leads to the correct expression of important regulators of development (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006; Pasini et al. 2007). Consistent with this, the expression of Wnt5a, Irx3, and Igf2 (Fig. 6A; Supplemental Fig. 4A) is induced during differentiation of ES cells (Supplemental Fig. 4D). Importantly, this induction correlates with Rbp2 and PRC2 displacement from the promoters (Fig. 6B; Supplemental Fig. 4B), increased H3K4me3 levels, and decreased H3K27me3 levels (Fig. 6C; Supplemental Fig. 4C). Moreover, the analysis of differentiated cells, such as mouse embryonic fibroblasts (MEFs) and mouse myoblasts C2C12, showed that both PRC2 and Rbp2 are lost from the same promoters (Fig. 6D; Supplemental Fig. 4E).
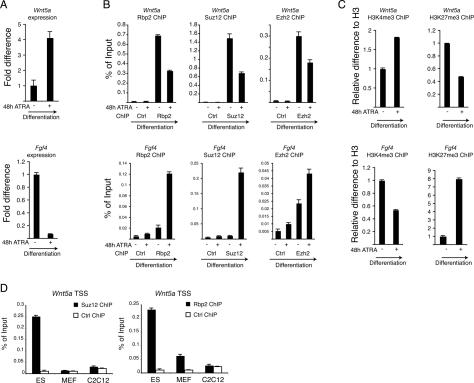
Figure 6.
Regulation of Rbp2 and PRC2 activities during ES cell differentiation. (A) Real-time quantitative expression analysis of Wnt5a and Fgf4 in ES cells before and 48 h after ATRA-induced (1 μM) differentiation. (B) ChIP assay in pluripotent and differentiated ES cells presented in A for Rbp2, Suz12, and Ezh2. ChIPs with antibody against HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal. (C) ChIP analysis as described in A using antibodies for histone H3, H3K4me3, and H3K27me3. ChIP enrichments for H3K4me3 and H3K27me3 are normalized to histone H3 density. (D) ChIP assays for Suz12 and Rbp2 in mouse ES, MEF, and C2C12 cells. ChIPs with antibody against the HA tag served as negative control. ChIP enrichments are presented as percentage (%) of bound/input signal.
Previously, we have also shown that the PRC2 complex is recruited to repress the expression of specific genes during ES cell differentiation (Pasini et al. 2007). To understand if Rbp2 is recruited together with the PRC2 complex to promoters that are repressed during ES cell differentiation, we performed ChIP analysis on the Fgf4 and Otx2 promoters. As shown in Figure 6A–C and Supplemental Figure 4A–C, Rbp2 is recruited together with the PRC2 complex to these promoters during ES cell differentiation, and the recruitment correlates with increased H3K27me3 levels, reduced H3K4me3 levels, and Fgf4 and Otx2 repression.
Taken together, these results demonstrate a functional role for the PRC2 complex and Rbp2 for the coordinated regulation of the histone methylation state of their target promoters during ES cell differentiation.
Discussion
A model for repression
In this study, we show that Rbp2 (1) binds to a significant number of PcG target genes in mouse ES cells, (2) binds to the PRC2 complex, (3) is a critical regulator of PRC2 target genes, and (4) is recruited to its target genes by the PRC2 complex during ES cell differentiation.
PcG proteins regulate the expression of a large number of factors that determine cell fate decisions during development (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006). The PcG proteins are required to maintain the silencing of specific target genes in ES cells, and it is believed that the correct transcriptional activation of a subset of these target genes upon ES cell differentiation is achieved, at least in part, through the dissociation of the PRC2-repressive activity from promoters (Boyer et al. 2006). In addition to this, we have previously shown that the PRC2 complex can also be “actively” recruited to target genes during differentiation to silence the expression of specific genes (Pasini et al. 2007). These results suggest that the PRC2 complex does not play only a “passive” role during differentiation and are consistent with the broad expression pattern of Ezh2 during development (O’Carroll et al. 2001).
