Abstract
Members of the Eph family of tyrosine kinase receptors have been implicated in the regulation of developmental processes and, in particular, axon guidance in the developing nervous system. The function of the EphA4 (Sek1) receptor was explored through creation of a null mutant mouse. Mice with a null mutation in the EphA4 gene are viable and fertile but have a gross motor dysfunction, which is evidenced by a loss of coordination of limb movement and a resultant hopping, kangaroo-like gait. Consistent with the observed phenotype, anatomical studies and anterograde tracing experiments reveal major disruptions of the corticospinal tract within the medulla and spinal cord in the null mutant animals. These results demonstrate a critical role for EphA4 in establishing the corticospinal projection.
Recent studies show that axons are guided to their targets by a system of guidance molecules, including Eph receptors and their ligands (1–3). The role of these molecules has been studied intensely in development of the visual system (4–6), where the reciprocal gradient expression of the Eph receptors in the retina and of their ligands in the optic tectum is the suggested basis for the formation of the retinotectal topographic map. Other observations pertinent to the role of these molecules in the developing nervous system include axonal fasciculation and establishing brain commissures (7–9).
The Eph family receptors can be divided into two groups, EphA and EphB, based on the sequence similarities of their extracellular domain (10). Each EphA receptor is able to bind several Ephrin A ligands that are associated with the membrane via a GPI-linkage; these receptors show little or no binding to the transmembrane Ephrin B ligands (11, 12). The EphB group of receptors show the reverse pattern, binding predominantly to Ephrin B ligands. An exception to this “rule” is the EphA4 (previously known as Sek1) receptor, which was found to bind significantly to some of the transmembrane ligands in addition to all of the GPI-linked ligands (11–13).
EphA4 expression during development shows a defined spatiotemporal pattern within the developing forebrain, hindbrain, and mesoderm (14, 15). In the final stages of embryogenesis, expression of EphA4 is found predominantly within regions of the central nervous system, including the cerebral cortex, striatum, thalamus, hippocampus, and ventral spinal cord. In the hindbrain, EphA4 shows restricted expression to rhombomeres 3 and 5 (14), which suggests a role of this receptor in establishing boundaries during embryogenesis. This notion is supported by overexpression of dominant negative, truncated EphA4 receptor in zebrafish embryos. The resultant mutant embryos were found to have disruption in the rhombomere boundaries and an expansion of the developing retina into the diencephalon (16, 17). The expression pattern of EphA4 coupled with the unique binding characteristics of this receptor suggests that loss of function of the EphA4 gene may affect neural patterning events.
This report describes the generation of mice deficient in the EphA4 receptor. The EphA4 mutant mice displayed a gross motor abnormality in the hindlimbs. Anatomical analyses and anterograde tracing of cortical neurons demonstrated a severe disruption of the corticospinal tract (CST) in these animals. The CST is the single longest axonal projection in the mammalian central nervous system (18). CST neurons arise from layer V in the neocortex and extend their axons through the forebrain, midbrain, and hindbrain and terminate at various levels of the spinal cord. In primates, the CST axons predominantly synapse directly with the spinal motor neurons whereas in the rodent, most of the cortical axons synapse with interneurons, which then connect to the spinal motor neurons. The EphA4 null mutant mice showed specific defects in the CST both at the level of the medulla and the spinal cord, which indicates that EphA4 is required for the correct formation of the CST, possibly by guidance of CST axons.
MATERIALS AND METHODS
Targeted Disruption of EphA4 Gene.
For homologous recombination, 5′ HindIII-SacI 3-kilobase (kb) sequence and 3′ Eco47III-BamHI 5.5-kb sequence flanking exon III were subcloned into the pKJ1 vector (Fig. 1). The vector contains the neomycin-resistance gene (neo) with the phosphoglycerate kinase (pgk) promoter and pgk polyadenylation signal. The W9.5 embryonic stem cell line was electroporated with the SalI linearized targeting construct and was selected with G418 for 10 days. A total of 480 surviving clones were expanded, and homologous recombinants were identified by Southern blot analysis of genomic DNA from single clones digested with EcoRI. Two isolated clones with a single targeted mutation of EphA4 gene each were injected into (C57BL/6 × C57BL/10)F2 blastocysts. Chimeras were mated to C57BL/6 mice to produce heterozygotes. Southern blot analysis of tail DNA was used for genotyping the offspring.
