Abstract
Brown fat is a specialized tissue that can dissipate energy and counteract obesity through a pattern of gene expression that greatly increases mitochondrial content and uncoupled respiration. PRDM16 is a zinc-finger protein that controls brown fat determination by stimulating brown fat-selective gene expression, while suppressing the expression of genes selective for white fat cells. To determine the mechanisms regulating this switching of gene programs, we purified native PRDM16 protein complexes from fat cells. We show here that the PRDM16 transcriptional holocompex contains C-terminal-binding protein-1 (CtBP-1) and CtBP-2, and this direct interaction selectively mediates the repression of white fat genes. This repression occurs through recruiting a PRDM16/CtBP complex onto the promoters of white fat-specific genes such as resistin, and is abolished in the genetic absence of CtBP-1 and CtBP-2. In turn, recruitment of PPAR-γ-coactivator-1α (PGC-1α) and PGC-1β to the PRDM16 complex displaces CtBP, allowing this complex to powerfully activate brown fat genes, such as PGC-1α itself. These data show that the regulated docking of the CtBP proteins on PRDM16 controls the brown and white fat-selective gene programs.
Keywords: PRDM16, adipogenesis, CtBP, PGC-1, PPAR-γ, resistin
Adipose tissues in mammals occur in two types: white and brown. White adipose tissue (WAT) is specialized for the storage of excess energy, containing all of the enzymatic machinery necessary to build triglycerides from fatty acids synthesized de novo or from fatty acids imported from circulating lipoproteins. In addition, it is now appreciated that WAT plays a central role in energy balance through the synthesis and secretion of molecules that control various aspects of systemic metabolism (Rosen and Spiegelman 2006). Several adipokines, including TNFα, leptin, adiponectin, resistin, and angiotensinogen, are produced and released mainly or entirely from WAT in various physiological or pathophysiological states. Circulating levels of these adipokines correlate with obesity, insulin resistance (TNFα and resistin), and hypertension (angiotensinogen) observed in metabolic syndrome (Engeli et al. 2000; Massiera et al. 2001; Steppan et al. 2001; Banerjee et al. 2004; Steppan and Lazar 2004; Hotamisligil 2006). Leptin is also increased in obesity, where it can act to restrain further food intake and increase energy expenditure, and to improve insulin sensitivity in muscle or liver (Halaas et al. 1995; Pelleymounter et al. 1995; Friedman 2002).
In contrast, brown adipose tissue (BAT) is specialized to dissipate chemical energy in the form of heat in response to cold or excess feeding. Due to its remarkable oxidative capacity, modulation of BAT metabolism influences whole body energy balance, and counteracts obesity (Dulloo and Miller 1984; Lowell et al. 1993; Guerra et al. 1998; Lowell and Spiegelman 2000; Spiegelman and Flier 2001; Almind et al. 2007). This thermogenic function of BAT is due to a pattern of gene expression that results in a high mitochondrial content, and elevated cellular respiration that is largely uncoupled from ATP synthesis. This uncoupling occurs through the expression of UCP-1 (uncoupling protein-1), a brown fat-specific mitochondrial protein that promotes proton leak across the inner mitochondrial membrane. Unlike WAT, BAT is not believed to play an important role in systemic signaling via secretion of adipokines. In part, this is due to a substantially smaller mass of BAT compared with WAT, and the inability of BAT to express all of the adipokines (Cannon and Nedergaard 2004).
The transcriptional control of the adipocyte lineage has been studied extensively (Rosen and Spiegelman 2000; Farmer 2006; Gesta et al. 2007). PPARγ, a member of the nuclear hormone receptor superfamily, plays a dominant role in the differentiation of both white and brown fat cells (Tontonoz et al. 1994; Barak et al. 1999; Rosen et al. 1999; He et al. 2003; Nedergaard et al. 2005). Several C/EBP (CCAAT/enhancer-binding protein) members also play important roles, primarily in activating and maintaining the expression of PPARγ. PPARγ and C/EBPs cross-regulate each other’s expression and represent a transcriptional loop that maintains a stable differentiated state of the adipocyte (Wu et al. 1999; Rosen et al. 2002). Although PPARγ is required for differentiation of both white and brown fat cells, expression of the PPARγ or C/EBP factors in mesenchymal cells induces only a white, not brown fat phenotype.
Several transcription-related molecules have been reported to influence the brown or white fat phenotype selectivity including Rb (retinoblastoma protein), p107, TIF2 (transcriptional intermediary factor 2), 4E-BP1 (4E-binding protein1), and RIP140 (Tsukiyama-Kohara et al. 2001; Picard et al. 2002; Hansen et al. 2004; Leonardsson et al. 2004; Scime et al. 2005; Hansen and Kristiansen 2006; Powelka et al. 2006). These factors all act negatively to suppress the brown fat phenotype. A forkhead transcription factor family FOXC2, on the other hand, induces the emergence of brown fat cells in WAT in vivo. However, this factor has not been shown to work in a cell-autonomous fashion, and most likely induces a browning of white fat by inducing a state of chronic β adrenergic signaling, rather than working directly on cell type determination (Cederberg et al. 2001). PGC-1α (PPAR-γ-coactivator-1α) is highly induced in BAT of mice during cold exposure, and activates the thermogenic gene program of brown fat, including UCP-1, Dio2 (type 2 deiodinase), and PGC-1α itself (Puigserver et al. 1998; Tiraby et al. 2003). Genetic ablation of PGC-1α, however, has shown that this molecule does not determine the identity of brown fat cells (Lin et al. 2004; Uldry et al. 2006).
Most recently, PRDM16 has been shown to induce the brown fat phenotype, either from white fat precursors or fibroblasts expressing PPAR-γ. Importantly, it does this in a cell-autonomous manner (Seale et al. 2007). PRDM16 is a 140 kDa zinc-finger protein-containing PR-(PRD1-BF-1-RIZ1 homologous) domain. It was originally identified at a chromosomal breakpoint in human myeloid leukemia cells (Mochizuki et al. 2000; Nishikata et al. 2003). It is selectively expressed in brown fat relative to white fat cells (Seale et al. 2007). In cultured cells, PRDM16 turns on a large array of brown fat-selective genes, including PGC-1α and UCP-1. PRDM16 expression also represses a number of genes that are characteristic of white fat, including the adipokine resistin. Importantly, transgenic expression of PRDM16, driven by the aP2 promoter in white fat cells, results in the emergence of brown fat pockets within the fat depots that would otherwise be WAT. Hence, PRDM16 directs the white adipocyte lineage toward brown fat in vivo, by activating a robust pattern of brown fat gene program and simultaneously suppressing the white fat gene program (Seale et al. 2007).
PRDM16 has been shown to bind directly to a specific DNA sequence via two sets of zinc fingers (Nishikata et al. 2003). Intriguingly, abrogation of DNA binding by introducing a mutation in a zinc-finger domain does not substantially alter the ability of PRDM16 to induce the brown fat phenotype, when compared with the wild-type protein (Seale et al. 2007). Further study suggests that PRDM16 directly binds to both PGC-1α and PGC-1β, and increases their transcriptional activities. These results suggest that PRDM16 is a coregulatory protein that can function as a molecular switch in brown and white fat development, apparently through multiple protein–protein interactions. How PRDM16 simultaneously stimulates both gene activation and gene repressive events, however, is not clear. To address this question, we purified the native PRDM16 transcriptional holocomplex from fat cells. We have identified the C-terminal-binding proteins (CtBPs) as corepressor proteins that directly bind to PRDM16 in a regulated fashion. This interaction with CtBPs specifically mediates the suppressive effects of PRDM16 on white fat-selective gene expression. Notably, PGC-1α and PGC-1β can displace the CtBPs to form a PRDM16/PGC-1 complex that allows positive activation of brown fat-selective genes.
