Abstract
The time course of polysome formation was studied in a long-term wheat germ cell-free translation system using sedimentation and electron microscopy techniques. The polysomes were formed on uncapped luciferase mRNA with translation-enhancing 5′ and 3′ UTRs. The formation of fully loaded polysomes was found to be a long process that required many rounds of translation and proceeded via several phases. First, short linear polysomes containing no more than six ribosomes were formed. Next, folding of these polysomes into short double-row clusters occurred. Subsequent gradual elongation of the clusters gave rise to heavy-loaded double-row strings containing up to 30–40 ribosomes. The formation of the double-row polysomes was considered to be equivalent to circularization of polysomes, with antiparallel halves of the circle being laterally stuck together by ribosome interactions. A slow exchange with free ribosomes and free mRNA observed in the double-row type polysomes, as well as the resistance of translation in them to AMP-PNP, provided evidence that most polysomal ribosomes reinitiate translation within the circularized polysomes without scanning of 5′ UTR, while de novo initiation including 5′ UTR scanning proceeds at a much slower rate. Removal or replacements of 5′ and 3′ UTRs affected the initial phase of translation, but did not prevent the formation of the double-row polysomes during translation.
INTRODUCTION
Polyribosomes or polysomes are clusters of translating ribosomes that are held together by mRNA (1–5). According to the generally accepted model, ‘ribosomes attach themselves to one end of the polysomal cluster, and then gradually move along the messenger strand as the polypeptide chain increases in length by sequential addition of amino acids starting from the N-terminal end; at the end of the messenger chain, the ribosomes are believed to detach and the polypeptide chain is released’ (cited from Ref. 6). Somewhat after the discovery of polysomes the results of functional studies using both intact cells and cell-free systems revealed that eukaryotic polysomes displayed a slow rate of exchange with free ribosomes or their subunits and used preferentially the terminating ribosomal particles for re-entry into the same translating polysomes (7–9). ‘The most simple explanation for the slow rate of exchange is a topographical one; ribosomes that have completed one round of translation dissociate into RSU (ribosomal subunits) near the site where RSU will attach to mRNA and initiate a new round of translation. In this model, the 3′-end (termination site) of mRNA should be close to the 5′-end (initiation site) of the same mRNA’ (cited from Ref. 9). Indeed, the electron microscopy observations demonstrated that eukaryotic polysomes were often visible as circular and double-row structures (10–12). Based upon this observation it was suggested that the polyribosomes could be arranged ‘in closed circle configuration’ and ‘ribosomes, carrying the growing peptide chain, could move along the circular mRNA without being released’ (7). Thus, the model of the circularization of eukaryotic polysomes and the circular translation of mRNA was proposed as early as in the 1960s.
More recently the circular organization of eukaryotic polysomes has been confirmed by electron microscopy studies with various mRNAs (13–17). In the case of long polysomes (long mRNA) they often form the so-called ‘double rows’ and ‘hairpins’ which appeared to be topologically circular polysomes with the 5′- and 3′-halves laterally stuck together and the 5′- and 3′-ends near each other (16,17). On the other hand, the progress in understanding of the role of poly(A) tail in initiation of translation on eukaryotic mRNAs has led to hypothesis that the interaction between the 5′-cap structure and the poly(A) sequence via poly(A)-binding protein (PABP) bridge is a prerequisite for an efficient initiation and translation—the so-called closed-loop model (18). A functional ‘synergy’ between cap and poly(A) tail of eukaryotic mRNAs was proclaimed even earlier (19). The idea was further supported by the finding of a direct association of PABP with the large subunit (eIF4G) of cap-binding initiation factor eIF4F (20–22). The physical circularization of mRNA complexed with eukaryotic translation initiation factors eIF4E/eIF4G and PABP was visualized by atomic force microscopy (23). From these data, the model of the circularization of eukaryotic polysomes and the circular translation of mRNA, now based on functional protein-mediated interaction of the cap structure with poly(A) tail, was born anew (24). True, it has been reported that an exogenous poly(A) also stimulates initiation of translation to the same extent and thus mRNA circularization per se seems to be not the cause of the functional cap–poly(A) synergy (25).
The 5′ and 3′UTRs of viral RNAs and some exceptional cellular mRNAs [e.g. 5′UTR of hydrozoan obelin mRNA (26) and 3′UTR of mammalian histone mRNA (27)] can circumvent the requirement for capping and polyadenylation of eukaryotic mRNAs. The RNAs of many plant viruses with RNA genomes, such as tobacco mosaic virus (TMV), alfalfa mosaic virus (AMV), brome mosaic virus (BMV), turnip yellow mosaic virus (TYMV), are not polyadenylated and, instead, have pseudoknot/tRNA-like 3′UTRs (28–30). It has been demonstrated that these 3′UTRs can functionally replace poly(A)-tail in translation when attached to heterologous (non-viral) coding sequences both in plant and animal systems, in vivo and in vitro (31–34). The recombinant mRNA constructs containing translation-enhancing 5′UTRs and 3′UTRs derived from viral RNAs, such as TMV RNA, were reported to display ‘synergy’ between the ends (35) in a manner similar to the synergism between cap and poly(A) in cellular mRNAs (19). Recently we have shown that the non-capped 5′UTR derived from hydroid polyp (Obelia longissima) mRNA encoding for the light-emitting protein obelin is a powerful enhancer of translation in a wheat germ cell-free translation system. This 5′UTR in combination with some viral 3′UTRs, such as those from STNV RNA and TMV RNA, provided efficient translation of mRNAs with foreign coding sequences (26,36).
The recent progress in cell-free protein synthesis methodology has provided new possibilities to study the processes of translation and polysome formation in long-term in vitro systems, such as continuous-flow and continuous-exchange (CFCF and CECF) cell-free translation systems (37,38). These systems are capable of working for many hours and thus can accomplish up to hundreds rounds of translation of the same mRNA by ribosomes. It should be mentioned, however, that cap and poly(A) are not very compatible with the long-term cell-free systems because of the enzymatic decapping and deadenylation processes which take place in cell extracts; however, these elements can be successfully replaced by some viral RNA leaders and other enhancing 5′ sequences in combination with viral 3′UTRs (such as those of TMV RNA, STNV RNA, etc.) (36,39). Using the CECF system based on the wheat germ extract Madin et al. (40) managed to reproduce the formation of long heavy-loaded polysomes of the double-row type under in vitro conditions in the absence of poly(A) tail and cap structure in mRNA. However, the process and the mechanism of formation of the double-row polysomes have not been studied.
In this work we analyzed the process of formation of the double-row type polysomes using luciferase-coding mRNA with uncapped 5′UTR of obelin mRNA and the 3′UTR of TMV RNA in the wheat germ CECF system. We found that polysome formation is a long process, requiring many rounds of translation that proceeds via distinct phases, starting from short linear polysomes, followed by their transformation into short double-row clusters, until they ultimately grow into heavy densely packed double-row-type polysomes. These double-row-type polysomes exhibited only a slow exchange with free ribosomes and free mRNA, thus suggesting re-initiation of translation by terminating ribosomes within the polysomes (‘circular translation’). Such a circularization during translation was also shown for polysomes formed on the luciferase mRNA with replaced or removed UTRs. In contrast to initial phases of translation, the re-initiation in the double-row polysomes was shown to be resistant against AMP-PNP, an inhibitor of ATP-dependent scanning of 5′ UTR by initiating 43S ribosomal complex.
MATERIALS AND METHODS
Materials
Restriction endonucleases HindIII, EcoRI, NcoI, SmaI and XhoI, T4 DNA ligase, calf intestine alkaline phosphatase, T7 and SP6 RNA polymerases, creatine phosphokinase, Expand High Fidelity PCR System, total yeast tRNA, creatine phosphate, dNTPs and NTPs were purchased from Roche Diagnostics. Endonucleases Acc65I, Bsp120I and RiboLock RNase inhibitor were from MBI Fermentas. Amino acids and AMP-PNP were purchased from Sigma, spermidine was from Fluka. [14C]leucine (306 mCi/mmol) and sodium [3H]borohydrid (16.7 Ci/mmol) were purchased from Amersham/Pharmacia Biotech.