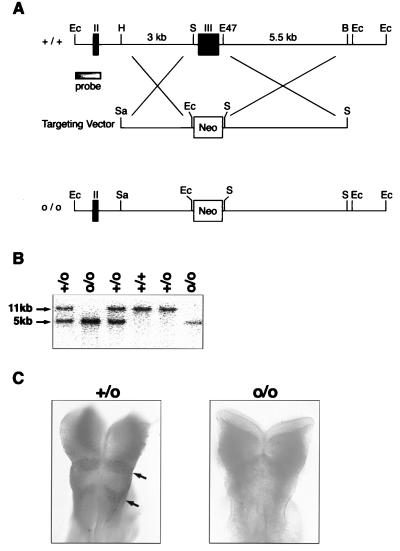
Figure 1.
Targeted disruption of EphA4 gene. (A) Partial map of the ephA4 genomic locus (+/+) with the targeting construct and the resulting targeted loci (o/o). The EphA4 targeting vector was designed to replace exon III (217–880 bp of EphA4 cDNA) (38) with the 1.8-kb neomycin selection gene. For homologous recombination, 5′ HindIII-SacI 3-kb sequence and 3′ Eco47III-BamHI 5.5-kb sequence flanking exon III were subcloned into the pKJ1 vector. Homologous recombination would cause a frame shift in the EphA4 gene, resulting in a null mutant protein. The probe used for all Southern blot analyses was a 1-kb genomic fragment containing exon II (149–216 bp) and EcoRI site. Ec, EcoRI; H, HindIII; S, SacI; E47, Eco47III; B, BamHI; Neo, neomycin gene; II, exon II; and III, exon III. (B) Genotype analysis of EphA4 homozygous (o/o), heterozygous (+/o), and wild-type (+/+) animals. Genomic DNA was isolated from 0.5 cm tail tissue (39), was digested with EcoRI, and was subjected to Southern blot analysis using the 5′ external probe shown in A. Alleles bearing the ephA4 mutation results in a 5-kb band whereas an 11-kb band was observed in the wild-type alleles. (C) Whole-mount immunocytochemistry of E 8.5 embryos by using anti-EphA4 antibody. EphA4 is expressed in rhombomeres 3 and 5 (arrows) in heterozygotes (+/o), but no EphA4 protein is detected in homozygous (o/o) mutants. The embryos were genotyped by PCR from yolk sac DNA.
Whole-Mount and Tissue Immunocytochemistry and PCR Genotyping of Embryos.
Whole-mount immunocytochemistry was performed with anti-EphA4 antibody (provided by D.G. Wilkinson of National Institute of Medical Research, Mill Hill, U.K.) as described (19), and color detection was carried out by using 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (Promega) as substrate. For tissue sections, tissues were fixed for 24 hours in 4% paraformaldehyde and then another 24 hours in fixative containing 30% sucrose. Frozen tissue was sectioned serially 50 μm thick. Immunohistochemistry was performed by using anti-EphA4 antibody, and the same protocol as for whole mounts, except the ABC Elite detection system (Vector Laboratories) was used to detect color staining.
Embryos were genotyped by PCR of yolk sac DNA (20) by using primer pairs P1 CGTGCTACTTCCATTTGTCACGTCCTG and P2 TGCCGTGATAGCAAATTTGAG or P3 AGGAAGTGAGCATTATGGATGA and P4 TGCTCCTCGTGCCCAGCGTT. A 600-bp band was generated from the mutant allele between the neomycin primer P1 and ephA4 endogenous primer P2; a 645-bp product was generated from the wild-type allele between exon III primers P3 and P4. The PCR reaction was in a total volume of 50 μl and consisted of 50–500 ng DNA, 30 pmol of each primer, 2.0 mM MgCl, 100 μM dNTPs, and 1 unit Taq polymerase (Roche, Gipf-Oberfrick, Switzerland), with the appropriate reaction buffer supplied by the manufacturer. The cycling reaction was 15 cycles of 96°C for 30 sec, 70°C for 30 sec (−1°C per cycle), and 72°C for 1 min, followed by 20 cycles of 96°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min.
Histology.
Histological examination was carried out on EphA4 homozygous, heterozygous, and wild-type littermates of embryonic day (E) 16 and 8- and 24-day-old mice. Embryos and adult tissues were fixed overnight in 10% formalin, were paraffin-embedded, and were sectioned serially 4 μm thick. Sections were stained with either haematoxylin and eosin or luxol fast blue.
In Situ Hybridization.
For EphA4 mRNA expression, tissues were fixed overnight in 10% formalin, were paraffin-embedded, and were sectioned serially 4 μm thick. In situ hybridization was performed as described (21) by using 33P-radiolabeled complimentary EphA4 RNA probe. The antisense probe was synthesized with T7 polymerase from the HindIII-linearized plasmid Bluescript KS, containing a 1.5-kb EcoRI fragment of 3′ untranslated and C-terminal coding sequences of EphA4 (provided by D.G. Wilkinson).