Results
PRDM16 transcriptional holocomplex contains CtBP-1 and CtBP-2
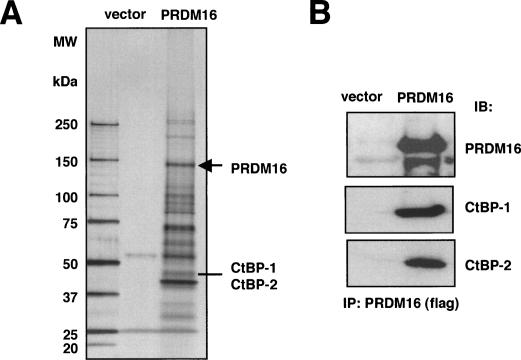
To investigate mechanisms used by PRDM16 to control the program of brown and white fat-selective gene expression, we purified the native PRDM16 transcriptional complex to homogeneity from fat cells stably expressing a Flag-tagged protein. We have shown previously that expression of PRDM16 in fibroblasts, together with PPARγ, is sufficient to stimulate the brown fat gene program and suppress white fat-selective gene expression (Seale et al. 2007). At day 4 of differentiation in these cultures, the PRDM16 holocomplex was purified from the nuclear extracts, and separated by SDS-PAGE followed by silver staining (Fig. 1A). Several prominent bands were observed, and by a combination of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometric fingerprinting and MALDI-TOF/TOF mass spectrometric sequencing, we identified CtBP-1 and CtBP-2 in high amounts relative to PRDM16. In addition, several other proteins were identified, which will be described elsewhere. Our attention here is focused on CtBP-1 and CtBP-2 because (1) the interaction of PRDM16 and CtBPs is near stoichiometric, and (2) CtBPs are known as potential coregulatory factors, often with transcriptional repressor activity (Chinnadurai 2007). To confirm this interaction, we immunoprecipitated PRDM16 from brown fat cells and detected endogenous CtBP-1 and CtBP-2 by Western blotting (Fig. 1B).
Figure 1.
Purification and identification of CtBP-1 and CtBP-2 in the native PRDM16 transcriptional complex. (A) Silver staining of the PRDM16-associated proteins. Nuclear extracts prepared from adipocytes expressing either vector (left lane) or PRDM16 (right lane) were purified using Flag M2 agarose. After extensive washing, the PRDM16 complex was eluted with 3xFlag peptide and separated by 4%–12% gradient SDS-PAGE. (B) PRDM16 complex was immunopurified from the immortalized brown fat cells and separated by SDS-PAGE. Endogenous CtBP-1 and CtBP-2 were detected by Western blotting.
PRDM16 directly interacts with CtBP-1 and CtBP-2 through its PLDLS motif
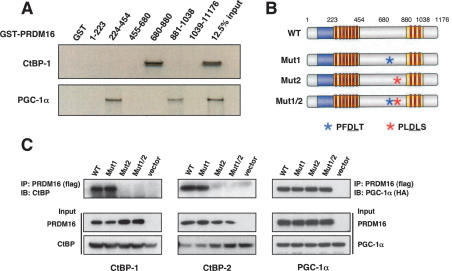
Structural aspects of the interaction between PRDM16 and CtBPs were further examined in vitro, using various GST fusions of PRDM16 fragments. As shown in Figure 2A, one region of PRDM16 (amino acids 680–1038) bound strongly to in vitro-translated CtBP-1. Similar results were observed with CtBP-2 (data not shown). In contrast, two domains of PRDM16 (224–454; zinc-finger domain at the N terminus and 881–1038; zinc-finger domain at the C terminus) interacted with PGC-1α (Fig. 2A, bottom panel). Within the 680–1038 region of PRDM16, there are two putative sequences (PFDLT at 774–778 and PLDLS at 804–808), which have been identified previously as motifs that bind CtBP in a PR domain-containing protein, EVI-1 (Chinnadurai 2007). These two motifs in PRDM16 are evolutionarily conserved in several species including human, chicken, dog, and Xenopus (SupplementalFig. S1).
Figure 2.
PRDM16 directly interacts with CtBP-1 and CtBP-2 through its PLDLS motif. (A) Full-length CtBP-1 (top) or PGC-1α (bottom) were 35S-labeled by in vitro translation and incubated with various GST fusion fragments of PRDM16. GST beads were washed and separated by 4%–12% gradient SDS-PAGE. (B) Schematic illustration of wild-type PRDM16 (WT) and mutant forms of PRDM16. PFDLT was mutated to PFAST in Mut1, PLDLS was mutated to PLASS in Mut2, and both motifs were mutated in Mut1/2 by site-directed mutagenesis. The blue box represents the PR domain, and the red boxes show the zinc fingers of PRDM16. (C) Flag-tagged PRDM16 was transiently expressed along with CtBP-1 (left), CtBP-2 (middle), or PGC-1α (right) in COS-7 cells. PRDM16 was immunoprecipitated using Flag M2 agarose, separated by SDS-PAGE, and CtBP-1, CtBP-2, or PGC-1α was detected by Western blotting. The inputs of each assay are shown in the bottom panels.
To determine whether the binding of PRDM16 to CtBP is required for the action of PRDM16 in the program of brown fat determination, we created three mutant forms of PRDM16 in which potential CtBP-binding motifs were disrupted. As illustrated in Figure 2B, two putative CtBP-binding motifs at 774–778 (PFDLT) and at 804–808 (PLDLS) were mutated to PFASL (Mut1) and PLASS (Mut2), respectively, using site-directed mutagenesis. Both motifs were mutated in Mut1/2. Wild-type and mutant forms of PRDM16 were transiently expressed in COS-7 cells to test their ability to interact with coexpressed CtBP-1 or CtBP-2. Coimmunoprecipitation assays showed that wild-type PRDM16 and Mut1 interacted normally with CtBP-1 and CtBP-2. However, Mut2 and Mut1/2 did not bind CtBPs (Fig. 2C, left panel for CtBP-1 and middle panel for CtBP-2). Therefore, the PLDLS at 804–808 is required for the physical interaction between PRDM16 and the CtBPs. Importantly, these mutant forms of PRDM16 still interacted with PGC-1α (Fig. 2C, right panel) with similar affinity as the wild-type protein, suggesting that these mutations did not significantly impact the overall architecture of the protein.
The interaction between PRDM16 and CtBPs mediates the repression of the white fat-selective gene program
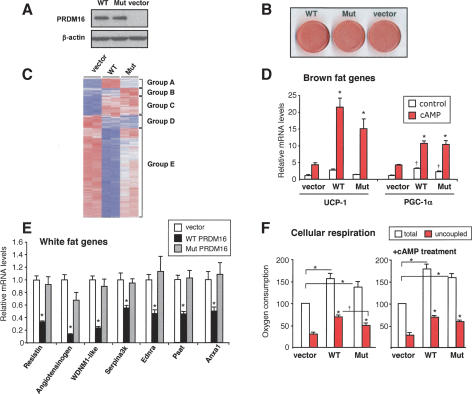
To examine the functional role of the interaction between PRDM16 and the CtBPs in the brown fat gene program, wild-type and the CtBP-binding-deficient mutant of PRDM16 (Mut2) were ectopically expressed in 3T3-F442A cells, an authentic white fat cell line. Both proteins were expressed at equivalent levels with retroviral vectors (Fig. 3A). At day 6 of differentiation, nearly 100% of the cells showed a fully differentiated phenotype with accumulation of copious lipid droplets; no morphological differences were observed between cells infected with the wild-type and mutant PRDM16 vectors (Fig. 3B). In addition, the expression levels of three genes common to both white and brown fat cells, adiponectin, PPARγ, and aP2, were similar between these groups (Supplemental Fig. S2A). Thus, ectopic expression of wild-type or mutant PRDM16 did not alter the adipogenic program of 3T3-F442A cells per se.