Plasmid constructions
Plasmids were constructed from the pUC19 vector (Novagen). In the first step the expression vector pObeTMV was made so that it could be used to introduce a target open reading frame (ORF) between obelin mRNA 5′UTR (5′UTRObelin) (GenBank accession number U07128) and TMV 3′UTR (3′UTRTMV) (GenBank accession number NC_001367), this allowing efficient expression in a wheat germ cell-free system. The ObeTMV insert was prepared using 3-primer PCR with primers T7-Obe, Obe-TMV and TMV-r in molar ratio 20:1:20, and pYGFP plasmid (kindly provided by Dr T. Metzler, Roche Diagnostics, Penzberg, Germany) was used as a template for (His6-tag)-3′UTRTMV sequence. T7-Obe primer (5′TGCCAAGCTTAATACGACTCACTATAGATCTAACCAAACAACTCAGCTCACAGCTACTGAACAAC) contained HindIII site (underlined), T7 promoter sequence and 5′ part of obelin 5′UTR sequence (italics); Obe-TMV primer (5′CTCACAGCTACTGAACAACTCTTGTTGTGTACAATCACCATGGTACCCGGGGGGGGTTCTC) contained the 3′-part of obelin 5′UTR sequence (italics), start codon and NcoI and SmaI sites for inserting of ORF; TMV-r primer (5′CAGTGAATTCCGCATATATGGGC) containing EcoRI site was complementary to the 3′-end of TMV 3′UTR sequence in pYGFP. The generated DNA fragment was treated with HindIII and EcoRI restriction endonucleases and subcloned into similarly digested and dephosphorylated pUC19 to produce the pObeTMV vector for high yield expression of the cloned ORF in a wheat germ cell-free translation system. In the next step the plasmid pObeLucTMV was constructed by insertion of firefly luciferase coding sequence (GenBank accession number M15077) into NcoI/SmaI sites of pObeTMV vector. The luciferase coding sequence was taken from the plasmid pTZ10ΩLuc (kindly provided by K.S. Vassilenko, Institute of Protein Research RAS, Pushchino, Russia) by digesting it with XhoI, blunting with T4 DNA polymerase, and finally cutting with NcoI. The inserted fragment contained the complete luciferase coding sequence with stop-codon and 52 nt sequence of natural 3′UTR of luciferase mRNA.
pObeGFP-TMV plasmid was prepared by insertion of the DNA fragment containing the coding sequence of GFP with His6-tag flanked with 5′UTRObelin and 3′UTRTMV sequences into HindIII/EcoRI sites of pUC19. DNA fragment was generated in 3-primer PCR with T7-Obe, Obe-GFP (5′GCTCACAGCTACTGAACAACTCTTGTTGTGTACAATCACCATGACTAGCAAAGGAGAAGAAC) and TMV-r primers (20:1:20), and pYGFP plasmid was used as a template for GFP-(His6-tag)-3′UTRTMV sequence. Plasmid constructs were verified by restriction analysis and DNA sequencing.
pTZ10ΩLuc plasmid described in (41) contained luciferase coding sequence flanked with 5′UTR (Ω) and 3′UTR sequences of TMV RNA. Plasmid pTZ-gaLuc (kindly provided by K.S. Vassilenko) was the derivative of pTZ10ΩLuc where the 5′UTR (Ω) sequence was removed and the SP6 promoter sequence was added. In order to do this the DNA fragment was generated with primers SP-Luc (5′atcaagcttatttaggtgacactataGAATGGAAGACGCCAAAAAC) and Luc-Sph-r (5′CTGGCATGCGA GAATCTGAC) using pTZ10ΩLuc plasmid as template. The fragment was digested with HindIII and SphI and then inserted into similarly digested pTZ10ΩLuc plasmid. The resulting pTZ-gaLuc plasmid encoded Luc-3′UTRTMV mRNA where the start codon was preceded with only 2 nt (GA).
In vitro transcription
mRNAs were prepared by in vitro transcription of the linearized plasmids. pObeLucTMV digested with Bsp120I or Acc65I was used for synthesis of 5′UTRObelin-Luc-3′UTRTMV and 5′UTRObelin-Luc mRNAs, respectively; pTZ10ΩLuc digested with Bsp120I or XhoI was used for preparation of 5′UTRTMV-Luc-3′UTRTMV and Luc-3′UTRTMV mRNAs, respectively; pTZ-gaLuc digested with Bsp120I or XhoI was used for preparation of Luc-3′UTRTMV mRNA and mRNA without specific 5′ and 3′ UTRs (Luc), respectively. Transcription was performed in 100 μl aliquots in a reaction mixture containing 120 mM HEPES-KOH pH 7.6, 20 mM MgCl2, 20 mM dithiotreitol, 4 mM ATP, CTP, UTP and GTP each, 2 mM spermidine, 50 U of RNase inhibitor, 2 μg of the linearized plasmid and 300 U of T7 RNA polymerase (42). The reaction mixture was incubated for 2 h at 37°C. RNA was deproteinized with phenol-chloroform and precipitated with 3 M LiCl. The RNA pellet was then dissolved in MilliQ water, the solution was adjusted to 2.5 M NH4OAc and RNA was reprecipitated with ethanol (3 v/v). The transcripts homogeneity was checked by 4% polyacrylamide/urea gel electrophoresis.
mRNA translation in a wheat germ cell-free system (batch format)
Wheat germ extract (200 OU260/ml, ribosome concentration 140 OU260/ml) was prepared and cell-free translation reactions were performed according to protocols described in (36) and given in Supplementary Data. Briefly, translation mixture contained 30% (v/v) of wheat germ extract, 500 U/ml of human placental RNase inhibitor, 100 μg/ml of creatine phosphokinase, 50 µg/ml of yeast total tRNA, 0.2 mM each of 20 amino acids, 1 mM ATP, 0.4 mM GTP, 16 mM creatine phosphate and 25 mM HEPES-KOH buffer pH 7.6 with 3 mM Mg(OAc)2, 85 mM KOAc, 3 mM NaN3, 1.6 mM DTT, 0.25 mM spermidine, 2% glycerol. The reaction mixture was first preheated for 2 min at 25°C before initiating the reaction at 25°C by the addition of mRNA (final concentration 300 nM or as indicated). Luciferase synthesis was monitored by measuring the luminescence in situ or by analyzing 2 µl aliquots supplemented with 0.01 mg/ml cycloheximide.
mRNA translation in a wheat germ CECF system
Translation in the wheat germ continuous-exchange cell-free (CECF) system was performed according to protocols described in (36) and given in detail in Supplementary Data. The reaction mixture (100 µl) was prepared in the same way as described earlier and incubated in the reactor equipped with a reaction chamber of 1 mm thickness covered with a flat cellulose membrane of 12000–14000 kDa cut-off (Servapor, Serva), and a feeding solution chamber filled with 1 ml of feeding solution containing the same components as the reaction mixture but without the wheat germ extract, mRNA, tRNA, creatine phosphokinase and RNase inhibitor. The assembled reactor was incubated at 25°C with continuous stirring. The aliquots and samples taken for sedimentation, luciferase activity test or EM analysis were supplemented with 0.01 mg/ml cycloheximide to stop translation.
Luciferase activity test
Luciferase synthesis was monitored by an activity test with luciferin. Briefly, a 2 µl aliquot of the translation reaction mixture was added to 20 µl of the luciferase reaction buffer [50 mM HEPES-KOH pH 7.6, 10 mM Mg(OAc)2, 70 mM KOAc, 2 mM ATP, 0.1 mM luciferin, 1 mM DTT], the mixture was incubated for 2 min at 25°C, and then the luminescence was recorded using TRIATHLER luminometer (Hidex, Finland). Alternatively, in the case of in situ monitoring (43), luciferin was added to the translation reaction mixture to the concentration of 0.1 mM, and the luminescence in the reaction volume (10 ml) was recorded in 2 min intervals during translation in a luminometer cell kept at 25°C. The luminescence data were directly collected by computer in tabular form and further processed in the IGOR Pro® package (WaveMetrics).