Our results show that the Rbp2 H3K4 demethylase is important for the correct regulation of PcG target genes. Taken together with previously published data, we have summarized our findings in two models (Fig. 7): In the “derepression model,” Rbp2 together with PRC2 contributes to the silencing of target genes in ES cells. Differentiation signals lead to their dissociation and recruitment of transcriptional activators such as H3K4 histone methyltransferases and H3K27 demethylases (Agger et al. 2007; M.G. Lee et al. 2007b). In the “repression model,” PRC2 and Rbp2 are recruited to their target promoters during ES cell differentiation, while transcriptional activators are dissociated. In other words, our results show that methylation of H3K27 and demethylation of H3K4 are coordinately regulated by PRC2 and Rbp2 to maintain transcriptional repression, whereas previous results have shown the coordinated regulation of H3K4 methylation and H3K27 demethylation to maintain transcriptional activation. The mechanisms leading to the recruitment of PRC2 to target genes are still unknown. The fact that Rbp2 is not completely displaced from target genes in the absence of the PRC2 complex suggests, as is highlighted in the models (Fig. 7), that other factors may be involved in recruiting and or maintaining Rbp2 and PRC2 associated to target genes.
Figure 7.
Models for Rbp2–PRC2-repressive mechanisms during ES cell differentiation.
Rbp2 interaction with PRC2
In this work, we show that endogenous levels of Rbp2 and PRC2 interact and that Rbp2 and members of the PRC2 complex coelute and interact in higher molecular weight fractions (≥1 MDa). Moreover, we show that RBP2 interacts with high affinity with the PRC2 complex when coexpressed in insect cells. Taken together, our results suggest that a substantial fraction of Rbp2 binds to PRC2 and that a substantial fraction of PRC2 binds to Rbp2. However, these data suggest that Rbp2 is not a core subunit of the PRC2 complex.
Implication of Rbp2 recruitment at ‘bivalent domains’
Interestingly, it has recently been proposed that genes involved in cell fate decisions during embryonic development are featured by the coexistence of H3K27me3 and H3K4me3 marks, and that this correlates with a poised repressed state in ES cells (Bernstein et al. 2006). This “bivalent mark” may provide plasticity to the gene; i.e., the gene can either be activated or maintained repressed in specific cell lineages during differentiation. The fact that both PRC2 and Rbp2 associate with these “bivalent domains” could appear counterintuitive, but our data, together with previous publications showing that the H3K4me3 MLL methyltransferase complex specifically associates with the H3K27me3 UTX demethylase (Agger et al. 2007; Lan et al. 2007; M.G. Lee et al. 2007b), suggest that the cooperation between KMTs and KDMs is a common feature in epigenetic regulation of transcription. The fact that Rbp2 associates with promoters enriched for H3K4me3 and that UTX associates with promoters enriched for H3K27me3 (Agger et al. 2007; Lan et al. 2007; M.G. Lee et al. 2007b) strongly suggests that these activities are not present to completely “erase” the methylation marks from promoters, but instead are required for “fine-tuning” the regulation of the transcriptional activity of the target genes. Consistent with this, the complete loss of specific modifications during different cellular processes does not appear to be solely dependent on an “active” process catalyzed by demethylases, but to a high degree are also dependent on the specific loss of KMT activities, as documented for the H3K27me3 mark during replicative senescence of MEFs and differentiation of myotubes (Caretti et al. 2004; Bracken et al. 2007).
