Abstract
Granulocyte macrophage-colony stimulating factor (GM-CSF) is produced by T cells, but not B cells, in response to immune signals. GM-CSF gene activation in response to T-cell stimulation requires remodelling of chromatin associated with the gene promoter, and these changes do not occur in B cells. While the CpG methylation status of the murine GM-CSF promoter shows no correlation with the ability of the gene to respond to activation, we find that the basal chromatin environment of the gene promoter influences its ability to respond to immune signals. In unstimulated T cells but not B cells, the GM-CSF promoter is selectively marked by enrichment of histone acetylation, and association of the chromatin-remodelling protein BRG1. BRG1 is removed from the promoter upon activation concomitant with histone depletion and BRG1 is required for efficient chromatin remodelling and transcription. Increasing histone acetylation at the promoter in T cells is paralleled by increased BRG1 recruitment, resulting in more rapid chromatin remodelling, and an associated increase in GM-CSF mRNA levels. Furthermore, increasing histone acetylation in B cells removes the block in chromatin remodelling and transcriptional activation of the GM-CSF gene. These data are consistent with a model in which histone hyperacetylation and BRG1 enrichment at the GM-CSF promoter, generate a chromatin environment competent to respond to immune signals resulting in gene activation.
INTRODUCTION
Highly dynamic changes in gene expression are a feature of the immune system, where the orchestration of an immune response to invading pathogens relies on rapid induction of cytokine gene expression. However, a key feature of cytokine gene expression is that many cytokines are expressed in a cell-type restricted fashion. For example, granulocyte macrophage-colony stimulating factor (GM-CSF) which plays a key role in myeloid cell production and function is induced in a range of cell types including T cells, macrophages, endothelial cells and fibroblasts, but not B cells, in response to immune or inflammatory signals (1). The combination of transcription factor-binding sites found in the promoters and enhancers of cytokine genes and the availability of these transcription factors in particular cell types is an important factor in the cell-type specific expression patterns of cytokines. However, it is clear that the chromatin environment of cytokine genes can also dictate the ability of transcription factors to access the control regions of genes (2,3), and therefore provides an additional level at which cell-type specific gene expression can be controlled.
DNA methylation, histone modifications and variants and also the location and density of nucleosomes can all contribute to the generation of a specific epigenetic environment that can determine the competency of a gene to respond to activating signals (4). It is now well established that many cytokine genes are found in a relatively inaccessible chromatin environment and the activation of gene expression requires the reorganization of chromatin structure to a more accessible form (2,3). For example, it has long been known that activation of both the GM-CSF and IL-2 genes following T-cell activation is accompanied by the appearance of DNase I hypersensitive (DH) sites in promoter and enhancer regions (5–7). The appearance of these DH sites correlates well with the ability of the GM-CSF gene to be activated in different cell types (8), and therefore it is likely that these chromatin remodelling events contribute to the cell-type specificity of gene expression. It is now clear that the appearance of DH sites across the IL-2 promoter and GM-CSF promoter and enhancer reflect highly localized changes in chromatin structure, involving the selective disruption of one or two nucleosomes (9–12), and these events were recently shown to involve the depletion of histones from these regions (13,14). However, the mechanisms involved in eviction of histones from the GM-CSF and IL-2 promoters in response to T-cell activating signals remain to be elucidated. Furthermore, it is not clear how these histones are specifically targeted for depletion in T cells and whether they are differentially marked in particular cell types to enable their remodelling.
Changes in chromatin structure which facilitate gene activation can be brought about by two general mechanisms. First, histone proteins are subject to a range of modifications, including acetylation, phosphorylation and methylation, which may alter higher order chromatin structure directly or act as binding sites for non-histone proteins that are able to modify chromatin structure and function (15,16). The association of histone acetylation with gene activation has been well documented at many individual genes, e.g. (17,18) as well as in a number of genome-wide studies (19–21). Secondly, a number of different complexes that are able to harness the energy from ATP hydrolysis to remodel nucleosomes have been described (22). The best characterized of these is the SWI/SNF complex, which has been implicated in the activation of a number of inducible genes. For example, the SWI/SNF complex is required for the activation of the myogenin gene during myoD-induced muscle differentiation (23) and has been demonstrated to contribute to activation of a number of pro-inflammatory genes following macrophage activation (24).
The importance of histone modifications and chromatin remodelling complexes in bringing about changes in chromatin structure is now well established and the precise series of events involved in remodelling nucleosomes to facilitate inducible gene transcription are understood in detail for several genes. The best described of these are the response of the IFN-β gene to viral infection, and the yeast Pho5 and Pho8 genes to low phosphate concentrations. In both these cases, changes in chromatin structure are facilitated by stimulus-driven local increases in histone acetylation and recruitment of the SWI/SNF remodelling complex (25–28).
Activation of the GM-CSF cytokine gene in response to T-cell activating stimuli is accompanied by the selective depletion of histones from the promoter (13), reminiscent of that observed at the Pho5 promoter (29,30). Here we demonstrate a role for local histone acetylation and the SWI/SNF ATPase BRG1 in activation of the murine GM-CSF gene. However, in contrast to the genes described above, BRG1 is not recruited to the promoter in response to T-cell activating signals, but rather is poised at the GM-CSF promoter in unstimulated EL-4 T cells. Similarly, histone acetylation does not increase at the GM-CSF promoter in response to T-cell activation, but is already enriched at the GM-CSF promoter in unstimulated T cells. These marks are not present at the GM-CSF promoter in B cells, in which gene expression is not inducible. These data suggest that both BRG1 and histone acetylation are involved in generating a transcriptionally competent environment at the GM-CSF promoter and this environment contributes to the cell-type specific expression of GM-CSF in immune cells.