Sedimentation analysis of polysomes
Polysome profiles were analyzed by sedimentation in a concave 15–50% sucrose gradient prepared in buffer containing 15 mM HEPES-KOH pH 7.6, 5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA and 0.01 mg/ml cycloheximide. The concave gradient was found to be optimal for providing the sufficiently high resolution of different size polysome peaks and for preventing heavy-loaded polysome pellet formation. The concave gradients were formed as follows: 7.1 ml of 15% sucrose solution was placed in mixer, 50% sucrose was pumped into the mixer at 0.25 ml/min while the gradient solution was pumped out through the capillary inserted into a centrifuge tube at 0.55 ml/min. Fifty microliters aliquots of the translation reaction mixture supplemented with 0.01 mg/ml cycloheximide were loaded on the gradient and the tubes were centrifuged in SW-41 rotor (Beckman) at 37 000 rpm for 80 min at 5°C. UV-absorbance (254 nm) profile along the gradient was recorded by pumping the content of the tube through the Uvicord SII flow-photometer (LKB) connected to AD-converter E-24 (L-CARD), and 0.5 ml fractions were collected. Absorbance profile recorded in blank gradient was subtracted. PowerGraph® and IGOR Pro® software were used for plotting and analysis of polysome profiles. When radioactive amino acids were present in the reaction mixture the radioactivity in gradient fractions was analyzed by hot TCA precipitation and scintillation counting. Isolation of polysomes for EM analysis was done by sedimentation of 100 µl sample in linear glycerol gradient (20–50%, SW-55, 37 000 rpm, 80 min at 5°C).
Electron microscopy
Specimens for EM studies were prepared by the surface spreading technique (44) in micro-scale variant (40) with some modifications. This technique provides spreading of macromolecules by surface tension within a protein monolayer on an aqueous surface. A 2 µl drop of translation mixture was placed on a Teflon film, and after 2 min a sample was taken from the drop with platinum loop (diameter 1.5 mm) and placed onto the surface of hypophase (10 mM HEPES-KOH pH 7.6, 5 mM MgCl2, 100 mM KCl) in 5 cm Teflon dish. The specimen was then mounted as usual on the EM grid with carbon film, washed twice with the buffer and negatively stained with 2% aqueous uranyl acetate. Specimens of polysomes isolated by glycerol gradient centrifugation were mounted on the EM grid directly from samples of the gradient fractions and negatively stained using single-layer carbon technique (45) or shadowed by carbon-platinum at an angle of tan1/3 using an electron gun (46). The samples were examined in JEM 100C microscope operating at 80 kV.
Preparation of 3H-labeled ribosomes by reductive methylation
Ribosomes were isolated from wheat germ extract by pelletting through a 20% glycerol cushion (30 µl) containing 40 mM HEPES-KOH pH 7.6, 5 mM Mg(OAc2), 400 mM KOAc, 4 mM dithiotreitol in a TLA100 rotor (Beckman) at 67 000 rpm for 90 min at 5°C. The dissolved pellets were dialyzed against buffer containing 10 mM HEPES-KOH pH 7.6, 5 mM Mg(OAc2), 50 mM KOAc. For reductive methylation the ribosome solution was diluted to 2 mg/ml with the reaction buffer [50 mM HEPES-KOH pH 8.9, 5 mM Mg(OAc)2, 50 mM KOAc], and then 5 µl of 80 mM formaldehyde was added to 100 µl of the ribosome solution on ice. After a 2.5 min incubation 300 nmol (5 mCi) of sodium [3H]borohydride was added and the mixture was incubated for 20 min on ice. Ribosomes were purified from reactants by passing through an NAP-5 column (Amersham/Pharmacia Biotech) equilibrated in WGE buffer (40 mM HEPES pH 7.6, 5 mM Mg(OAc)2, 100 mM KOAc, 4 mM dithiotreitol) followed by sedimentation in TLA100.2 rotor at 100 000 rpm for 40 min at 5°C. Ribosome pellets were dissolved in WGE buffer and the solutions were clarified by low-speed centrifugation (20 min at 14 000 rpm in a table-top centrifuge). The specific radioactivity of [3H]ribosome preparations was 250 000 cpm/OU260 (in TCA-precipitated material). Translational activity of 3H-labeled ribosomes was tested and found to be the same as that of non-labeled ribosomes, and the 3H-labeled ribosomes were shown to be incorporated into translating polysomes in cell-free systems.
Polysomes-[3H]ribosomes exchange experiment
Translation of luciferase mRNA was performed in five parallel 100 µl CECF reactors for 1 h at 25°. Following this initial translation step, the reaction mixtures were combined in one 500 µl batch and 1.7 OU260 of [3H]ribosomes (approximately 8% of the ribosome amount in the reaction mixture) were added. Translation was continued, and 80 µl aliquots were taken at 0, 10, 20, 30, 40 and 60 min after the addition of [3H]ribosomes and layered onto a 15–45% sucrose gradient with 0.01 mg/ml cycloheximide. The gradients were centrifuged in SW-41 rotor at 37 000 rpm for 120 min at 5°C and fractionated as described. Radioactivity in fractions was determined after precipitation with 10% TCA using scintillation counting on CF/F filters. The content of [3H]ribosomes in polysomes was calculated as radioactivity per UV-absorbance unit. In control experiment [3H]ribosomes were added at the start of translation in CECF system to estimate their content in polysomes at equilibrium.
AMP-PNP inhibition test
Translation reaction was started with 5′UTRObelin-GFP-3′UTRTMV mRNA in batch format as described earlier but with 0.1 mM concentration of ATP and 2.1 mM of Mg(OAc)2. At indicated time points (10 or 60 min after start of translation) equimolar mixture of AMP-PNP and Mg(OAc)2 was added up to 2 mM final concentration. In control experiments ATP-Mg(OAc)2 mixture was added instead; in a blank control the reaction was performed essentially as described for standard batch reaction. The mRNA coding for GFP was used in place of luciferase mRNA because AMP-PNP impeded the ATP-dependent luciferase activity test. GFP synthesis was monitored by measuring the fluorescence of 2 µl aliquots in RF5301 spectrofuorometer (Shimadzu) operating at 395 nm excitation and 510 nm emission wavelengths. Calibration curve was obtained using standard GFP solution.
RESULTS
Translation of luciferase mRNA (5′UTRObelin-Luc-3′UTRTMV) in a wheat germ cell-free system
The mRNA, designated 5′UTRObelin-Luc-3′UTRTMV, was constructed by inserting the firefly luciferase coding sequence between obelin mRNA 5′UTR and TMV RNA 3′UTR, as described in ‘Materials and Methods’ section. The pair of these UTRs has been reported to enhance expression of coding sequences in a wheat germ cell-free translation system (36).
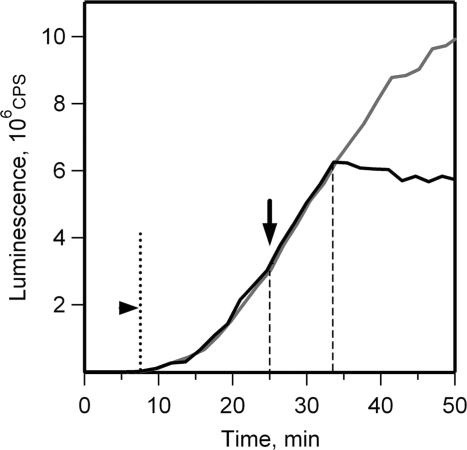
The initial phase of translation was monitored in the reaction mixture (batch format) containing luciferin using the in situ luciferase activity assay (43). In this assay the duration of one round of translation can be defined as the time after which the first luciferase luminescence signal is registered. During this time the first ribosomes, which initiated on added mRNA, complete translation, and then terminate and release the first active luciferase molecules. Figure 1 shows that in our experiments (25°C) the first active luciferase molecules appeared in 7–8 min after the start of translation. In order to determine the duration of one round of translation at later stages in the translation reaction an inhibitor of eukaryotic translation initiation, antibiotic edeine, was added to the incubation mixture and the time, after which the release of activity stops, was recorded. As shown in Figure 1, this time was 8 min. These data indicate that the ribosomes in the cell-free translation system read out the full sequence of luciferase mRNA for 7–8 min (at 25°C), both at the beginning and at the later stages of the translation reaction.
Figure 1.
Determination of the time of one round of translation for luciferase mRNA in a wheat germ cell-free system. Luciferase synthesis was monitored by in situ measurement of the luminescence at 2 min intervals in batch reaction mixture containing 0.1 mM luciferin. Arrowhead indicates the time at which the first active luciferase molecules appear. Edeine was added to 1 µM concentration at the 25th minute (indicated by arrow) to block translation initiation (black line); control reaction was without edeine addition (grey line).