Figure 3.
PRDM16/CtBP interaction mediates the repression of the white fat-selective gene program. 3T3-F442A cells expressing retroviral wild-type PRDM16, CtBP-binding-deficient mutant PRDM16 (Mut2), or an empty vector, were differentiated to mature adipocytes. (A) Protein expression of wild-type or mutant PRDM16 detected by Western blotting. (B) Oil-red O staining of 3T3-F442A cells at day 6 of differentiation-expressing wild-type PRDM16 (WT), mutant PRDM16 (Mut), or vector control (vector). (C) Microarray analysis of the differentiated cells expressing vector (left), wild-type PRDM16 (WT, middle), or mutant PRDM16 (Mut, right). See the text for details on each group. (D) mRNA levels of brown fat-selective genes (UCP-1 and PGC-1α) analyzed by real-time PCR. The cells were treated with or without cAMP (forskolin, 10 μM) for 4 h. (*) P < 0.01 relative to control with cAMP treatment; (†) P < 0.05 relative to control without cAMP treatment. (E) mRNA levels of white fat-selective genes (as indicated). (F) Cellular respiration of the differentiated 3T3-F442A cells expressing wild-type PRDM16, mutant PRDM16, or vector control. Total mitochondrial oxygen consumption and uncoupled respiration were measured from cells with or without cAMP treatment (0.5 mM dibutyryl cAMP) for 12 h (n = 4). (*) P < 0.01; (†) P < 0.05.
Next, we used Affymetrix microarray chips to analyze the global patterns of gene expression from the differentiated fat cells transduced with wild-type or mutant PRDM16. As shown in Figure 3C, we identified a total of 231 genes that were significantly elevated or reduced by PRDM16, which could be clustered into five groups: (1) genes induced only by wild-type, but not by mutant PRDM16 (Group A); (2) genes induced only by mutant PRDM16 (Group B); (3) genes induced both by wild-type and by mutant PRDM16 (Group C); (4) genes reduced by both wild-type and mutant PRDM16 (Group D); and (5) genes reduced only by wild-type PRDM16 (Group E). For instance, UCP-1 and PGC-1α, critical components of the thermogenic program in brown fat and major gene targets of PRDM16, were similarly induced by both wild-type and mutant PRDM16 (Group C) (Fig. 3D). In particular, these genes are highly cyclic AMP (cAMP)-responsive (Uldry et al. 2006; Seale et al. 2007). Forskolin treatment (10 μM) for 4 h significantly enhanced the ability of PRDM16 to induce the expression of both genes, with no statistical difference between the wild-type and mutant-expressing cells. Cytochrome c oxidase subunit 8b (Cox8b) and cell death-inducing DNA fragmentation factor, α subunit-like effector A (Cidea) are also BAT-selective genes and have been shown to be positively regulated by PRDM16 (Seale et al. 2007). Both wild-type and mutant PRDM16 significantly induced these genes compared with vector control; however, the mutant had a significantly lower potency (Supplemental Fig. S2B).
White fat cells express a specific gene program that is either not expressed or significantly reduced in brown fat cells. Microarray data analysis, combined with a previous data set comparing WAT and BAT tissues and cells (Seale et al. 2007), lead to the identification of 43 WAT-enriched genes (more than fivefold) that are suppressed by PRDM16 (Supplemental Table S1). We validated the expression of 12 of these genes by quantitative real-time PCR (n = 6) (Supplemental Fig. S3). Strikingly, nearly this entire set (41 out of 43, 95.3%) of WAT-selective genes was not suppressed by the mutant PRDM16 that does not bind CtBPs (Group E). For example, resistin, a well-known WAT-selective adipokine that promotes insulin resistance (Banerjee et al. 2004), was significantly repressed by wild-type PRDM16 by 70%. However, the CtBP-binding-deficient mutant of PRDM16 completely failed to suppress resistin gene expression (Fig. 3E). Protein levels of resistin released from cells also followed this pattern (Suppemental Fig. S4). Angiotensinogen, a critical component of the renin-angiotensin system, also belongs to the set of highly WAT-selective genes (86-fold enrichment in WAT vs. BAT). Wild-type PRDM16 strongly repressed angiotensinogen gene expression by 90%, but this repression was greatly reduced in cells expressing the mutant PRDM16 (30% reduction). In addition to the previously reported WAT-selective genes, such as Serpina3k and Psat, several newly identified WAT-selective genes such as endothelin receptor type A (Ednra), WDNM1-like protein, and annexin A1 (Anxa1) were also repressed by PRDM16 in a CtBP-dependent manner. We also found that 26 genes were repressed by both wild-type and by mutant PRDM16 with similar efficacy (CtBP-independent repression). Among them, only two genes (endonuclease domain-containing 1 and procollagen type Vα1) were WAT-selective. Similar results were obtained in mouse embryonic fibroblasts (MEFs) expressing either wild-type or mutant PRDM16 together with PPARγ2 (Supplemental Fig. S5). Altogether, these data suggest the requirement for a PRDM16/CtBP complex for the appropriate silencing of critical white fat-selective genes.
An important characteristic of brown fat cells is their extraordinarily high rates of respiration, particularly uncoupled respiration. We asked if the disrupted suppression of the white fat gene program by the mutation in PRDM16 affected this aspect of brown fat cell physiology. We therefore measured oxygen consumption in the differentiated 3T3-F442A cells expressing either wild-type or the Mut2 mutant of PRDM16. As shown in Figure 3F, wild-type and mutant PRDM16 robustly increased total respiration (by 57% and 39%, respectively) and uncoupled respiration by 2.2- and 1.7-fold, respectively, relative to control cells (n = 4, P < 0.001). There was a small but statistically significant difference in uncoupled respiration between cells expressing wild-type and mutant PRDM16 (P = 0.046). When the cells were stimulated for 12 h with 0.5 mM dibutyryl cAMP, cells expressing both wild-type and mutant PRDM16 significantly increased total respiration (by 68% and 52%, respectively) and also showed a large increase (2.5- and 2.2-fold) in uncoupled respiration, relative to control cells. Thus, the CtBP-binding-deficient mutant of PRDM16 is still able to robustly induce the respiratory function of brown fat cells. These results suggest that the interaction of PRDM16 with CtBPs is not required for the respiratory function of recipient fat cells, but is specifically required for the PRDM16-mediated repression of white fat gene program.