Rbp2 recruitment at target genes
Interestingly, our results showed that Rbp2 binding to target genes is dependent on the PRC2 complex. Moreover, using a heterologous reporter system, we show that the PRC2 complex can direct recruitment of Rbp2 to target genes. Consistent with this, Rbp2 association and displacement from promoters during ES cell differentiation always correlate with PcG binding. Rbp2 contains an Arid/Bright domain that potentially retains the ability to directly bind DNA sequences. The DNA-binding specificity of this domain is promiscuous, and while for some proteins it was shown to have preference for AT-rich motifs (Gregory et al. 1996), this specificity was not observed for other proteins (Herrscher et al. 1995; Huang et al. 1996; Whitson et al. 1999). In addition, recent studies have shown that the Arid domain of JARID1B/PLU1 has a preferential affinity for GC-rich motifs (Scibetta et al. 2007), suggesting that Arid domains have DNA-binding affinities with either low or no conserved sequences. The fact that Rbp2 down-regulation does not affect PRC2 binding, and that Rbp2 is displaced from promoters in the absence of PcG proteins, suggests that the putative DNA-binding ability of Rbp2 may not be required for PRC2 and Rbp2 recruitment to target genes. However, it is possible that several factors contribute to the recruitment and stabilization of the PRC2–Rbp2 complex to promoters such as unidentified transcription factors, the affinity of the Arid domain for DNA, the ability of RbAp48/46 (a subunit of the PRC2 complex) to bind histone tails (Verreault et al. 1998), and the affinity of the PRC2 complex for H3K27me3 (K.H. Hansen, A.P. Bracken, D. Pasini, N. Dietrich, A. Monrad, and K. Helin, in prep.).
Rbp2 and development
H3K4 demethylase activity is essential for the development of a number of different organisms such as Drosophila and Caenorhabditis elegans (Gildea et al. 2000; Christensen et al. 2007). The fact that Rbp2 knockout mice are viable and display only minor defects in the hematopoietic system (Klose et al. 2007) suggests that three other members of the Jarid1 family compensate for the lack of Rbp2. Therefore, it would be interesting to investigate if other Jarid1 proteins play a role in the regulation of PcG target genes. That this is a likely scenario is suggested by the fact that SMCY (JARID1D), like Rbp2 (Supplemental Table 1) and the PRC2 complex (Boyer et al. 2006), associates with the Engrailed promoter and represses its expression through association with the Ring6a/MBLR Polycomb-like protein (M.G. Lee et al. 2007a).
PcG proteins are essential for development and regulate important cellular and epigenetic processes such as proliferation, differentiation, X-chromosome inactivation, and imprinting (Delaval and Feil 2004; Pasini et al. 2004a; Heard and Disteche 2006; Schwartz and Pirrotta 2007). The identification of Rbp2 as a functional important interaction partner of PRC2 provides important knowledge as to how two distinct enzymatic activities regulate transcription in ES cells. However, many important and challenging questions remain to be addressed in the future, such as the identification of the proteins that mediate the recruitment of Rbp2 and PRC2, the stimuli that can modulate their activities, and the mechanisms by which histone post-translational modifications affect transcription.
Materials and methods
Cell culture
293T, MEF, and C2C12 cells were grown in DMEM (Gibco) supplemented with 10% FBS (Gibco), Glutamax (Gibco), and Pen/Strep (Gibco). Mouse ES cells were grown on 0.1% gelatin-coated (Sigma) tissue culture plates (Nunc) in ES cell media as described (Pasini et al. 2007) in the presence of 103 U/mL LIF (ESGRO). To induce ES cell differentiation, 1.5 × 106 cells per 10-cm dish were plated and induced to differentiate 16 h later by removal of LIF and stimulation with 1 μM All Trans Retinoic Acid (ATRA) (Sigma) for the indicated times. Gal4-EED-inducible 293T cells are described elsewhere (K.H. Hansen, A.P. Bracken, D. Pasini, N. Dietrich, A. Monrad, and K. Helin, in prep.). The expression of Gal4-EED was induced by the addition of 1 μg/mL tetracycline (Sigma) to the cell media. For the siRNA experiment, the GAL4-EED and parental 293T cells clones were transfected with oligofectamine (Invitrogen) in the presence of 50 nM Cyclophilin B (Dharmacom) or RBP2 (GCACAAGGAT GAACATTCT) siRNA oligos. Cells were transfected using the same conditions 48 h after the first transfection and supplemented with 40 ng/mL tetracycline (Sigma). Cells were harvested 48 h later and analyzed both for protein expression and for Luciferase activity with Passive Lysis Buffer (Promega).