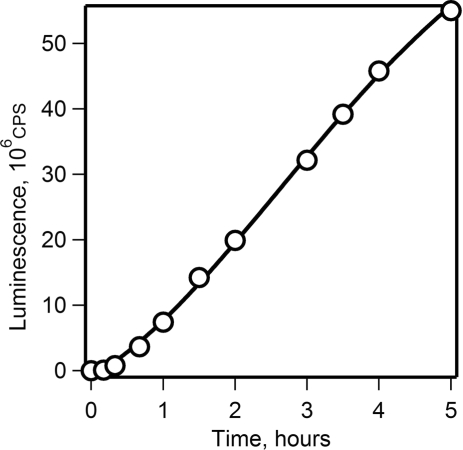
In contrast to the batch format, the continuous format of cell-free translation system provides for the steady-state conditions of translation reaction and therefore maintains the constant rate of translation for many hours (37,38). Figure 2 shows that the rate of protein synthesis remained unchanged for 5 h when translation of the luciferase mRNA was performed in the CECF translation system (the dialysis mode).
Figure 2.
Time course of translation of luciferase mRNA in a long-term wheat germ CECF system. Luciferase synthesis was monitored by measuring the luminescence in 2 µl aliquots taken at the indicated time points.
Time course of polysome formation in the cell-free system: sucrose gradient centrifugation analysis
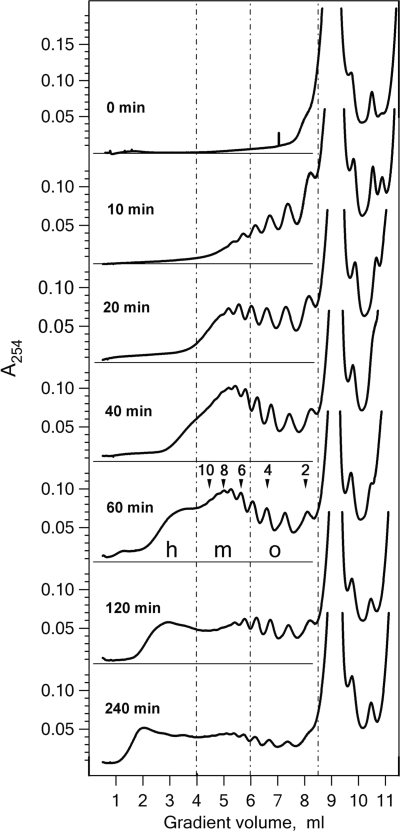
The polysome profiles at different time points of the luciferase synthesis in the wheat germ CECF system was analyzed by sucrose gradient centrifugation. Fifty microliters aliquots of translation mixture were withdrawn from the CECF reactor at indicated time points, the reaction was stopped by addition of cycloheximide, and the samples were layered onto sucrose gradients. A concave 15–50% sucrose gradient and short centrifugation time were employed in order to detect the heavy polysome species (see ‘Materials and Methods’ section). Figure 3 shows that after 10 min translation, when the first round of translation was completed, mainly short oligosomes were detected. During the following 30 min the fraction of medium size polysomes (6–12 ribosomes) increased and heavy polysomes containing more than 12 ribosomes began to appear. After 60 min of translation 12 peaks of short and medium size polysomes could be clearly distinguished, and the fraction of heavy polysomes accumulated in lower fractions significantly increased. During subsequent hours of translation the heavy polysomes became a major fraction while the amount of medium size polysomes decreased. The rate of luciferase synthesis remained constant during the entire reaction (Figure 2).
Figure 3.
Sucrose gradient sedimentation profiles of polysomes formed in the wheat germ CECF system with luciferase mRNA. Fifty microliters aliquots of the reaction mixture were taken at the indicated incubation time and analyzed in concave 15–50% sucrose gradient (see ‘Materials and Methods’ section). Gradient zones containing oligosomes (o), medium size (m) and heavy-loaded (h) polysomes are indicated by dashed lines. Optical densities in polysome-containing zones were integrated with IGOR Pro® package. In the presented sedimentation profiles the medium-size polysomes constituted 25, 44, 49, 43, 36, 35%, and heavy-loaded polysomes 5, 19, 24, 35, 42, 53% of total polysome amount at the 10th, 20th, 40th, 60th, 120th and 240th min of incubation time, respectively.
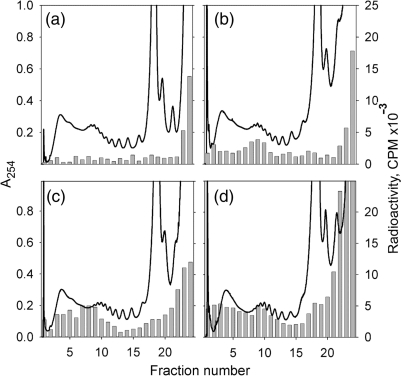
The translational activity of polysome fractions after 2 h of incubation was examined. A radioactive amino acid was added to the cell-free system after 2 h of translation and the radioactivity incorporation into synthesized protein was monitored in the fractions of sucrose gradient at different time intervals. As shown in Figure 4, the radioactive amino acid was incorporated in the medium-loaded and heavy-loaded polysomes and the radioactivity level approached the maximum within the first 8 min (the time of one round of translation), thus demonstrating that the polysomes from both fractions had normal activity.
Figure 4.
Activity of polysomes formed during 2 h of luciferase mRNA translation in the wheat germ CECF system. After 2 h incubation in the dialysis (CECF) mode [14C]leucine was added to the concentration of 40 µM and the reaction was continued in the batch mode. Fifty microliters aliquots of the reaction mixture were taken at the points of 0 min (a), 4 min (b), 8 min (c) and 24 min (d) after the label addition and subjected to sedimentation analysis in sucrose gradient. UV-absorbance was monitored at 254 nm (line); radioactivity incorporation into TCA-precipitated material from sucrose gradient fractions was analyzed (bars).
Time course of polysome formation in the cell-free system: electron microscopy analysis
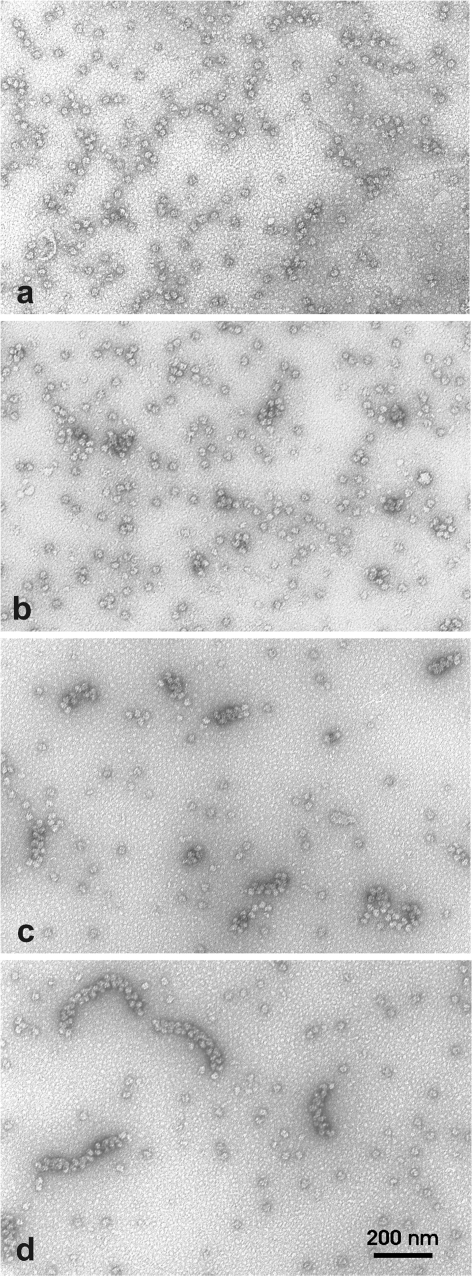
Formation of polysomes programmed with the luciferase mRNA was examined by EM as described in ‘Materials and Methods’ section. In full correspondence with the sucrose gradient centrifugation profile, after the first 10 min of incubation only short polysomes containing three to six ribosomes were observed on electron micrographs (Figure 5a). These polysomes appeared as linear strings of ribosomes. During the next 10 min of translation the formation of longer linear polysomes was not observed. Instead, we observed close-packed clusters composed of mostly four to six ribosomes (Figure 5b). During further incubation the close-packed clusters evolved into double-row structures. After 40 min of translation the close-packed double-row polysomes constituted the major component of the polysome population (Figure 5c). The average number of ribosomes within these polysomes was 8 to 10. At the same time the number of monosomes in the preparation had significantly decreased. Furthermore, the size of the close-packed double-row polysomes continued to increase without any change in their general morphology. After 2 h of incubation the average size of heavy-loaded polysomes was approximately 30 ribosomes (Figure 5d). The longest of them, the ‘super heavy’ polysomes, contained up to 40 ribosomes.
Figure 5.