For expression of EphrinB3 mRNA, digoxigenin-labeled in situ hybridization was performed on frozen 20-μm tissue sections as described (22). To generate the Ephrin B3 probe, Ephrin B3 cDNA was amplified by PCR from adult mouse brain cDNA by using primers TTAGAATTCCCCGAGGAGGAGCTGTAC and CTAGAATTCTGCAGTCCCACCACCCCG. The PCR product, which spans 551 to 953 bp of Ephrin B3 cDNA (13), was cloned into EcoRI site of Bluescript SK and was sequenced. The antisense probe then was synthesized with T3 polymerase from the HindIII-linearized plasmid.
Surgery, Anterograde Tracing, and Tissue Processing.
Corticospinal axons and their terminal projections were labeled in 5-week-wold mice by using the anterograde tracer biotinylated dextran amine (15%; (Molecular Probes). Two wild-type, one heterozygous, and three homozygous EphA4 mutant mice were used for these studies. The animals were anesthetized by injecting i.p. (10 μl/g of body weight) a 1:1:6 ratio mixture of Hypnorm (Janssen), Hypernovel (Roche), and distilled H2O. Anesthetized animals had their head positioned in a stereotaxic frame, and a craniotomy (3–4 mm in diameter) was made to expose the rostral half of the left cerebral hemisphere. Seven injections of 0.3 μl of tracer were made into the cerebral cortex at a depth of 0.5–1.0 mm below its surface by using a glass pipette (tip diameter of 50 μm) attached to a Hamilton syringe (23). The injections covered the whole sensorimotor region of the cerebral cortex. The number of injections, the injection sites, and the amount of tracer used per injection were kept consistent between control and mutant animals. The brain and spinal cord were perfused 7 days after the injection with 0.9% phosphate buffered saline and 4% paraformaldehyde in phosphate buffer. The tissue was postfixed for 24 hours in 30% sucrose in buffered fixative.
The free-floating sections were processed according to the method as described (24) to visualize the axons and terminals labeled by biotinylated dextran amine. Phosphate buffer (0.1 M) was the vehicle for the immunoreagents and for rinsing after each of the following steps: (i) incubation in 0.3% hydrogen peroxide in methanol for 20 min to block any endogenous peroxidase activity; (ii) incubation in Avidin-peroxidase (Sigma) diluted 1:5,000 in 0.1 M phosphate buffer and 0.75% Triton X-100 for 2 hours; and (iii) processing for horseradish peroxidase histochemistry by using cobalt-enhanced diaminobenzidine reaction (25) for 8–10 min. This process stained the axons and terminals labeled with biotinylated dextran amine black. Transverse spinal cord sections were counterstained with haematoxylin.
RESULTS
Generation of EphA4 Homozygous Mice.
EphA4 deficient mice were generated by using targeted mutagenesis and embryonic-stem cell technology (26). The gene targeting strategy (Fig. 1A) replaced exon III with a neomycin selection gene, thereby introducing a frame shift and stop codon in the ephA4 gene. To demonstrate that the EphA4 mutation results in a null mutation, whole-mount immunohistochemistry was performed on E 8.5 embryos (Fig. 1C). In wild-type and heterozygous embryos, EphA4 was expressed in rhombomeres 3 and 5 (arrows), as described (14). In contrast, no staining was observed in the EphA4 homozygous embryos. The antibody recognizes the C terminus of the intracellular domain (2783–3195 residues) of EphA4 (19), and, thus, the lack of staining observed in the homozygous embryos implies that no EphA4 protein is produced in these mutant mice. EphA4 null mutant mice generated from two independent embryonic-stem cell lines were viable and fertile. The number of EphA4 homozygous mice in litters born from crossing heterozygotes showed a normal Mendelian ratio (25%), indicating no lethality of the mutation during embryogenesis.
EphA4 Homozygous Mice Display an Abnormal Hopping Gait.
The EphA4 null mice exhibited locomotor abnormalities with impairment of the coordinated movement of the limbs. Both mouse strains showed hesitation in initiating locomotion, and, once they began to move, there was lack of the normal synchronous movement of each forelimb with the contralateral hindlimb. Most striking was an abnormal, synchronous (kangaroo-like) movement of the hindlimbs while reciprocal movement of the forelimbs was maintained. In contrast, the heterozygous mice showed no abnormality.
Tests of neurological function were performed to further characterize the defects in these animals. The hesitation to move and lack of coordination in the hindlimbs was reflected in open field activity tests (27), which showed the distance traveled by the EphA4 homozygotes was only 30% of the heterozygote value (EphA4 homozygotes crossed 18 ± 24 grids per 5 min compared with heterozygous littermates, which crossed 60 ± 34 grids; n = 15, P < 0.0005). In addition, the EphA4 null mutant animals showed placing deficits of both hindlimbs, suggesting a defect in corticospinal projections (28, 29), whereas sensory tests were within normal limits.