Plasmids
shRNA constructs with the target sequences Rbp2 (5′-GAAGT TAGCTAAAGAAGAA-3′) and Suz12 (5′-GCTGTTACCAA GCTCCGAG-3′) were generated as described (Ivanova et al. 2006). pCMVHA-RBP2 wild type and pCMVHA-RBP2 (H483G/ E485Q) mutant have been described (Christensen et al. 2007).
Lentiviral transduction
Lentiviral particles were produced in 293FT cells grown in ES media. Medium containing viral particles was filtered (0.45 μm; Millipore) and supplemented with 10 μg/mL Polybrene (Sigma) and 103 U/mL LIF (ESGRO). ES cells were transduced with lentiviral particles for 16 h, washed, incubated, and selected with 200 μg/mL Hygromycin B (Invitrogen) 24 h after the transduction.
RNA preparation and qPCR
RNA was extracted from whole-cell pellets using the RNeasy extraction kit (Qiagen) according to the manufacturer’s recommendation. RNA was retrotranscribed using the TaqMan RT–PCR kit (Applied Biosystem). qPCR was carried out with 5–10 ng of retrotranscribed RNA per reaction.
qPCR primers
Primers for qPCR were designed using Primer Express software with an optimal annealing temperature of 60°C. Primer sequences are available as Supplemental Table 2.
ChIP
ChIP assays were performed as described (Pasini et al. 2007).
Genome-wide location analysis
Sample preparation for genome-wide location analysis was performed as described (Bracken et al. 2006). In brief, ChIP DNA was amplified by ligation-mediated PCR and purified with a PCR purification kit (Qiagen). DNA was hybridized on a mouse promoter array (MM8 RefSeq promoter) manufactured by NimbleGen Systems, Inc. Sample labeling, hybridization, and data analysis were carried out by NimbleGen Systems, Inc. Database handling and statistical analysis were carried out with GeneSpring software (Agilent).
Antibodies
The following antibodies were used for immunoblotting: rabbit anti-Rbp2 (Christensen et al. 2007), mouse anti-Ezh2 (BD43) (Pasini et al. 2004b), mouse anti-EED (Bracken et al. 2003), rabbit anti-SUZ12 (Upstate Biotechnologies), rabbit anti-β-Tubulin (Santa Cruz Biotechnologies); rabbit anti-hemagglutinin (HA) (BabCo); rabbit anti-Gal4 (Santa Cruz Biotechnologies).
The following antibodies were used for IP analysis: mouse anti-HA (12CA5), mouse anti-EZH2 (AC22) (Bracken et al. 2006); mouse anti-SUZ12 (2AO9) (Villa et al. 2007); rabbit anti-RBP2 (Christensen et al. 2007); rabbit anti-HA (Y11) (Santa Cruz Biotechnologies).
The following antibodies were used for ChIP analysis: rabbit anti-Rbp2 (Christensen et al. 2007), rabbit anti-SUZ12 (Abcam), mouse anti-EZH2 (AC22) (Bracken et al. 2006); rabbit anti-histone H3 (Abcam); rabbit anti-H3K4me3 (Abcam); rabbit anti-H3K27me3 (Upstate Biotechnologies); rabbit anti-HA (Y11) (Santa Cruz Biotechnologies).
IP assays and size exclusion chromatography
IP assays were performed in IP(150) buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5% glycerol, 0.2% Igepal [Sigma], Aprotein, Leupeptin, 100 mM PMSF, 1 mM DTT). Ethidium bromide (20 μg/μL) was added when indicated. Size exclusion chromatography was performed with ES cell nuclear extracts in a Superose-6 10/300 gel filtration column (GE Healthcare) on an AKTA purifier system (GE Healthcare) in IP(300) buffer (50 mM Tris-HCl at pH 7.5, 300 mM NaCl, 5% glycerol, 0.2% Igepal [Sigma], Aprotein, Leupeptin, 100 mM PMSF, 1 mM DTT).