EM analysis of polysome formation during translation of luciferase mRNA in the wheat germ CECF system. Aliquots of the reaction mixture were withdrawn after 10 min (a), 20 min (b), 40 min (c) and 120 min (d) of incubation. EM samples were prepared by surface spreading and negatively stained with uranyl acetate.
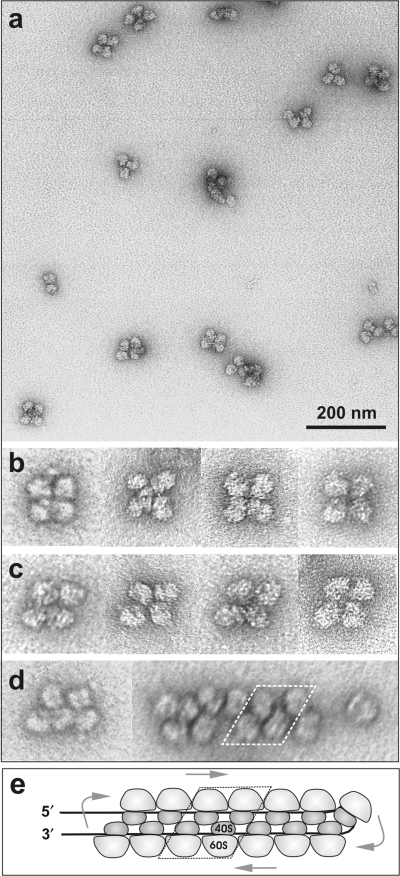
For a closer inspection of the arrangement of ribosomes in polysomes, the fractions of polysomes were isolated by glycerol gradient centrifugation. In Figure 6 the EM images of polysomes from the tetrasome fraction after 20 min of translation are shown. The fraction contained predominantly the tetrasomes with close-packed ribosomes. It can be seen that the ribosomes contact each other by their small (40S) subunits faced to the center of a tetrasome. Two forms of tetrasomes, rectangular (Figure 6b) and rhomb-shaped (Figure 6c), were observed in approximately equal proportions. (It can be imagined that the adjacent pairs of ribosomes in a tetrasome may slide relative each other, this shift providing an easy transformation of one form to the other). It should be mentioned that in longer polysomes the contacts characteristic of the rhomb-shaped tetrasomes were predominantly realized (Figure 6d).
Figure 6.
EM micrographs of particles from the tetrasome fraction isolated by glycerol gradient centrifugation. Negative staining with uranyl acetate. (a) A field of tetrasomes. (b and c) Selected images of rectangular and rhomb-shaped tetrasomes at higher magnification. (d) Individual images of an oligosome and a medium-size double-row polysome. (e) The model of a circular polysome with two antiparallel halves of the circle being laterally stuck together (the double-row polysome). Small ribosomal subunits are viewed from their heads. Arrows indicate the path of ribosomes along the mRNA chain.
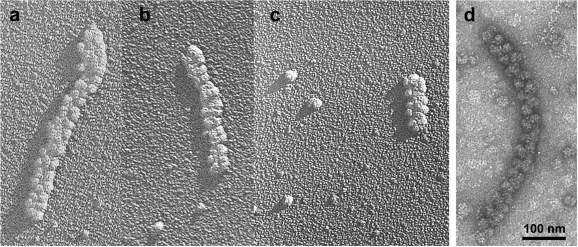
Figure 7 represents the EM micrographs of shadowed and negatively stained single double-row polysomes isolated by glycerol gradient centrifugation after 2 h of translation. The shadow casting technique was specially used in order to examine whether the two rows of ribosomes lay in the same plane. Indeed, the length of a polysome shadow in Figure 7a–c does not exceed the length of a single ribosome shadow. Hence, the height of a polysome is equal to that of a ribosome. The above observations indicate that the ribosomes within the densely packed double-row polysomes are regularly arranged, with the two rows of ribosomes contacting each other through their small subunits, being in the same plane and not forming a three-dimensional helix. Similarly packed double-row polysomes of smaller length (12–18 ribosomes) were observed during translation of 5′UTRObelin-GFP-3′UTRTMV mRNA with twice shorter coding sequence (data not shown).
Figure 7.
EM micrographs of shadowed (a–c) and negatively stained (d) single double-row-type polysomes isolated by glycerol gradient centrifugation. Polysomes were isolated after 2 h of translation of luciferase mRNA in the wheat germ CECF system. Fractions of heavy (a,b,d) and medium-size (c) polysomes are presented.
Functional properties of the double-row-type polysomes: slow exchange with free ribosomes and resistance to competitive mRNA
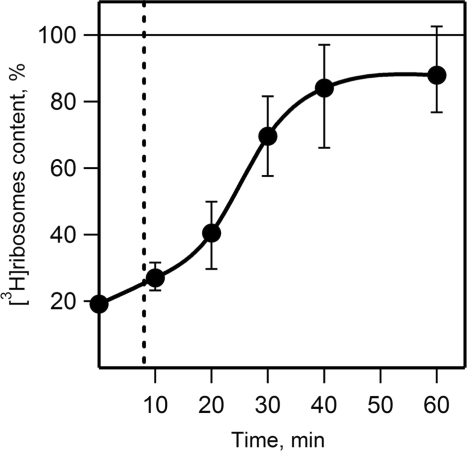
In order to test whether the double-row polysomes display a circular mode of translation the exchange of polysomal ribosomes with free ribosomes after 1 h translation was analyzed. At this time point the medium-loaded double-row polysomes constituted the major polysome fraction (Figures 3 and 5). The ribosomes exchange was examined by the addition of wheat germ 3H-labeled ribosomes to the cell-free system after formation of the double-row polysomes (after 1 h translation, see ‘Materials and Methods’ section). The time course of [3H]ribosomes incorporation into polysome peaks was monitored by sucrose gradient sedimentation analysis (Figure 8 and Figure 1S). In a control experiment the 3H-labeled ribosomes were added at the start of translation; the level of incorporation of the labeled ribosomes into polysomes in this control was taken as the equilibrium level (100% in Figure 8). Upon the addition of [3H]ribosomes the radioactive label was quickly observed in short polysomes and slowly accumulated in longer polysomes (see Supplementary Data, Figure 1S). As seen in Figure 8, the equilibrium level of radioactivity in the fraction of nona- to dodecasomes was reached only after 40 min of translation. The duration of one round of translation was measured after 1 h of translation in experiment with inhibition of initiation by edeine (see Supplementary Data, Figure 2S). We found that one round of translation took 7–8 min at this time point, such a duration being similar to that observed at the start of translation (Figure 1). Consequently, up to five rounds of translation of mRNA within polysomes was required to fully exchange the polysomal ribosomes with free ribosomes in the translation reaction mixture.
Figure 8.
Time course of incorporation of free 3H-labeled ribosomes into polysomes. [3H]ribosomes were added to the wheat germ system after 1 h translation of luciferase mRNA. Incorporation of [3H]ribosomes into medium-size polysomes was examined by sucrose gradient centrifugation of aliquots of the reaction mixture taken at indicated time points after addition of [3H]ribosomes and their content in polysomes was calculated as radioactivity per UV-absorbance unit in gradient fractions. The [3H]ribosome content in the sample where [3H]ribosomes were added at the start of translation was taken as 100% (indicated by horizontal line). Vertical dashed line indicates the time of one round of mRNA translation (8 min). See also Figure 1S in Supplementary Data. Error bars indicate the data spread in three independent experiments.