Disruption of Spinal Cord Architecture and the Anterior Commissure in EphA4 Homozygous Mice.
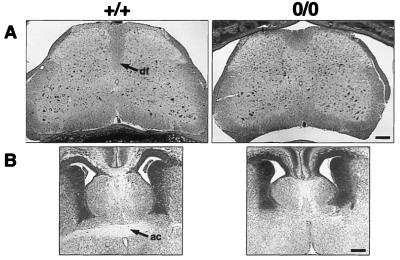
Anatomical studies were performed to determine whether there were major structural changes in the central nervous system of the EphA4 null mice. Although there was no macroscopic abnormality, histological analysis of spinal cord sections showed that the dorsal funiculus was markedly shallower in the EphA4 null animals compared with heterozygous and wild-type animals (Fig. 2A). The major motor pathway, the CST, descends through the dorsal funiculus in the rodent spinal cord. Anatomical studies revealed a further defect in the EphA4 null mutant mice, a loss of the anterior commissure (AC). This was observed in 12 of the 14 homozygous specimens examined (Fig. 2B), but the AC appeared normal in all heterozygous and wild-type mice. No other anatomical abnormalities were observed in the brains of EphA4 mutants, including within the motor cortex, midbrain, and medullary pyramids.
Figure 2.
Histological sections of EphA4 homozygous (o/o) and wild type (+/+) animals. (A) Transverse sections stained with luxol fast blue of lumbar spinal cord from adult mice. Area of the dorsal funiculus (df) appears to be shallower in EphA4 homozygotes. (Bar = 160 μm.) (B) Coronal sections stained with haematoxylin and eosin of E 16 embryo brains. A loss of the anterior commissure (ac) is observed in homozygotes. (Bar = 140 μm.)
Corticospinal Projection Is Aberrant in EphA4 Homozygous Mice.
Functional tests and the abnormality in the dorsal funiculus suggested that the CST may be disrupted or absent in EphA4 deficient mice. This possibility was explored by using dye tracing studies. Corticospinal axons were labeled anterogradely from their origin, layer V neurons in the motor cortex, to their terminal projections. Normally, CST axons descend through the internal capsule, basis pedunculi in the midbrain, pons, and medullary pyramids (Fig. 3). In the medulla, the CST fibers cross the midline (decussate), then descend in the dorsal funiculus of the spinal cord and terminate predominantly in the dorsal horn contralateral to the cells of origin.
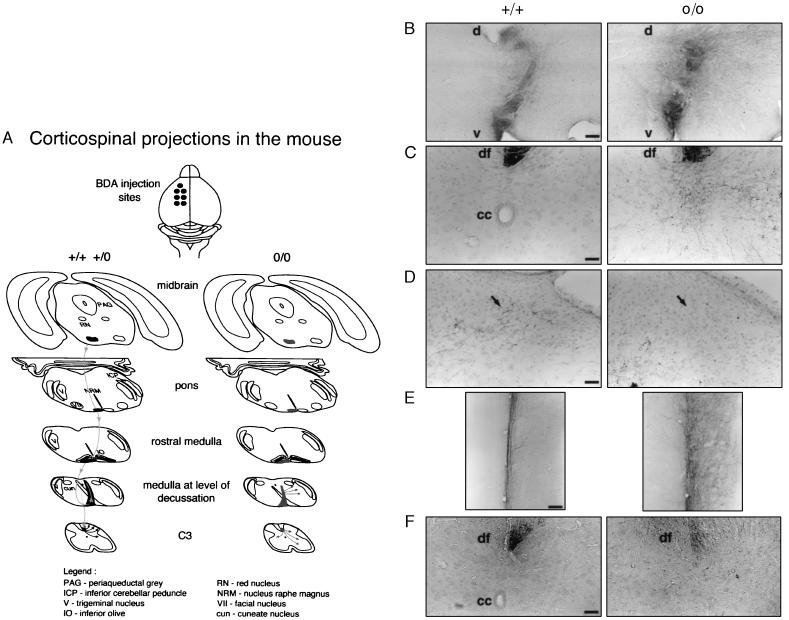
Figure 3.