Genetic requirement for the CtBPs in PRDM16-mediated suppression of white fat gene expression
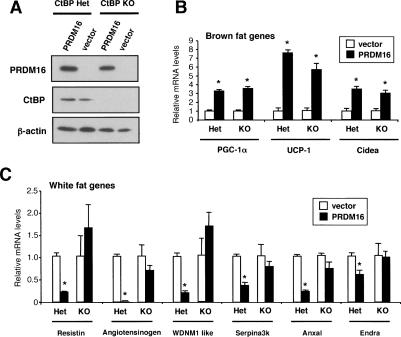
To determine the genetic requirement for CtBPs in PRDM16-mediated gene regulation, we used CtBP-1 and CtBP-2 double-deficient cells. CtBP-1 and CtBP-2 double-deficient mice die by embryonic day 10.5 (E10.5) due to aberrant extraembryonic development (Hildebrand and Soriano 2002), well before the development of adipose tissues (Cinti et al. 1999). We therefore employed MEFs derived from CtBP-1 and CtBP-2 double-deficient mice established by Hildebrand and Soriano (2002). MEFs derived from the double heterozygous embryos (CtBP-1+/− CtBP-2+/−) were used as controls (Hildebrand and Soriano 2002; Grooteclaes et al. 2003). These cells were converted into adipocytes by expression of PPARγ2 in CtBP-1+/− CtBP-2+/− (Het), and CtBP-1−/− CtBP-2−/− (KO) cells. PRDM16 was expressed using retroviral vectors, and Western blotting ensured the equivalent expression of the proteins in cells from both genotypes (Fig. 4A). As shown in Figure 4B, expression of brown fat-selective genes such as PGC-1α, UCP-1, and cidea were induced to similar extent by PRDM16 in both CtBP heterozygous and null cells. By marked contrast, the white fat-selective genes, including resistin and angiotensinogen, were repressed by PRDM16 in the CtBP heterozygous cells, but showed a complete loss of this repression in the genetic absence of CtBPs (Fig. 4C). Similar results for additional white fat-selective genes are shown in Supplemental Figure S6. These data establish a near absolute requirement for CtBPs in repression of white fat gene program by PRDM16.
Figure 4.
Genetic requirement for the CtBPs in the PRDM16-mediated suppression of white fat-selective gene expression. MEFs derived from CtBP-1 and CtBP-2 double-deficient (CtBP-1−/− CtBP-2−/−, KO) or double heterozygous (CtBP-1+/− CtBP-2+/−, Het) embryos stably expressing PRDM16 or vector control together with PPARγ2 were differentiated into mature adipocytes. (A) Protein expression of PRDM16 and CtBP detected by Western blotting. (B) mRNA levels of brown fat-selective genes (UCP-1, PGC-1α, and cidea) analyzed by real-time PCR. (C) mRNA levels of white fat-selective genes (as indicated). (*) P < 0.05.
PRDM16 recruits CtBP onto the promoters of white fat-selective genes
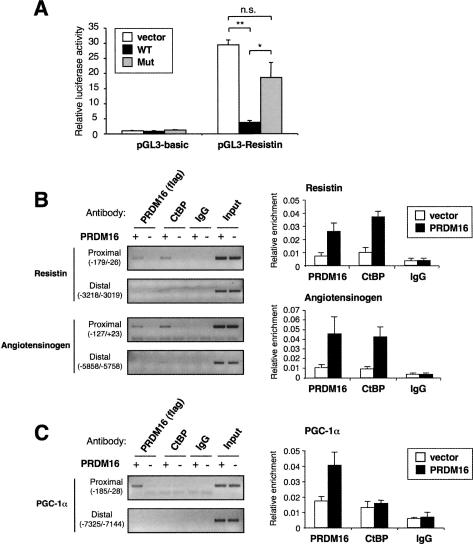
A key question is how the PRDM16/CtBP complex acts to repress expression of the white fat genes. We focused this question particularly on resistin because it is an adipokine with important biological activity that is selectively expressed and secreted by white fat cells. When PRDM16 was transiently expressed in differentiated 3T3-L1 cells, the activity of the resistin promoter was dramatically reduced by 90% (Fig. 5A). Consistent with the results observed with the endogenous mRNAs, the CtBP-binding-deficient mutant of PRDM16 failed to repress the activity of the resistin promoter. This result suggests that the inhibitory effect of PRDM16 on resistin gene transcription is, at least in part, through actions on its promoter.
Figure 5.
PRDM16 recruits CtBP onto the promoters of white fat-selective genes. (A) Transcriptional activity of resistin promoter in response to wild-type PRDM16 (WT), CtBP-binding-deficient mutant PRDM16 (Mut), or vector control in 3T3-L1 cells. Luciferase constructs and each expression plasmids were transfected into the differentiated 3T3-L1 cells by electroporation. After 48 h, cells were harvested and luciferase assay was performed. Each value was normalized by β-galactosidase activity (n = 3). (*) P < 0.05; (**) P < 0.01. (B) ChIP assay showing the recruitment of PRDM16 and CtBP onto the promoters of white fat genes (resistin and angiotensinogen). Differentiated brown fat cells expressing either PRDM16 or vector were immunoprecipitated with antibody against Flag (PRDM16), CtBP, or IgG (negative control). Protein-associated DNA was amplified by primer sets designed for the indicated regions of the promoters. PCR products were separated by 2% agarose gel. Graphs on the right show quantitative results in the proximal promoter regions using real-time PCR. (C) ChIP assay of PRDM16 and CtBP on the PGC-1α promoter.
We next investigated whether PRDM16 and CtBP are recruited to the resistin promoter. Chromatin immunoprecipitation (ChIP) experiments in differentiated brown fat cells showed that PRDM16 and CtBP were both enriched in the proximal region (amplified region by PCR; −179/−26), but not in the distal region (−3218/−3019) of the resistin gene promoter (Fig. 5, top panel). Quantitative results of the proximal region using real-time PCR are shown graphically in the right panel of Figure 5. Angiotensinogen is another white fat-selective gene suppressed by PRDM16, and its mRNA expression is also increased by depletion of endogenous PRDM16 in brown fat cells, indicating angiotensinogen as one of the direct targets of PRDM16 (Supplemental Fig. S7). We found significant coenrichment of PRDM16 and CtBP in the proximal region (−127/+23) of the angiotensinogen gene, but not in the distal region (−5858/−5758) (Fig. 6B, bottom panel). These results indicate that the PRDM16 and CtBP are both recruited to the promoters of the resistin and angiotensinogen genes.
Figure 6.
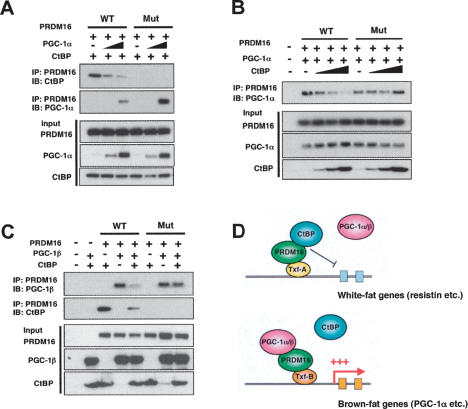
PGC-1α/β and CtBP directly compete for binding to the PRDM16 complex. (A) Fixed amounts of wild-type PRDM16 (WT) or CtBP-binding-deficient mutant PRDM16 (Mut) were expressed along with fixed amounts of CtBP (CtBP-1) and variable amounts of PGC-1α in COS-7 cells. Cell lysates were incubated with Flag M2 agarose (PRDM16) and separated by SDS-PAGE. The interaction of PRDM16 with CtBP or PGC-1α was detected by Western blotting. The inputs are shown in the bottom panels. (B) Fixed amounts of wild-type PRDM16 (WT) or mutant PRDM16 (Mut) were expressed along with fixed amounts of PGC-1α and variable amounts of CtBP. Interaction of PRDM16 with CtBP was detected as described above. (C) Wild-type PRDM16 (WT) or mutant PRDM16 (Mut) were coexpressed with PGC-1β or CtBP, and the interaction of PRDM16 with PGC-1β or CtBP was examined as described above. (D) A schematic model of brown fat determination by PRDM16. PRDM16 represses white fat-selective genes such as resistin by recruiting CtBP onto their promoters. Recruitments of PGC-1α and PGC-1β to PRDM16 trigger the dismissal of CtBP from the PRDM16 complex, leading to the robust activation of brown fat-selective genes.