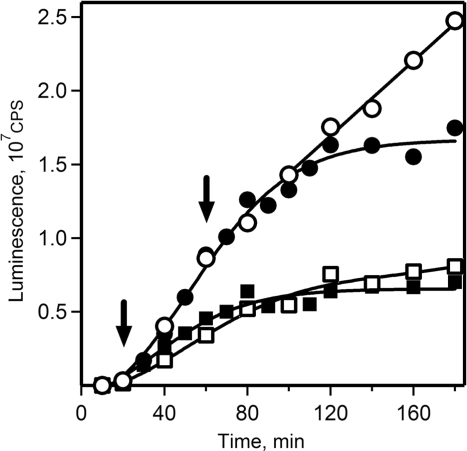
Thus, the above experiments demonstrated that the ribosomes of the double-row-type polysomes slowly exchange with free ribosomal particles, and most polysomal ribosomes reinitiate translation without being released into the common ribosomal pool. Similar conclusion was made earlier by Nelson and Winkler (47) who studied the kinetics of labeled histone mRNA entry into polysomes using nuclease-treated reticulocyte lysates. The question arised, however, as to whether the terminating polysomal ribosomes preferentially reinitiate on polysomal mRNA (‘circular translation’), or whether they are just in an activated functional state after termination that enable them to overcompete other ribosomes in the process of de novo initiation. In order to discriminate between these alternative interpretations, an experiment was performed in which an excess competitive mRNA (GFP mRNA with obelin RNA leader and TMV RNA tail) was added to the double-row polysomes formed during preceding 1 h translation (Figure 9). If such pre-activated ribosomes were released as a result of termination, we predict they should be trapped by the added competitive mRNA; as a consequence, the preformed polysomes should shorten and the synthesis of protein on them should decline. As shown in Figure 9, we did not observe the predicted effect: the synthesis of luciferase on the preformed polysomes was unaffected for as long as 40 min (five rounds of translation). On the contrary, the addition of the competitive mRNA at the 20th minute of translation triggered a decline of the rate of luciferase synthesis to the level similar to that in control sample when both mRNAs were added simultaneously at the start of translation. Thus, ribosomes in the fully formed double-row polysomes do not switch to the translation of competitive mRNA, while ribosomes of short polysomes can be readily distributed between polysomal and competitive mRNAs. From all this it follows that in the double-row-type polysomes terminating ribosomes preferentially reinitiate on the same polysomal mRNA. In other words, the double-row-type polysomes are found to be functionally circular polysomes.
Figure 9.
Effect of the addition of excess competitive mRNA on translation of luciferase mRNA in the wheat germ CECF system. Translation reaction was started with 250 nM 5′UTRObelin-Luc-3′UTRTMV mRNA alone (open circle, filled square, filled circle) or with the same amount of this mRNA and 1250 nM competitive 5′UTRObelin-GFP-3′UTRTMV mRNA (open square). The competitive 5′UTRObelin-GFP-3′UTRTMV mRNA was added to the reaction mixtures after 20 min (filled square) or 60 min (filled circle) of incubation as indicated by arrows. Luciferase synthesis was monitored by measuring the luminescence in 2 µl aliquots taken at the indicated time points.
Are 5′ and 3′ UTRs required for the formation of the double-row-type polysomes?
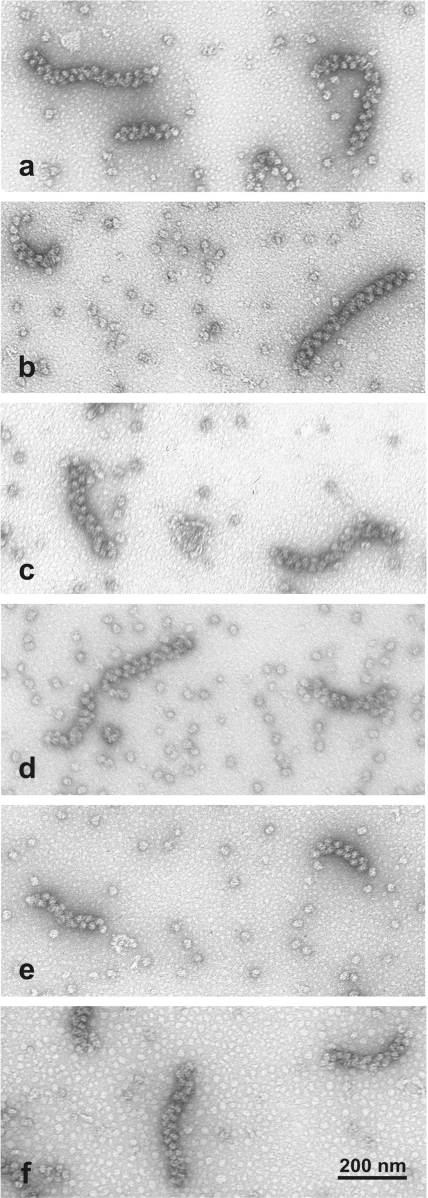
In order to control whether the interactions or synergism between 5′ and 3′ UTRs of mRNA could determine the formation of the functionally circular polysomes, we tested several other constructs of luciferase mRNA where either 5′ UTR, or 3′ UTR, or both were changed or deleted. It was found that despite the differences at the initial phase of translation with different combinations of 5′ and 3′ UTRs, the rates of translation upon the formation of polysomes became similar (see Supplementary Data, Figure 3S). The most exiting observation was that in all the cases tested, including those of leaderless mRNA, tailless mRNA and mRNA without both 5′ and 3′ enhancing sequences, the polysomes of the double-row type of the same appearance were visualized by elecrtron microscopy (Figure 10). Hence, neither functional synergism, nor direct physical interactions between 5′ and 3′ UTRs seem to be necessary for the formation of the double-row polysomes. From this we propose that the formation of the double-row polysomes and the consequent functional circularization could be determined by direct interactions between the two antiparallel rows of the translating double-row polysome.
Figure 10.
EM micrographs of double-row polysomes formed during 2 h of translation in the cell-free systems programmed with different luciferase mRNA constructs. (a, b) mRNAs with both 5′ and 3′ UTRs: 5′UTRObelin-Luc-3′UTRTMV, and 5′UTRTMV-Luc-3′UTRTMV, respectively; (c, d) mRNAs with only 5′UTR: 5′UTRObelin-Luc, and 5′UTRTMV-Luc (Ω-Luc), respectively; (e) mRNA with only 3′UTR: Luc-3′UTRTMV; (f) mRNA without specific 5′ and 3′ UTRs (Luc). Specimens were prepared by surface spreading and negatively stained with uranyl acetate.
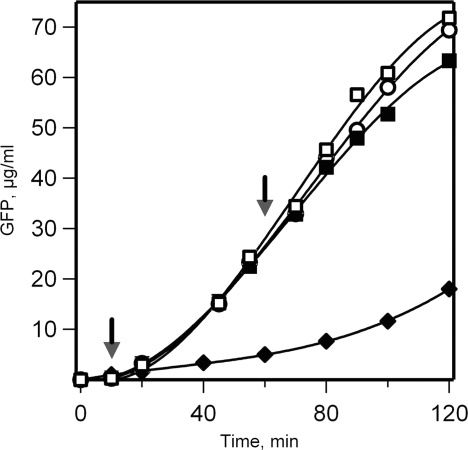
At the same time the fact of the independence of the double-row polysome formation on 5′ UTR rises the question whether 5′ end and the leader sequence are involved in the re-initiation of translation required for maintenance of the active polysome. The 5′ end and the leader sequence are known to be very important for the de novo translation initiation on free mRNA, an essential steps of this process being the association with the end and the subsequent ATP-dependent scanning of the leader sequence. As to the requirements for the intra-polysomal re-initiation process, nothing is known. In order to check whether the ATP-dependent scanning of 5′ UTR, when it is present, is involded in the re-initiation process, we inhibited the ATP-dependent RNA-helicase (eIF4A) by the addition of great excess AMP-PNP to the cell-free system. Figure 11 shows that, according to the expectation, the de novo translation was completely blocked by the inhibitor, whereas no significant inhibition of translation was observed when AMP-PNP was added after the formation of the double-row polysomes. This result indicates that the re-initiation in the double-row polysomes occurs without scanning of 5′ UTR sequnce, but rather via direct jumping of terminating ribosomes to initiation site.
Figure 11.
Effect of AMP-PNP, a non-hydrolyzable analog of ATP, on mRNA translation in a cell-free system. The wheat germ cell-free system programmed with 5′UTRObelin-GFP-3′UTRTMV mRNA (500 nM) was incubated in batch as described in ‘Materials and Methods’ section at the ATP concentration of 0.1 mM. AMP-PNP was added to the reaction mixture to the final concentration of 2 mM in 10 (filled diamond) or 60 (filled square) minutes after translation start. In control reactions the same amount of ATP was added after 10 min (open square), or the reaction was conducted without any addition (open circle). Arrows indicate the time of AMP-PNP or ATP addition. The synthesis of GFP was recorded by the measurement of fluorescence in 2 µl aliquots taken at the indicated time points.
DISCUSSION
In earlier studies the double-row-type polysomes formed on long eukaryotic mRNAs were observed both on ultrathin sections of animal tissues (12,17) and in the samples isolated from eukaryotic cells (11,16), as well as in a eukaryotic (wheat germ) cell-free translation system (40). In some electron microscopy images the double-row polysomes were visible as oblate circles (see e.g. 16,17,40). This allowed one to interpret the double rows as circularized polysomes with two ends of a read-out mRNA sequence being in close proximity and the two halves of the cycle laterally contacting each other.