Labeled CST in normal (+/+ and +/o) and EphA4 null mutant (o/o) mice. (A). Schematic representation of the corticospinal projection traced in mice. Multiple injections of the tracer were made in the motor cortex in the left cerebral hemisphere of adult mice. The labeled CST axons descend through the midbrain, pons, and pyramid in the medulla. In wild-type and heterozygous mice, the CST axons decussate at the medulla, crossing the midline traveling from left ventral to right dorsal, enter the dorsal funiculus of the spinal cord, and terminate predominantly in the dorsal horn contralateral to the tracer injections. In EphA4 null mutant mice, labeled CST axons appeared to terminate in the medulla and intermediate and ventral region of the spinal gray matter. Some labeled fibers were observed to recross the midline. (B) Transverse sections of medulla showing the decussation of labeled CST fibers travelling from left ventral (v) to right dorsal (d). In EphA4 o/o mice, many CST axons do not enter the dorsal column area. (Bar = 450 μm.) (C and D) Transverse sections of cervical spinal cord showing area of dorsal funiculus (C) and dorsal horn (D). In wild-type animals, labeled CST axons terminate in the right dorsal horn (arrow). In homozygotes, axons project predominantly into the intermediate and ventral regions of the gray matter, and no labeled axons were observed terminating in the dorsal horn. cc, central canal; df, dorsal funiculus. (Bar = 125 μm.) (E) Longitudinal sections of cervical spinal cord. CST axons in the right dorsal funiculus are seen in the midline. In homozygous animals, some CST fibers recross the midline and project to the gray matter ipsilateral to the tracer injections. (Bar = 300 μm.) (F) Transverse sections of lumbar spinal cord. A reduced number of labeled CST axons was observed in the dorsal funiculus of the null mutant mice compared with normal. (Bar = 125 μm.)
Anterograde labeling of corticospinal neurons in EphA4 null mice showed normal projection within the fore- and mid-brain (data not shown). However, the CST pathway within the medulla and spinal cord was clearly abnormal. It was observed in the medulla that, although many of the CST axons crossed the midline, a considerable number of axons appeared to terminate inappropriately at this level, so that a reduced number of axons descended in the dorsal column of the spinal cord (Fig. 3B). In addition, those axons that descended in the dorsal funiculus showed an aberrant pattern of termination within the gray matter of the spinal cord (Fig. 3 C and D), with terminal branches observed predominantly in the intermediate zone and ventral horn and very few terminals in the dorsal horn. A number of axons also recrossed the midline and terminated in the gray matter ipsilateral to the cortical tracer injection (Fig. 3E). In the lumbar cord, there was a significant reduction in the number of CST axons (Fig. 3F), making it difficult to demonstrate whether their pattern of termination was also aberrant at this level.
A small proportion of CST axons do not decussate in the medulla but continue to descend ipsilaterally into the spinal cord in the ventral funiculus (30). The ipsilateral CST found within the ventral funiculus did not appear to be notably different in homozygous, heterozygous, and wild-type animals (data not shown).
Expression of EphA4 and Ligand During CST Development.
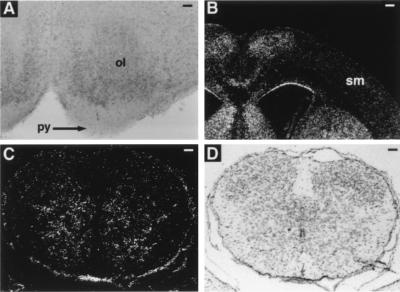
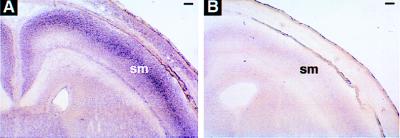
To determine whether EphA4 protein was expressed in the CST, immunohistochemical studies were undertaken on neonatal mouse brain tissues because it is during the neonatal period that the CST projects through the medulla and enters the spinal cord (refs. 31 and 32; J. Coonan and M.G., unpublished observations). EphA4 protein was not detected within the medullary pyramid or any other part of the CST at this age; however, it is expressed in the olivary nucleus, which is dorsal to the pyramidal tract (Fig. 4A). In addition, in situ hybridization studies were undertaken to determine whether EphA4 mRNA was detected within the motor cortex, which is where the cell bodies of the CST are localized. Consistent with the immunohistochemistry data, in situ hybridization analysis shows levels of EphA4 mRNA within the sensorimotor cortex that are not above background (Fig. 4B). However, a gradient expression of EphA4 mRNA was found within the spinal cord with high levels of expression detected in the intermediate and ventral regions of the spinal cord gray matter and low levels of expression in the dorsal horns (Fig. 4C). These data indicate that EphA4 is not expressed in the CST axons but is found expressed in surrounding structures.
Figure 4.