These results lead to an important question: If CtBP is part of the PRDM16 protein complex, how can PRDM16 powerfully activate brown fat-selective genes? Our previous study has shown that PRDM16 increased the activity of the −2 kb PGC-1α promoter in a manner dependent on the PGC-1α protein (Seale et al. 2007). The autoregulatory action of PGC-1α on its own promoter was shown earlier (Handschin et al. 2003). Thus, we investigated whether PRDM16 is recruited to the proximal regions of the PGC-1α promoter. As shown in Figure 6C, ChIP experiments showed an enrichment of PRDM16 in the proximal region (−185/−28), but not in the distal region (−7325/−7144) of the PGC-1α gene (Fig. 5C). Importantly, however, we did not observe any enrichment of CtBP within the region of the PGC-1α promoter with which PRDM16 is associated, implying that distinct mechanisms were used to induce brown fat gene expressions and to repress white fat-selective gene expression.
PGC-1s and CtBP directly compete for binding to the PRDM16 complex
PGC-1α and PGC-1β, powerful coactivators of oxidative metabolism, mitochondrial biogenesis and mitochondrial uncoupling, are partners of PRDM16 in the activation of brown fat gene expression (Seale et al. 2007). The direct binding of PRDM16 to PGC-1α and PGC-1β robustly increases their transcription activities (Seale et al. 2007). One rather simple idea as to how PRDM16 might activate brown fat and suppress white fat-selective genes is that the CtBPs and the PGC-1α/β might be in direct competition for binding to PRDM16. To test this notion, we conducted competitive binding assays by transiently expressing PRDM16 (wild-type and CtBP-binding-deficient mutant), PGC-1α/β, and CtBP (CtBP-1) in COS-7 cells. First, fixed amounts of PRDM16 and CtBP were coexpressed with variable amounts of PGC-1α, and the PRDM16/CtBP interaction was examined. As shown in Figure 6A, high expression of PGC-1α lead to docking of PGC-1α to PRDM16, and this apparently caused the loss of CtBP from the PRDM16 complex (one to three lanes from left). As expected, mutant PRDM16 bound to PGC-1α, but did not interact with CtBP (Fig. 6A, four to six lanes from left). We did not find any physical interaction between PGC-1α and CtBPs (data not shown). Conversely, when fixed amounts of PRDM16 and PGC-1α were expressed, along with increasing amounts of CtBP, the expression of CtBP displaced PGC-1α from the PRDM16 complexes (Fig. 6B, two to five lanes). Importantly, CtBP did not affect the interaction of the CtBP-binding-deficient mutant of PRDM16 with PGC-1α (Fig. 6B, six to nine lanes). Lastly, we tested the competition between PGC-1β and CtBP for occupancy in the PRDM16 complex. As shown in Figure 6C, PGC-1β also competed with CtBP for docking to wild-type PRDM16, but not to mutant PRDM16. These data strongly suggest that PGC-1α/β and CtBP are mutually exclusive, and directly compete for binding to PRDM16.
Discussion
Despite sharing the ability to accumulate triglycerides, the physiological roles of WAT and BAT are almost diametrically opposite. WAT stores most of the excess energy in mammals and also contributes to systemic energy balance through releasing a variety of adipokines in response to changes in energy status (Rosen and Spiegelman 2006). On the other hand, BAT plays an important role in energy expenditure by dissipating chemical energy in response to cold or excess feeding. BAT is not thought to play a substantial role in systemic signaling both because the mass of this tissue is much smaller than WAT and because not all the adipokines are expressed in BAT (Cannon and Nedergaard 2004). We show here that PRDM16 coordinately represses the expression of a wide range of WAT-selective genes in a CtBP-dependent manner, while simultaneously activating BAT-selective genes responsible for mitochondrial biogenesis and oxidative metabolism.
Notably, PRDM16 directly suppresses at least two important WAT-selective adipokines: resistin and angiotensinogen. Resistin is increased in diet-induced and genetic obesity, and inhibits the action of insulin on hepatic glucose production (Steppan et al. 2001; Banerjee et al. 2004; Steppan and Lazar 2004). Angiotensinogen, a unique substrate of rennin, is a precursor of the vasoactive peptide angiotensin II. WAT is a major extrahepatic synthesis site of angiotensinogen, which shows positive correlation with body mass index and can cause high blood pressure (Engeli et al. 2000; Massiera et al. 2001). In addition to these two well-known WAT-selective molecules, PRDM16 also represses serpina3k, a WAT-selective secreted protein that belongs to serpin (serine protease inhibitors) family. Although the function of serpina3k is unknown, two adipokines, PAI-1 (plasimogen inhibitor-1) and vaspin (visceral adipose tissue-derived serpin), are also serpin members, and are associated with obesity-linked cardiovascular disease and insulin resistance, respectively (Juhan-Vague et al. 2000; Hida et al. 2005).
We show a nearly absolute requirement for the CtBPs in the PRDM16-mediated repression of white fat genes by two independent lines of experimental evidence: (1) A mutant PRDM16 that no longer binds CtBPs loses the ability to repress nearly the entire set of white fat-selective genes regulated by wild-type PRDM16. (2) Genetic ablation of the CtBPs causes a loss of wild-type PRDM16’s suppressive effects on white fat-selective gene expression.
CtBP was originally identified as a phosphoprotein that bound the C-terminal region of the human adenovirus EIA protein (Boyd et al. 1993; Schaeper et al. 1995). It has been shown that CtBP functions predominantly as a transcriptional repressor by associating with other corepressors and histone modifying enzymes, including class I histone deacetylases HDAC1/2, histone methyltransferases (HMTs, G9a, and GLP/Eu-HMT1), histone lysine-specific demethylase (LSD1), and CoREST (Shi et al. 2003; Chinnadurai 2007). The repressive effects are thought to occur through coordinate histone modifications by HDACs and G9a/GLP actions on deacetylation and methylation of H3K9, and also through additive demethylation on H3K4 by LSD1 (Shi et al. 2003, 2004; Tachibana et al. 2005). Notably, PRDM16 also contains a SET domain, a structural hallmark of histone lysine methyltransferase shared by G9a, GLP, SETDB1, and Suv39h1/2 (Rea et al. 2000; Schultz et al. 2002; Trievel et al. 2002; Tachibana et al. 2005). An aberrant form of PRDM16 lacking PR-domain (SET domain) is expressed exclusively in adult T-cell leukemia cells (Yoshida et al. 2004). This short form of PRDM16, but not the regular form of PRDM16 possesses “oncogenic” potential (Du et al. 2005; Shing et al. 2007). The functional importance of the SET domain in PRDM16 for silencing gene expression in the context of brown fat determination remains unanswered.