Recombinant protein expression
Generation of baculoviruses for RBP2, EZH2, SUZ12, and EED has been described (Pasini et al. 2004b; Christensen et al. 2007). Recombinant proteins were produced in Sf9 cells infected with equal amounts of viral supernatants for RBP2, EZH2, SUZ12, and EED.
Acknowledgments
We thank Natalia Ivanova for providing the lentiviral vector for expressing the shRNAs. We thank members of the Helin laboratory for technical advice and support. D.P. was supported by a post-doctoral fellowship from the Danish Medical Research Council and P.A.C.C. was supported by a grant from the Benzon Foundation. The work in the Helin laboratory was supported by grants from the Danish Cancer Society, the Novo Nordisk Foundation, the Danish Medical Research Council, the Danish Natural Science Research Council, the Danish National Research Foundation, and the International Association for Cancer Research.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.470008.
References
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K., Christensen J., Cloos P.A., Helin K. The emerging functions of histone demethylases. Curr. Opin. Genet. Dev. 2008 doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya E.V., Murray H.L., Branton P., Young R.A., Kaelin W.G. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., et al. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes & Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M., et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Delaval K., Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., Lee M.G., Schneider J., Ilvarsonn A., Shiekhattar R., Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Bio. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- Faust C., Schumacher A., Holdener B., Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Francis N.J., Kingston R.E. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- Francis N.J., Kingston R.E., Woodcock C.L. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Gildea J.J., Lopez R., Shearn A.F. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S.L., Kortschak R.D., Kalionis B., Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 1996;16:792–799. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Disteche C.M. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes & Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Herrscher R.F., Kaplan M.H., Lelsz D.L., Das C., Scheuermann R., Tucker P.W. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: A B cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- Huang T.H., Oka T., Asai T., Okada T., Merrills B.W., Gertson P.N., Whitson R.H., Itakura K. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H.H., Whetstine J.R., Bonni A., Roberts T.M., Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D.G., Zhang Y., Kaelin W.G. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.G., Norman J., Shilatifard A., Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007a;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Di Croce L., Shiekhattar R. Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science. 2007b;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lee N., Zhang J., Klose R.J., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat. Struct. Mol. Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery N.D., Yee D., Chen A., Kalantry S., Chamberlain S.J., Otte A.P., Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- O’Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004a;3:396–400. [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004b;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L. Polycomb comes of age: Genome-wide profiling of target sites. Curr. Opin. Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Saurin A.J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Scibetta A.G., Santangelo S., Coleman J., Hall D., Chaplin T., Copier J., Catchpole S., Burchell J., Taylor-Papadimitriou J. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol. Cell. Biol. 2007;27:7220–7235. doi: 10.1128/MCB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L., Eisenman R.N. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Raible F., Mollaaghababa R., Guyon J.R., Wu C.T., Bender W., Kingston R.E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat. Rev. Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Simon J.A., Tamkun J.W. Programming off and on states in chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Mei P., Fang R., Leonor T., Rutenberg M., Shimizu F., Li J., Rao A., Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P.D., Kobayashi R., Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Villa R., Pasini D., Gutierrez A., Morey L., Occhionorelli M., Vire E., Nomdedeu J.F., Jenuwein T., Pelicci P.G., Minucci S., et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Whitson R.H., Huang T., Itakura K. The novel Mrf-2 DNA-binding domain recognizes a five-base core sequence through major and minor-groove contacts. Biochem. Biophys. Res. Commun. 1999;258:326–331. doi: 10.1006/bbrc.1999.0643. [DOI] [PubMed] [Google Scholar]
- Yamane K., Tateishi K., Klose R.J., Fang J., Fabrizio L.A., Erdjument-Bromage H., Taylor-Papadimitriou J., Tempst P., Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]