Analysis of EphA4 expression in wild-type neonatal mouse tissues by immunohistochemistry (A) and in situ hybridization (B and C). (A) Coronal section of the medulla stained with anti-EphA4 antibody. EphA4 was detected in the inferior olivary nucleus (ol), but not in the pyramidal tract (py). (Bar = 125 μm.) (B) Dark-field photomicrograph showing a coronal section of brain hybridized with radiolabeled-antisense EphA4 probe. The level of EphA4 mRNA within the sensorimotor cortex (sm) region is not above background. (Bar = 420 μm.) (C) Dark-field and (D) bright-field photomicrograph of cervical spinal cord transverse section hybridized with antisense EphA4 probe. EphA4 mRNA was found expressed within the intermediate and ventral regions of the spinal cord gray matter. df, dorsal funiculus. No signal was observed in equivalent tissue sections stained with radiolabeled-sense probe (data not shown). (Bars = 150 μm.)
To determine whether a ligand for EphA4 may be expressed in the CST, we analyzed the expression of Ephrin B3 within E 18.5 mouse brain tissue (Fig. 5). Of the transmembrane ligands, Ephrin B3 binds to EphA4 with the highest affinity (12, 13). In situ hybridization with digoxigenin-labeled Ephrin B3 antisense probe detected strong expression within the sensorimotor cortex region (Fig. 5A), thereby suggesting that Ephrin B3 is expressed in the motor neurons of the CST during its development.
Figure 5.
Analysis of EphrinB3 expression in wild-type E 18.5 mouse tissue by in situ hybridization. Coronal sections of whole head were hybridized with digoxigenin-labeled (A) antisense Ephrin B3 and (B) sense riboprobes. An intense signal of Ephrin B3 mRNA was detected within the sensorimotor (sm) cortex region. (Bar = 400 μm.)
DISCUSSION
The EphA4 null mutant mice were the first Eph receptor null mice to display a motor phenotype (7–9). This motor defect was more marked in the hindlimbs and the animals have an abnormal hopping gait. Analysis of the CST in these animals reveal a reduced number of CST axons in the lower spinal cord segments and an abnormal pattern of termination at higher segments of the spinal cord and medulla. This progressive diminution of the CST, relative to normal animals, along the length of the cord is consistent with the more marked motor defect observed in the lower limbs of these animals. There may be other motor defects in the mutant animals; however, preliminary tracing studies on spinal motor neurons have not detected aberrant projections to muscle (J. Coonan and M.G., unpublished observations). Additionally, it has been observed that some rats that have had their CST disrupted by transection also show a phenotype with a hopping gait similar to the EphA4 null mutants (B. Bregman, personal communication). Thus, a defective CST can account for the motor defect, albeit other unrecognized defects in the motor system also may be present in these mice.
The perturbation of the CST in null mutant animals establishes that EphA4 is required for CST development. During CST development, the first pioneering axons to advance down the spinal cord are those that will innervate the lumbar segments, and these then are followed by a bulk of later-arriving fasciculating CST fibers projecting to upper cord segments (18). As the primary growth cones of corticospinal axons continue to elongate down the midline of the spinal cord, the brainstem and spinal cord targets are contacted by collateral branches sprouted along the corticospinal axon shafts (32). The paucity of CST axons observed within the lumbar spinal cord regions in the EphA4 mutants is presumably caused by misguidance of the primary cortical axons. It is possible that guidance of the collateral branches along the whole CST also are disrupted; however, this is yet to be determined. Altogether, these data strongly suggest that EphA4 regulates axon guidance in the CST.
The immunohistochemistry and in situ hybridization data suggest that EphA4 is not expressed by cortical motor neurons or in the CST during its development. However, EphA4 was found highly expressed within the intermediate and ventral regions of the spinal cord, which is the region where the CST axons do not normally terminate. This is consistent with the notion that EphA4 is expressed on structures surrounding the CST where it acts as a signal for CST axons bearing Ephrin ligands to be guided appropriately. Also consistent with this model, EphrinB3 mRNA was detected within the sensorimotor cortex at E 18.5, which suggests that this transmembrane ligand is expressed on CST axons as they extend through the brain and spinal cord. EphA4 binds to EphrinB3 with high affinity, and the transmembrane Ephrin ligands have been shown to induce signaling on receptor binding (12, 13, 33, 34).
Both in vitro and in vivo studies have suggested that the Eph receptor family regulate axon guidance through mechanisms of contact repulsion rather than attraction (5, 6, 35, 36). For example, in EphB2 receptor-null mice, the posterior tract of the AC innervates the floor of the brain aberrantly (7). EphB2 normally is expressed in areas ventral to the commissure, and the commissural axons express a ligand for EphB2, Ephrin-B1. This, therefore, suggests that EphB2 repels AC axons from entering this ventral area via Ephrin-mediated signals (33, 34). Our findings are consistent with a similar mechanism relating to guidance of the CST. A full understanding of the molecular mechanisms of CST guidance mediated by EphA4 will require a detailed temporal and spatial expression analyses of EphA4 and its ligands as the CST extends through the medulla and into the spinal cord.