In this work the process of formation and the functional properties of the double-row-type polysomes were studied for the first time. The polysomes were formed on a luciferase-encoding mRNA (1650 nt coding sequence) containing 5′ and 3′ translation-enhancing UTRs in a wheat germ CECF translation system. The use of the CECF system allowed the translation process to proceed with steady concentrations of substrates over many hours, thus more effectively mimicking in vivo conditions. The time course of polysome formation in this translation system was monitored by sucrose gradient sedimentation technique for quantitative analysis of the polysomes size distribution, and by electron microscopy for visualization of the structural organization of polysomes. The duration of one round of translation was measured and used as a scaling factor to evaluate the time course of polysome growth; the time of one round of translation of mRNA by a single ribosome was determined to be equal to 7–8 minutes in our system.
We have found that the formation of the heavy-loaded double-row polysomes is a long step-wise process that requires many rounds of mRNA translation. The process has been shown to proceed via several intermediate stages. During the first round of translation, short linear polysomes containing three to six ribosomes were formed (Figure 5a). Subsequently, the packing of polysomal ribosomes into short two-row clusters was observed; it began as early as at the stage of tetrasomes (Figure 5b). During the next rounds of translation the initial small clusters of ribosomes grew into double-row arrays. By the 40th minute, equivalent to five rounds of translation, most polysomes acquired the double-row structure and the linear forms disappeared (Figure 5c). The length of the double-row polysomes increased during further translation, and after 2 h the heavy-loaded polysomes containing about 25 ribosomes became abundant (Figure 5d). In whole, the process of the heavy-loaded polysome formation approached the completion only by 90–120 min of translation and thus included up to 15 rounds of translation.
It is noteworthy that the presence of 5′ and 3′ UTRs in mRNA was found to be not strictly required for the formation of the double-row polysomes: the mRNAs without 5′ UTR, or 3′ UTR, or both were still capable of forming the double-row-type polysomes. The hypothesis may be proposed that direct interactions between polysomal ribosomes could well contribute to the formation of the double-row polysomes and thus to the circular translation of mRNA. It can be assumed that the tetrasome stage is critical for the formation of the polysomal double-row arrays: at this stage the possibility for cooperative antiparallel interaction between ribosomes of two halves of a polysome string first appears, and such folded tetrasomes act as nucleators for the subsequent growth of the double-row polysomes.
The schematic model of a circular polysome with antiparallel halves of the cycle stuck together into a double-row structure is presented in Figure 6e. As already mentioned, a characteristic feature of the double-row polysomes often visible on EM micrographs is that the two rows contact each other via small ribosomal subunits. This observation is consistent with topological circularity of the double-row polysomes if ribosomes are universally arranged on mRNA with small subunits facing inside the cycle. Moreover, in order to provide the shortest path for mRNA chain between ribosomes all small subunits must be in a ‘head-to-head’ orientation. It is noteworthy that the ‘head-to-head’ orientation of small subunits was earlier found in tetramers of prokaryotic ribosome crystals (48,49) and in dimers of 70S ribosomes (50), and also seen in the atomic force microscopy image of a eukaryotic disome particle (16).
The heavy-loaded double-row polysomes contained 25–35 ribosomes (Figure 7a and d). As the coding sequence of luciferase mRNA is 1650 nt residues long, each ribosome covered a 50–70 nt section. In an extended RNA chain 1 nt spans a distance of 0.59 nm (51). This implies that the translating ribosomes are arranged along the extended mRNA chain at the distance of 30–40 nm between their centers. It should be noted that for very densely stacked clusters of ribosomes caused by the pausing of the leading ribosome in eukaryotic polysomes, an even smaller distance of 27–29 nt between the ribosome centers was reported (52).
As already mentioned, the growth of the double-row polysomes proceeds slowly. In accordance to this, it was directly shown that the ribosomes of the double-row polysomes exchanged with free ribosomes of the incubation mixture at a relatively slow rate. The complete replacement of ribosomes from the formed double-row polysomes by newly entered radioactive-labeled ribosomes required about 40 min that is equivalent to five rounds of translation (Figure 8). This means that only 1–2 ribosomes are added to the polysome during each round of translation. At the same time, during all the stages of the double-row polysome formation they are normally active in protein synthesis, with each polysomal ribosome running over the full length of the coding sequence within ∼8 min.
Correspondingly, the addition of free competitive mRNA to the fully formed double-row polysomes did not cause immediate switching of polysomal ribosomes to newly added mRNA: the translation rate in polysomes remains unaffected for more than five rounds of translation (Figure 9). These data imply that the rate of re-initiation on the polysomal mRNA greatly exceeds the rate of de novo initiation on free mRNA. However, if competitive mRNA was added to polysomes at the initial stage of their formation, before the formation of the double rows, translation in polysomes decreased after one round of translation manifesting that at this stage the re-initiation in polysomes was not yet prevailing over the de novo initiation.
In the previous publication from our laboratory the phenomenon of acceleration of protein synthesis during the first rounds of translation of non-capped luciferase mRNA in a wheat germ cell-free system was demonstrated (41). The acceleration observed was discussed in terms of the model of two different initiation pathways: translation starts with slow initiation at 5′ UTR, and after somewhat loading with translating ribosomes the polysome rearranges in such a way that the second mechanism, namely re-initiation of terminating ribosomes, switches on. Here this model is further supported. The model implies that in the formed polysomes the termination site of mRNA is close to the initiation site of the same mRNA, and thus the polysomes are arranged in closed circle configuration. The circular organization of polysomes may be attained via formation of the double-row polysomes during translation, probably due to inter-ribosomal interactions between two antiparallel halves of the polysomal cycle. Three facts give evidence that the re-initiation of terminated ribosomes proceeds without ATP-dependent scanning of 5′ UTR, but rather via a direct shunting from termination site to initiation site of mRNA: (1) 5′UTR is not strictly required for the double-row polysome formation during translation; (2) after the formation of the double-row polysomes the rates of translation become independent of the sequence and the presence of 5′UTR; (3) translation in the formed double-row polysomes is not inhibited by AMP-PNP, an inhibitor of RNA-helicase that is strictly required for the scanning.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Funding was provided by The Program on Molecular and Cellular Biology of Russian Academy of Sciences; Russian Foundation for Basic Research (grants # 06-04-49152 to V.S., and 01-04-49720 to V.V.) We are very grateful to V.A. Kolb, L. Runnel, A.G. Ryazanov and K.S. Vassilenko for fruitful discussions and critical reading of the manuscript. We thank T. Metzler and K.S. Vassilenko for providing us with the plasmids. We greatly appreciate the skillful technical assistance of E. Arutyunyan and S. Proshkina. Funding to pay the Open Access publication charges for this article was provided by the Institute of Protein Research, Sussian Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Warner JR, Rich A, Hall C. Microscope studies of ribosomal clusters synthesizing hemoglobin. Science. 1962;138:1399–1403. doi: 10.1126/science.138.3548.1399. [DOI] [PubMed] [Google Scholar]
- 2.Warner JR, Knopf PM, Rich A. A multiple ribosomal structure in protein synthesis. Proc. Natl Acad. Sci. USA. 1963;49:122–129. doi: 10.1073/pnas.49.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gierer A. Function of aggregated reticulocyte ribosomes in protein synthesis. J. Mol. Biol. 1963;6:148–157. doi: 10.1016/s0022-2836(63)80131-x. [DOI] [PubMed] [Google Scholar]