PGC-1α and PGC-1β are critical molecules in the brown fat phenotype. PGC-1α is a major coactivator of the thermogenic program of BAT in response to cAMP signaling, but is not required for the identity of brown fat cells. Double deficiency in PGC-1α and PGC-1β, however, causes a near total loss of the molecular identity of brown fat cells, suggesting an absolute requirement for PGC-1 coactivators in the brown fat program (Lin et al. 2004; Uldry et al. 2006). Our previous study has shown that PRDM16 robustly induced brown fat gene expression at least in part by directly binding to and stimulating both PGC-1α and PGC-1β (Seale et al. 2007). In this study, we also show that PGC-1α and/or β directly compete with CtBP for binding to the PRDM16 complex. Although PGC-1s and CtBP bind to the distinct domains of the PRDM16 molecule in the primary sequence (Fig. 2), they presumably compete at the level of three-dimensional structure of PRDM16. These results suggest a model as depicted in Figure 6D: PRDM16 represses white fat-selective gene expression including resistin and angiotensinogen by recruiting CtBP onto their promoters. Recruitment of PGC-1α/β to PRDM16 complex displaces CtBP, leading to the activation of the brown fat-selective genes such as PGC-1α itself. The binding of PGC-1s or CtBP to PRDM16 is mutually exclusive. Obviously, there must be at least some other components, most likely a transcription factor, onto which PRMD16 is recruited and determines whether PRDM16 will bind either to CtBP or PGC-1 coactivators.
It also seems likely that this switching mechanism might be affected by signal transduction pathways. Since BAT is highly innervated by the sympathetic nerve system, activation of β-adrenergic receptors by catecholamines is essential for many aspects of brown fat development and thermogenic program, including UCP-1 gene expression (Lowell and Spiegelman 2000; Bachman et al. 2002; Cannon and Nedergaard 2004; Hansen and Kristiansen 2006). Upon stimulation, an increase in intracellular cAMP levels activates cAMP-dependent protein kinase A (PKA) and its downstream p38MAPK (mitogen-activated protein kinase), which leads to the phosphorylations of PGC-1α and ATF-2/CRE. This phosphorylation event activates the recruitment of PGC-1α and ATF-2/CRE onto the regulatory regions of the PGC-1α gene itself and the UCP-1 gene, inducing their transcription (Puigserver et al. 2001; Cao et al. 2004). We have also shown previously that cAMP signaling augmented the activity of PRDM16 to induce both PGC-1α and UCP-1 gene expression in vivo and in vitro (Seale et al. 2007). A recent study reported that PKA directly phosphorylated CtBP-1 at T144 and modulated the association with GCN5 and SF-1 (Dammer and Sewer 2008). Thus, it is conceivable that modulations of PRDM16 by cAMP signaling and other pathways may influence the ability of PRDM16 to form complexes with PGC-1α/β and/or other coregulator proteins, to fine tune the activation of the brown fat gene program. Investigating this aspect will be necessary for fully understanding the transcriptional mechanisms of brown fat determination.
Materials and methods
Cell culture
Immortalized brown fat preadipocytes and PPAR-γ-deficient MEFs have been described elsewhere (Rosen et al. 2002; Uldry et al. 2006; Seale et al. 2007). 3T3-F442A cells were from H. Green (Green and Kehinde 1979). CtBP-1+/− CtBP-2+/− and CtBP-1−/− CtBP-2−/− MEFs were kind gifts from Dr. P. Soriano and Dr. J.D. Hildebrand (Hildebrand and Soriano 2002). COS-7 cells and 3T3-L1 cells were obtained from American Type Culture Collection. Adipocyte differentiation was induced by treating confluent cells in DMEM containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 μM dexamethasone, 850 nM insulin, 1 nM T3, and 1 μM rosiglitazone. Two days after the induction, cells were switched to the maintenance medium containing 10% FBS, 850 nM insulin, 1 nM T3, and 1 μM rosiglitazone. To stimulate respiration, cells were incubated with 0.5 mM dibutyryl cAMP or 10 μM forskolin for 4 h. All chemicals for cell culture were obtained from Sigma unless otherwise indicated.
Plasmids and viruses
Full-length Flag-tagged PRDM16 (Seale et al. 2007) was used as a template to create mutant forms of PRDM16 that lack the CtBP-binding motif (mut1, mut2, and mut1/2) by site-directed mutagenesis (QuikChange, Stratagene). These constructs were subcloned into pMSCV-puro retroviral vector (Stratagene) or pcDNA3.1 (Invitrogen). Various fragments of PRDM16 (1–223, 224–454, 455–680, 680–880, 881–1038, and 1039–1176) were amplified by PCR and cloned into the EcoRI or BamH1/XhoI sites of the bacterial expression vector pGEX-4T-2 vector (GE Healthcare). CtBP-1 and CtBP-2 constructs cloned in pcDNA3.1 were kind gifts from Dr. P. Puigserver.
The resistin promoter luciferase construct was generated by inserting the −13,580/+243 fragment of the mouse resistin promoter/enhancer into MluI and XhoI sites of the pGL3-basic vector (Promega) employing a recombineering strategy (Liu et al. 2003). Briefly, SW102 cells (kindly provided by Dr. N. Copeland) were made electrocompetent and transformed with 1 μg of a resistin BAC. These cells were treated to induce the recombination proteins exo, bet, and gam and made electrocompetent. To make a targeting vector, homology arms A (504 base pairs [bp]) and B (428 bp) were amplified by PCR using the resistin BAC as a template and subcloned into MluI and XhoI sites of the pGL3-basic vector. This targeting vector was linearized with EcoRI and purified from agarose gel. Approximately 50 ng of DNA were transformed into the SW102 cells carrying resistin BAC by a Bio-Rad electroporator under the following condition: 1.75 kV, 25 μF, with the pulse controller set at 200 Ω. Positive colonies were screened on ampicillin plates and verified by enzyme digestion and sequencing.
For retrovirus production, Phoenix packaging cells (Kinsella and Nolan 1996) were transfected at 70% confluence by calcium phosphate method with 10 μg of retroviral vectors. After 48 h, the viral supernatant was harvested and filtered. Cells were incubated overnight with the viral supernatant, supplemented with 8 μg/mL polybrene. Retroviral constructs of PRDM16 shRNA were described previously (Seale et al. 2007).
Affinity purification of the PRDM16 transcriptional complex and protein identification by mass spectrometry (MS)
PPAR-γ-deficient MEFs stably expressing Flag-tagged PRDM16 or an empty vector together with a vector expressing PPARγ2, were grown to confluence and were induced differentiation as described above. At day 4 of differentiation, cells were homogenized in a hypotonic solution (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, protein inhibitors) and spun at 1000g for 15 min to pellet the nuclei. The nuclei were resuspended in a hypertonic solution (20 mM HEPES at pH 7.9, 400 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM EDTA, 20% glycerol, protein inhibitors) for 30 min to extract nuclear proteins. Samples were spun at 16,000g for 30 min and dialyzed against a binding buffer containing 20 mM HEPES (pH 7.9), 150 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, and 0.1 mM PMSF.
The nuclear extracts were incubated overnight with Flag M2 agarose (Sigma), washed in a binding buffer (250 mM KCl), and then eluted by incubating with 3xFlag peptide (0.2 mg/mL). The eluted materials were TCA-precipitated, separated in a 4%–12% acrylamide gradient gel, and visualized by silver stain or commassie blue dye. Gel-resolved proteins were excised, digested with trypsin, and individually analyzed by MALDI reflectron time-of-flight (MALDI-reTOF) MS (UltraFlex TOF/TOF; BRUKER) as described before (Erdjument-Bromage et al. 1998; Sebastiaan Winkler et al. 2002). For peptide mass fingerprinting (PMF), experimental masses (m/z) combined from both MALDI-reTOF experiments were used to search a nonredundant rodent protein database (NR; ∼163,469 entries; NCBI), using the PeptideSearch (Matthias Mann, Max-Planck Institute for Biochemistry, Martinsried, Germany) algorithm. A molecular mass range of up to twice that predicted was covered, with a mass accuracy restriction of better than 40 ppm and a maximum of one missed cleavage site allowed per peptide. To confirm PMF results with scores ≤40, mass spectrometric sequencing of selected peptides was done by MALDI-TOF/TOF (MS/MS) analysis on the same prepared samples, using the UltraFlex instrument in “LIFT” mode. Fragment ion spectra were taken to search NR using the MASCOT MS/MS Ion Search program (Perkins et al. 1999), version 2.2. for Windows (Matrix Science Ltd.). Any tentative confirmation (Mascot score ≥30) of a PMF result thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
Protein interaction analysis
For coimmunoprecipitation experiments, immortalized brown fat cells stably expressing Flag-tagged PRDM16, or COS-7 cells were used. Total cell lysates were incubated overnight at 4°C with Flag M2 agarose, washed, and eluted with 3xFlag peptide. The eluted materials were then analyzed by Western blottings using the indicated antibodies. Antibodies used were anti-CtBP-1 (Upstate Biotechnologies), anti-CtBP-2 (BD Transduction Laboratories), anti-Flag M2 (Sigma), and anti-HA (Roche).