The commissural defect in the EphA4 mutants is reminiscent of those found in other Eph null animals (7, 8). In the EphA4 null animals, the lateral projection of the AC across the midline is disrupted whereas in EphB2-deficient mice, only the posterior tract of the AC is disrupted (7). The EphB2 commissural defect is more severe in mutant mice deficient in both EphB2 and EphB3 receptors, although the AC appeared normal in the single EphB3 mutants (8). Mice deficient in the EphA8 receptor also revealed a commissural defect whereby tectal axons failed to project from the superior colliculus of the midbrain to the contralateral inferior colliculus and instead projected to the ipsilateral cervical spinal cord (9). Taken together these results suggest a requirement for multiple Eph family members to function in commissural tract formation.
Another molecule recently found to be involved in CST development is the neural cell adhesion molecule L1. In mice deficient in L1, many of the CST axons failed to decussate at the medulla, passing ipsilaterally into the dorsal columns (31). Similar to EphA4 null mice, the number of CST axons within the dorsal funiculus of the spinal cord was reduced, and these axons did not project beyond the cervical levels. It was proposed that the interaction of L1 on the axons with CD24 (expressed in the midline) may modify the response of CST axons to midline inhibitory cues, thereby allowing the axons to cross the midline. Another molecule shown to act as a guidance cue for CST axons is Netrin-1 (37). It was shown that the pathfinding of CST axons from the cortex to the internal capsule of the forebrain may be mediated by the chemoattractive activity of Netrin-1. Taken together with our data on the EphA4 null mice, these data show that CST axons are guided by the combined actions of a number of attractive and repulsive guidance cues.
Acknowledgments
We are grateful to D. G. Wilkinson for providing anti-EphA4 antibody, S. Mihajlovic for technical assistance in histology, and J. Scott for blastocyst microinjection. We thank J. Scott, J. De Winter, B. Shore, and coworkers for their dedicated animal husbandry.
ABBREVIATIONS
- AC
anterior commissure
- CST
corticospinal tract
- kb
kilobase
- E
embryonic day
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. 65138).
References
- 1. Brambilla R, Klein R. Mol Cell Neurosci. 1995;6:487–495. doi: 10.1006/mcne.1995.0001. [DOI] [PubMed] [Google Scholar]
- 2.Tessier-Lavigne M. Cell. 1995;82:345–348. doi: 10.1016/0092-8674(95)90421-2. [DOI] [PubMed] [Google Scholar]
- 3.Friedman G C, O’Leary D D M. Curr Opin Neurobiol. 1996;6:127–133. doi: 10.1016/s0959-4388(96)80018-3. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H J, Nakamoto M, Bergemann A D, Flanagan J G. Cell. 1995;82:371–381. doi: 10.1016/0092-8674(95)90426-3. [DOI] [PubMed] [Google Scholar]
- 5.Drescher U, Kremoser C, Handwerker C, Loschinger J, Noda M, Bonhoeffer F. Cell. 1995;82:359–370. doi: 10.1016/0092-8674(95)90425-5. [DOI] [PubMed] [Google Scholar]
- 6.Nakamoto M, Cheng H J, Friedman G C, McLaughlin T, Hansen M J, Yoon C H, O’Leary D D M, Flanagan J G. Cell. 1996;86:755–766. doi: 10.1016/s0092-8674(00)80150-6. [DOI] [PubMed] [Google Scholar]
- 7.Henkemeyer M, Orioli D, Henderson J T, Saxton T M, Roder J, Pawson T, Klein R. Cell. 1996;86:35–46. doi: 10.1016/s0092-8674(00)80075-6. [DOI] [PubMed] [Google Scholar]
- 8.Orioli D, Henkemeyer M, Lemke G, Klein R, Pawson T. EMBO J. 1996;15:6035–6049. [PMC free article] [PubMed] [Google Scholar]
- 9.Park S, Frisen J, Barbacid M. EMBO J. 1997;16:3106–3114. doi: 10.1093/emboj/16.11.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orioli D, Klein R. Trends Genet. 1997;13:354–359. doi: 10.1016/s0168-9525(97)01220-1. [DOI] [PubMed] [Google Scholar]