- 4.Wettstein FO, Staehelin T, Noll H. Ribosomal aggregate engaged in protein synthesis: characterization of the ergosome. Nature. 1963;197:430–435. doi: 10.1038/197430a0. [DOI] [PubMed] [Google Scholar]
- 5.Penman S, Scherrer K, Becker Y, Darnell J. Polyribosomes in normal and poliovirus-infected HeLa cells and their relationship to messenger-RNA. Proc. Natl Acad. Sci. USA. 1963;49:654–662. doi: 10.1073/pnas.49.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rich A, Warner JR, Goodman HM. The structure and function of polyribosomes. Cold Spring Harbor Symp. Quant. Biol. 1963;28:269–285. [Google Scholar]
- 7.Philipps GR. Haemoglobin synthesis and polysomes in intact reticulocytes. Nature. 1965;205:53–56. doi: 10.1038/205567a0. [DOI] [PubMed] [Google Scholar]
- 8.Adamson SD, Howard GA, Herbert E. The ribosome cycle in a reconstituted cell-free system from reticulocytes. Cold Spring Harbor Symp. Quant. Biol. 1969;34:547–554. doi: 10.1101/sqb.1969.034.01.062. [DOI] [PubMed] [Google Scholar]
- 9.Baglioni C, Vesco C, Jacobs-Lorena M. The role of ribosomal subunits in mammalian cells. Cold Spring Harbor Symp. Quant. Biol. 1969;34:555–565. doi: 10.1101/sqb.1969.034.01.063. [DOI] [PubMed] [Google Scholar]
- 10.Mathias AP, Williamson R, Huxley HE, Page S. Occurrence and function of polysomes in rabbit reticulocytes. J. Mol. Biol. 1964;9:154–167. doi: 10.1016/s0022-2836(64)80097-8. [DOI] [PubMed] [Google Scholar]
- 11.Shelton E, Kuff EL. Substructure and configuration of ribosomes isolated from mammalian cells. J. Mol. Biol. 1966;22:23–31. [Google Scholar]
- 12.Dallner G, Siekevitz P, Palade GE. Biogenesis of endoplasmic reticulum membranes. I. Structural and chemical differentiation in developing rat hepatocyte. J. Cell Biol. 1966;30:73–96. doi: 10.1083/jcb.30.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu MT, Coca-Prodos M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 14.Ladhoff AM, Uerlings I, Rosenthal S. Electron microscopic evidence of circular molecules in 9-S globin mRNA from rabbit reticulocytes. Mol. Biol. Rep. 1981;7:101–106. doi: 10.1007/BF00778739. [DOI] [PubMed] [Google Scholar]
- 15.Christensen AK, Kahn LE, Bourne CM. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Amer. J. Anat. 1987;178:1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida T, Wakiyama M, Yazaki K, Miura K-I. Transmission electron and atomic force microscopic observation of polysomes on carbon-coated grids prepared by surface spreading. J. Electron Microscopy (Japan) 1997;46:503–506. doi: 10.1093/oxfordjournals.jmicro.a023550. [DOI] [PubMed] [Google Scholar]
- 17.Christensen AK, Bourne CM. Shape of large bound polysomes in cultured fibroblasts and thyroid epithelial cells. Anat. Record. 1999;255:116–129. doi: 10.1002/(SICI)1097-0185(19990601)255:2<116::AID-AR2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 19.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 20.Tarun SZJ, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 21.Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 22.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Well SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 24.Preiss T, Hentze MW. From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 25.Borman AM, Michel YM, Malnou CE, Kean KM. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J. Biol. Chem. 2002;277:36818–36824. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- 26.Shaloiko LA, Granovsky IE, Ivashina TV, Ksenzenko VN, Shirokov VA, Spirin AS. Effective non-viral leader for cap-independent translation in a eukaryotic cell-free system. Biotech. Bioeng. 2004;88:730–739. doi: 10.1002/bit.20267. [DOI] [PubMed] [Google Scholar]
- 27.Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol. Cell. Biol. 2002;22:7853–7867. doi: 10.1128/MCB.22.22.7853-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haenni A-L, Joshi S, Chapeville F. tRNA-like structures in the genomes of RNA viruses. Prog. Nucleic Acid Res. Mol. Biol. 1982;27:85–104. doi: 10.1016/s0079-6603(08)60598-x. [DOI] [PubMed] [Google Scholar]
- 29.Mans RMV, Pleij CWA, Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur. J. Biochem. 1991;201:303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- 30.Florentz C, Giegé R. tRNA-like structures in plant viral RNAs. In: Söll D, RajBhandary UL, editors. tRNA. Structure, Biosynthesis, and Function. Washington, DC: ASM Press; 1995. pp. 141–163. [Google Scholar]
- 31.Gallie DR, Walbot V. RNA preudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 1990;4:1149–1157. doi: 10.1101/gad.4.7.1149. [DOI] [PubMed] [Google Scholar]
- 32.Gallie DR, Feder JN, Schimke RT, Walbot V. Functional analysis of the tobacco mosaic virus tRNA-like structure in cytoplasmic gene regulation. Nucleic Acids Res. 1991;19:5031–5036. doi: 10.1093/nar/19.18.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryabova LA, Torgashov AF, Kurnasov OV, Bubunenko MG, Spirin AS. The 3′-terminal region of alfalfa mosaic virus RNA4 facilitates the RNA entry into translation in a cell-free system. FEBS Lett. 1993;326:264–266. doi: 10.1016/0014-5793(93)81804-9. [DOI] [PubMed] [Google Scholar]
- 34.Zeyenko VV, Ryabova LA, Gallie DR, Spirin AS. Enhancing effect of the 3′-untranslated region of tobacco mosaic virus RNA on protein synthesis in vitro. FEBS Lett. 1994;354:271–273. doi: 10.1016/0014-5793(94)01126-5. [DOI] [PubMed] [Google Scholar]
- 35.Leathers V, Tanguay R, Kobayashi M, Callie DR. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirokov VA, Kommer A, Kolb VA, Spirin AS. Continuous-exchange protein-synthesizing systems. In: Grandi G, editor. Methods in Molecular Biology, Vol. 375: In Vitro Transcription and Translation Protocols. 2nd. Totowa, NJ: Humana Press Inc; 2007. pp. 19–55. [DOI] [PubMed] [Google Scholar]
- 37.Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB. A continuous cell-free translation system capable of producing polypeptides in high yield. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 38.Spirin AS. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotech. 2004;22:538–545. doi: 10.1016/j.tibtech.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Gudkov AT, Ozerova MV, Shiryaev VM, Spirin AS. 5′-poly(A) sequence as an effective leader for translation in eukaryotic cell-free systems. Biotech. Bioeng. 2005;91:468–473. doi: 10.1002/bit.20525. [DOI] [PubMed] [Google Scholar]
- 40.Madin K, Sawasaki T, Kamura N, Takai K, Ogasawara T, Yazaki K, Takei T, Miura K-I, Endo Y. Formation of circular polyribosomes in wheat germ cell-free protein synthesis system. FEBS Lett. 2004;562:155–159. doi: 10.1016/S0014-5793(04)00221-2. [DOI] [PubMed] [Google Scholar]
- 41.Alekhina OM, Vassilenko KS, Spirin AS. Translation of non-capped mRNAs in a eukaryotic cell-free system: acceleration of initiation rate in the course of polysome formation. Nucleic Acids Res. 2007;35:6547–6559. doi: 10.1093/nar/gkm725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pokrovskaya ID, Gurevich VV. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal. Biochem. 1994;220:420–423. doi: 10.1006/abio.1994.1360. [DOI] [PubMed] [Google Scholar]
- 43.Kolb VA, Makeyev EV, Spirin AS. Folding of firefly luciferase during translation in a cell-free system. EMBO J. 1994;13:3631–3637. doi: 10.1002/j.1460-2075.1994.tb06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinschmidt AK. Monolayer techniques in electron microscopy of nucleic acids molecules. Methods Enzymol. 1968;12:361–377. [Google Scholar]
- 45.Valentine RC, Shapiro BM, Stadtman ER. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry. 1968;7:2143–2152. doi: 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- 46.Vasiliev VD, Koteliansky VE. Freeze-drying and high-resolution shadowing in electron microscopy of Escherichia coli ribosomes. Methods Enzymol. 1979;59:612–629. doi: 10.1016/0076-6879(79)59117-4. [DOI] [PubMed] [Google Scholar]
- 47.Nelson EM, Winkler MM. Regulation of mRNA entry into polysomes. Parameters affecting polysome size and the fraction of mRNA in polysomes. J. Biol. Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- 48.Selivanova OM, Vassilenko OV, Shirokov VA, Agalarov SCh, Vasiliev VD. Planar tetragonal microcrystals of 70S ribosomes from Thermus thermophilus: tetramer structure. Dokl. Akad. Nauk (Moscow) 1997;356:272–274. [English translation: Doklady Biophysics (1997), 352–357, 65–67.] [Google Scholar]
- 49.Yusupova GZh, Yusupov MM, Cate JHD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 50.Lake JA. Packing of ribosomes in dimers formed at low ionic strength. J. Mol. Biol. 1982;160:369–373. doi: 10.1016/0022-2836(82)90182-6. [DOI] [PubMed] [Google Scholar]
- 51.Sundaralingam M. Evolution of conformational principles in nucleic acids. Intern. J. Quantum Chem. Quantum Biol. Symposium. 1974;1:81–91. [Google Scholar]
- 52.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.