For in vitro binding assays, various fragments of the GST-PRDM16 fragments described above were expressed in BL21 by isopropyl thiogalactoside induction for 3 h at 30°C and purified on sepharose beads containing glutathione. [35S]-labeled proteins were made with a TNT reticulocyte lysate kit (Promega). Equal amounts of GST fusion proteins (2 μg) were incubated overnight at 4°C with in vitro translated proteins in a binding buffer containing 20 mM HEPES (pH 7.7), 300 mM KCl, 2.5 mM MgCl2, 0.05% NP40, 1 mM DTT, and 10% glycerol. The sepharose beads were then washed five times with the binding buffer. Bound proteins were separated by SDS-PAGE and analyzed by autoradiography.
Gene expression analysis
Total RNA was isolated from cells or tissues using the Trizol reagent (Invitrogen) following the manufacturer’s protocol. RT reactions were performed using IScript cDNA synthesis kit (Bio-Rad) or cDNA reverse transcription kit (Applied Biosystems). Primers were designed using the Primer Express 2.0 (Applied Biosystems). The sequences of primers used in this study are found in a previous study (Seale et al. 2007) or Supplemental Table 2. Quantitative real-time PCR was performed with SYBR green fluorescent dye using an ABI9300 PCR machine. TATA-binding protein (TBP) served as an internal control to normalize samples.
Microarray analysis
Total RNA was isolated from the 3T3-F442A cells transduced with wild-type PRDM16, mutant PRDM16, or vector control at day 6 of differentiation. Array hybridization and scanning were performed by the Dana-Farber Cancer Institute Core Facility using Affymetrix GeneChip Mouse Genome 430 2.0 arrays according to established methods (Lockhart et al. 1996). The array data were analyzed using the DNA-Chip Analyzer (dChip) software (Li and Wong 2001). The statistical significance of differences in gene expression was assessed by unpaired t-test (P < 0.05).
Reporter gene assay and ELISA
Mature 3T3-L1 adipocytes at days 8–10 of differentiation were detached from culture dishes with 0.25% trypsin, washed twice with PBS, and resuspended in electroporation buffer (solution V, AMAXA). Approximately 1 × 106 cells were electroporated (Nucleofector II, AMAXA, electroporation program T-020) after addition of 1 μg of reporter vector, 0.1 μg of expression vector, and 0.3 μg of β-galactosidase expression vector, and seeded into three wells of 24-well plates. After 48 h of incubation, the cells were harvested and luciferase activities were determined by a luciferase assay kit (Promega). Light units were normalized to the cotransfected β-galactosidase expression plasmid. Fold activations relative to the pGL3-basic and pcDNA3.1 were calculated and the results of triplicate samples were plotted. The resistin concentrations in the culture medium were measured using mouse resistin immunoassay (R&D Systems, Inc.) following the manufacturer’s protocol.
Cellular respiration assay
3T3-F442A cells transduced with retroviral wild-type PRDM16, mutant PRDM16, or vector control were grown to confluence and induced differentiation. At day 6 or 7 of differentiation, oxygen consumption was measured in fat cells as described previously (St-Pierre et al. 2003). For cAMP-induced respiration assays, fully differentiated fat cells were incubated with 0.5 mM dibutyryl cAMP for 12 h prior to measuring oxygen consumption.
ChIP assay
Immortalized brown cells were made, stably expressing either retroviral Flag-tagged PRDM16 or an empty vector. The cells were cross-linked with 1.5 mM ethylene glycolbis (Pierce) for 20 min, followed by 1% formaldehyde for additional 10 min (Zeng et al. 2006). The nuclear extracts from the cells were sonicated to shear the chromatin, and immunoprecipitated overnight at 4°C using M2 Flag antibody, anti-CtBP (Santa Cruz), or IgG (Upstate Biotechnology). After extensive washing, the immunoprecipitants were eluted with 2% SDS in 0.1 M NaH2CO3. Cross-linking was reversed by heating overnight at 65°C and was treated with proteinase K (Roche) for 1 h at 45°C. Input DNA and immunoprecipitated DNA were purified by PCR purification kit (Qiagen) and analyzed by quantitative PCR using SYBR green fluorescent dye (ABI). The protein-bound DNA was calculated as a ratio to input DNA. For visualizing the PCR products, DNA was amplified by PCR with 30–32 cycles of 30 sec at 94°C, 30 sec at 58°C, and 1 min at 72°C. PCR products were subsequently separated in 2% agarose gel. Primer sequences used in the ChIP assays were provided in Supplemental Table 2.
Acknowledgments
We are grateful to Dr. P. Soriano (Fred Hutchinson Cancer Research Center, Seattle, WA) and Dr. J.D. Hildebrand (University of Pittsburgh, Pittsburgh, PA) for providing cell lines, Dr. N.G. Copeland (National Cancer Institute, Frederick, MD) for providing M.A.L. materials, and Dr. P. Puigserver (Dana-Farber Cancer Institute, Boston, MA) for providing regents and for critical reading of the manuscript. We also thank Adah Levens for her editorial assistance. S.K. is supported by a fellowship from the Japan Society for Promotion of Science. J.L.R. is supported by a fellowship from the Wenner-Gren foundation, Sweden. This work is supported by grants from the Picower Foundation and NIH Merit award R37DK31405 to B.M.S. and DK49210 to M.A.L., and NCI Cancer Center Support Grant P30 CA08748 to P.T.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1666108.