- 11.Gale N W, Holland S J, Valenzuela D M, Flenniken A, Pan L, Ryan T E, Henkemeyer M, Strebhard K, Hirai H, Wilkinson D G, et al. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- 12.Gale N W, Flenniken A, Compton D C, Jenkins N, Copeland N G, Gilbert D J, Davis S, Wilkinson D G, Yancopoulos G D. Oncogene. 1996;13:1343–1352. [PubMed] [Google Scholar]
- 13.Bergemann A D, Zhang L, Chiang M, Brambilla R, Klein R, Flanagan J G. Oncogene. 1998;16:471–480. doi: 10.1038/sj.onc.1201557. [DOI] [PubMed] [Google Scholar]
- 14.Nieto M A, Gilardi-Hebenstreit P, Charnay P, Wilkinson D G. Development (Cambridge, UK) 1992;116:1137–1150. doi: 10.1242/dev.116.4.1137. [DOI] [PubMed] [Google Scholar]
- 15.Mori T, Wanaka A, Taguchi A, Matsumoto K, Tohyama M. Brain Res Mol Brain Res. 1995;29:325–335. doi: 10.1016/0169-328x(94)00263-e. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Alldus G, Holder N, Wilkinson D G. Development (Cambridge, UK) 1995;121:4005–4016. doi: 10.1242/dev.121.12.4005. [DOI] [PubMed] [Google Scholar]
- 17.Xu Q, Alldus G, Macdonald R, Wilkinson D G, Holder N. Nature (London) 1996;381:319–322. doi: 10.1038/381319a0. [DOI] [PubMed] [Google Scholar]
- 18.Stanfield B B. Prog Neurobiol. 1992;38:169–202. doi: 10.1016/0301-0082(92)90039-h. [DOI] [PubMed] [Google Scholar]
- 19.Irving C, Nieto M A, DasGupta R, Charnay P, Wilkinson D G. Dev Biol. 1996;173:26–38. doi: 10.1006/dbio.1996.0004. [DOI] [PubMed] [Google Scholar]
- 20.Robb L, Lyons I, Ruili L, Hartley L, Kontgen F, Harvey R P, Metcalf D, Begley C G. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyons I, Parsons L M, Hartley L, Li R, Andrews J E, Robb L, Harvey R P. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 22.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 23.Galea M, Darian-Smith I. J Comp Neurol. 1997;381:282–306. [PubMed] [Google Scholar]
- 24.Rees S, Rawson J, Nitsos I, Brumley C. Brain Res. 1994;642:185–198. doi: 10.1016/0006-8993(94)90921-0. [DOI] [PubMed] [Google Scholar]
- 25.Adams J. J Histochem Cytochem. 1981;29:775. doi: 10.1177/29.6.7252134. [DOI] [PubMed] [Google Scholar]
- 26.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 27.DeFries J, Gervais M, Thomas E. Behav Genet. 1978;8:3–13. doi: 10.1007/BF01067700. [DOI] [PubMed] [Google Scholar]
- 28.Bregman B S, Goldberger M E. Science. 1982;217:553–555. doi: 10.1126/science.7089581. [DOI] [PubMed] [Google Scholar]
- 29.Bregman B S, Kunkel-Bagden E, Schnell L, Dai H N, Gao D, Schwab M E. Nature (London) 1995;378:498–501. doi: 10.1038/378498a0. [DOI] [PubMed] [Google Scholar]
- 30.Casale E J, Light A R, Rustioni A. J Comp Neurol. 1988;278:275–286. doi: 10.1002/cne.902780210. [DOI] [PubMed] [Google Scholar]
- 31.Cohen N R, Taylor J S H, Scott L B, Guillery P, Soriano P, Furley A J W. Curr Biol. 1997;8:26–33. doi: 10.1016/s0960-9822(98)70017-x. [DOI] [PubMed] [Google Scholar]
- 32.Bastmeyer M, O’Leary D D M. J Neurosci. 1996;16:1450–1459. doi: 10.1523/JNEUROSCI.16-04-01450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland S J, Gale N W, Mbamalu G, Yancopoulos G D, Henkemeyer M, Pawson T. Nature (London) 1996;383:722–725. doi: 10.1038/383722a0. [DOI] [PubMed] [Google Scholar]
- 34.Brückner K, Pasquale E B, Klein R. Science. 1997;275:1640–1643. doi: 10.1126/science.275.5306.1640. [DOI] [PubMed] [Google Scholar]
- 35.Ohta K, Iwamasa H, Drescher U, Terasaki H, Tanaka H. Mech Dev. 1997;64:127–135. doi: 10.1016/s0925-4773(97)00056-7. [DOI] [PubMed] [Google Scholar]
- 36.Wang H U, Anderson D J. Neuron. 1997;18:383–396. doi: 10.1016/s0896-6273(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 37.Richards L J, Koester S E, Tuttle R, O’Leary D D M. J Neurosci. 1997;17:2445–2458. doi: 10.1523/JNEUROSCI.17-07-02445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilardi-Hebenstreit P, Nieto M A, Frain M, Mattei M G, Chestier A, Wilkinson D G, Charnay P. Oncogene. 1993;8:1103. [PubMed] [Google Scholar]
- 39.Laird P W, Ziderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]