References
- Almind K., Manieri M., Sivitz W.I., Cinti S., Kahn C.R. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proc. Natl. Acad. Sci. 2007;104:2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman E.S., Dhillon H., Zhang C.Y., Cinti S., Bianco A.C., Kobilka B.K., Lowell B.B. βAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- Banerjee R.R., Rangwala S.M., Shapiro J.S., Rich A.S., Rhoades B., Qi Y., Wang J., Rajala M.W., Pocai A., Scherer P.E., et al. Regulation of fasted blood glucose by resistin. Science. 2004;303:1195–1198. doi: 10.1126/science.1092341. [DOI] [PubMed] [Google Scholar]
- Barak Y., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M. PPAR γ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Boyd J.M., Subramanian T., Schaeper U., La Regina M., Bayley S., Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B., Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cao W., Daniel K.W., Robidoux J., Puigserver P., Medvedev A.V., Bai X., Floering L.M., Spiegelman B.M., Collins S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 2004;24:3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederberg A., Gronning L.M., Ahren B., Tasken K., Carlsson P., Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int. J. Biochem. Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Cinti S. The adipose organ. Editrice Kurtis; Milano, Italy: 1999. [Google Scholar]
- Dammer E.B., Sewer M.B. Phosphorylation of CtBP1 by PKA modulates induction of CYP17 by stimulating partnering of CtBP1 and 2. J. Biol. Chem. 2008;83:6925–6934. doi: 10.1074/jbc.M708432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Jenkins N.A., Copeland N.G. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulloo A.G., Miller D.S. Energy balance following sympathetic denervation of brown adipose tissue. Can. J. Physiol. Pharmacol. 1984;62:235–240. doi: 10.1139/y84-035. [DOI] [PubMed] [Google Scholar]
- Engeli S., Negrel R., Sharma A.M. Physiology and pathophysiology of the adipose tissue renin-angiotensin system. Hypertension. 2000;35:1270–1277. doi: 10.1161/01.hyp.35.6.1270. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H., Lui M., Lacomis L., Grewal A., Annan R.S., McNulty D.E., Carr S.A., Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Farmer S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.M. The function of leptin in nutrition, weight, and physiology. Nutr. Rev. 2002;60:S1–S14. doi: 10.1301/002966402320634878. [DOI] [PubMed] [Google Scholar]
- Gesta S., Tseng Y.H., Kahn C.R. Developmental origin of fat: Tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Green H., Kehinde O. Formation of normally differentiated subcutaneous fat pads by an established preadipose cell line. J. Cell. Physiol. 1979;101:169–171. doi: 10.1002/jcp.1041010119. [DOI] [PubMed] [Google Scholar]
- Grooteclaes M., Deveraux Q., Hildebrand J., Zhang Q., Goodman R.H., Frisch S.M. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. 2003;100:4568–4573. doi: 10.1073/pnas.0830998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Koza R.A., Yamashita H., Walsh K., Kozak L.P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaas J.L., Gajiwala K.S., Maffei M., Cohen S.L., Chait B.T., Rabinowitz D., Lallone R.L., Burley S.K., Friedman J.M. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- Handschin C., Rhee J., Lin J., Tarr P.T., Spiegelman B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.B., Kristiansen K. Regulatory circuits controlling white versus brown adipocyte differentiation. Biochem. J. 2006;398:153–168. doi: 10.1042/BJ20060402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.B., Jorgensen C., Petersen R.K., Hallenborg P., De Matteis R., Boye H.A., Petrovic N., Enerback S., Nedergaard J., Cinti S., et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc. Natl. Acad. Sci. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida K., Wada J., Eguchi J., Zhang H., Baba M., Seida A., Hashimoto I., Okada T., Yasuhara A., Nakatsuka A., et al. Visceral adipose tissue-derived serine protease inhibitor: A unique insulin-sensitizing adipocytokine in obesity. Proc. Natl. Acad. Sci. 2005;102:10610–10615. doi: 10.1073/pnas.0504703102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D., Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Juhan-Vague I., Alessi M.C., Morange P.E. Hypofibrinolysis and increased PAI-1 are linked to atherothrombosis via insulin resistance and obesity. Ann. Med. 2000;32 (Suppl. 1):78–84. [PubMed] [Google Scholar]
- Kinsella T.M., Nolan G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Leonardsson G., Steel J.H., Christian M., Pocock V., Milligan S., Bell J., So P.W., Medina-Gomez G., Vidal-Puig A., White R., et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wong L. Emerging patterns and gene expression data. Genome Inform. 2001;12:3–13. [PubMed] [Google Scholar]
- Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Lowell B.B., Spiegelman B.M. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- Lowell B.B., S- V., Hamann A., Lawitts J.A., Himms-Hagen J., Boyer B.B., Kozak L.P., Flier J.S. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Massiera F., Bloch-Faure M., Ceiler D., Murakami K., Fukamizu A., Gasc J.M., Quignard-Boulange A., Negrel R., Ailhaud G., Seydoux J., et al. Adipose angiotensinogen is involved in adipose tissue growth and blood pressure regulation. FASEB J. 2001;15:2727–2729. doi: 10.1096/fj.01-0457fje. [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Shimizu S., Nagasawa T., Tanaka H., Taniwaki M., Yokota J., Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- Nedergaard J., Petrovic N., Lindgren E.M., Jacobsson A., Cannon B. PPARγ in the control of brown adipocyte differentiation. Biochim. Biophys. Acta. 2005;1740:293–304. doi: 10.1016/j.bbadis.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Nishikata I., Sasaki H., Iga M., Tateno Y., Imayoshi S., Asou N., Nakamura T., Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003;102:3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- Pelleymounter M.A., Cullen M.J., Baker M.B., Hecht R., Winters D., Boone T., Collins F. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Picard F., Gehin M., Annicotte J., Rocchi S., Champy M.F., O’Malley B.W., Chambon P., Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Powelka A.M., Seth A., Virbasius J.V., Kiskinis E., Nicoloro S.M., Guilherme A., Tang X., Straubhaar J., Cherniack A.D., Parker M.G., et al. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M. PPAR γ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Hsu C.H., Wang X., Sakai S., Freeman M.W., Gonzalez F.J., Spiegelman B.M. C/EBPα induces adipogenesis through PPARγ: A unified pathway. Genes & Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper U., Boyd J.M., Verma S., Uhlmann E., Subramanian T., Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher F.J. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scime A., Grenier G., Huh M.S., Gillespie M.A., Bevilacqua L., Harper M.E., Rudnicki M.A. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1α. Cell Metab. 2005;2:283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Seale P., Kajimura S., Yang W., Chin S., Rohas L.M., Uldry M., Tavernier G., Langin D., Spiegelman B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiaan Winkler G., Lacomis L., Philip J., Erdjument-Bromage H., Svejstrup J.Q., Tempst P. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- Shi Y., Sawada J., Sui G., Affar E.B., Whetstine J.R., Lan F., Ogawa H., Luke M.P., Nakatani Y., Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shing D.C., Trubia M., Marchesi F., Radaelli E., Belloni E., Tapinassi C., Scanziani E., Mecucci C., Crescenzi B., Lahortiga I., et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Invest. 2007;117:3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman B.M., Flier J.S. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- St-Pierre J., Lin J., Krauss S., Tarr P.T., Yang R., Newgard C.B., Spiegelman B.M. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Steppan C.M., Lazar M.A. The current biology of resistin. J. Intern. Med. 2004;255:439–447. doi: 10.1111/j.1365-2796.2004.01306.x. [DOI] [PubMed] [Google Scholar]
- Steppan C.M., Bailey S.T., Bhat S., Brown E.J., Banerjee R.R., Wright C.M., Patel H.R., Ahima R.S., Lazar M.A. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes & Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D., Langin D. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPAR γ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Trievel R.C., Beach B.M., Dirk L.M., Houtz R.L., Hurley J.H. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Poulin F., Kohara M., DeMaria C.T., Cheng A., Wu Z., Gingras A.C., Katsume A., Elchebly M., Spiegelman B.M., et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Wu Z., Rosen E.D., Brun R., Hauser S., Adelmant G., Troy A.E., McKeon C., Darlington G.J., Spiegelman B.M. Cross-regulation of C/EBP α and PPAR γ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Nosaka K., Yasunaga J., Nishikata I., Morishita K., Matsuoka M. Aberrant expression of the MEL1S gene identified in association with hypomethylation in adult T-cell leukemia cells. Blood. 2004;103:2753–2760. doi: 10.1182/blood-2003-07-2482. [DOI] [PubMed] [Google Scholar]
- Zeng P.Y., Vakoc C.R., Chen Z.C., Blobel G.A., Berger S.L.2006In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation Biotechniques 41694 , 696, , 698 [DOI] [PubMed] [Google Scholar]