MATERIALS AND METHODS
Plasmids
The BRG1 construct pBJ5-BRG1K/R was provided by Dr G. Crabtree and the mouse GM-CSF constructs, AOGM and pGM0.2, were provided by Dr P. Cockerill, and have been described previously (31,32).
Cell culture
Murine EL-4 T cells were cultured in RPMI as described previously (9). Murine A20 and WEHI231 B cells were cultured in DMEM supplemented with 10% fetal bovine serum (JRH Biosciences, USA), 2 mM l-glutamine (CSL, Australia), 100 U/ml penicillin, 100 μg/ml streptomycin (CSL) and 0.05 mM 2-mercaptoethanol (Sigma-Aldrich, USA) at 37°C and 5% CO2. Cells were stimulated with 20 ng/ml phorbol 12-myristate 13-acetate (PMA, Boehringer Mannheim, Germany) and 1 μM calcium ionophore (A23187; Sigma–Aldrich). Cells were pre-treated with 200 ng/ml trichostatin A (TSA, Sigma–Aldrich) for 4–16 h, and were treated with 2-deoxy-5-azacytidine (Sigma–Aldrich) at 0.25 µM for 24 h, at which point the treatment was removed and the cells grown for a further 24 h.
Primary cells were extracted from the spleens of 8-week-old male C57B6 mice. Spleens were homogenized, and leucocytes were then isolated by density gradient centrifugation in Lympholyte®-M (Cedarlane, USA) according to manufacturer's instructions. CD4+ T cells and CD19+ B cells were magnetically separated from the leucocytes using CD4 (L3T4) or CD19 MicroBeads and MS MACS® columns (Miltenyi Biotec, Germany) following the manufacturer's instructions. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (JRH Biosciences), 2 mM l-glutamine (CSL), 100 U/ml penicillin, 100 μg/ml streptomycin (CSL), 1 mM sodium pyruvate, 10 mM HEPES (Sigma–Aldrich) and 0.05 mM 2-mercaptoethanol (Sigma–Aldrich) at 37°C and 5% CO2. Cells were stimulated as described above for cell lines.
Transfection and luciferase reporter assay
EL-4 and A20 cells (4.5 × 106) were transfected with 5 µg of pGL3-Control (Promega, USA) or 10 µg of GM0.2 plasmid at 270 V and 975 µF using a Bio-Rad Gene Pulser X Cell as previously described (33). At 24 h post-transfection, cells were stimulated with PMA and calcium ionophore for 8 h. Protein was isolated, quantitated by Bradford assay (Bio-Rad, USA) and 30 µg of protein analysed for luciferase activity (Luciferase assay kit, Promega) using a Turner Biosystems Veritas™ Microplate Luminometer. CpG methylated pGM0.2 plasmid was prepared for transfections by incubating 150 µg of plasmid with 200 U of M.SssI methylase (New England Biolabs, USA) and 640 µM S-adenosylmethionine for 3 h at 37°C, followed by phenol–chloroform extraction and ethanol precipitation.
Sorting transfected cells
EL-4 T cells were co-transfected by electroporation with 8 μg of either pBJ5 or pBJ5-BRG1K/R plasmids along with 3 μg of KKII selection plasmid (Miltenyi Biotech). Transfected cells were enriched 24 h post-transfection using the MACSelect transfected cell selection kit according to the manufacturer's instructions (Miltenyi Biotech).
Nuclear extracts and western blotting
Nuclear extracts were prepared by a modification of the method of Schreiber et al. (34), as described previously (35). Nuclear proteins were separated by SDS–PAGE through 12% polyacrylamide, transferred onto nitrocellulose membrane and subjected to western blot analysis using anti-HA (Sigma–Aldrich), anti-AcH3 (Upstate Biotech, USA), anti-Sp1, anti-p65, anti-cRel and anti-BRG1 (Santa Cruz Biotechnology, USA) antibodies and the corresponding peroxidase-conjugated secondary antibodies (DAKO, Denmark). Proteins were visualized using the Supersignal West Pico Chemiluminescent kit (Pierce, USA).
RNA isolation and real-time PCR analysis
Total RNA was isolated using Tri-Reagent (Sigma) and reverse transcribed using Superscript II Reverse Transcriptase (Life Technologies, USA), and SYBR Green PCR amplification performed on the Rotor-Gene 2000 real-time cycler (Corbett Research, Australia) using the QuantiTect SYBR Green PCR kit (Qiagen, USA), as described previously (35) PCR was conducted using GM-CSF primer set +II and in parallel using the GAPDH primers (Table 1) to normalize for differences in cDNA synthesis and RNA input. To correlate the threshold (Ct) values from the amplification plots to copy number, a standard curve was generated using the mouse GM-CSF plasmid AOGM (31) and pCR2.1-GAPDH plasmid (35). PCR product melt curves were analysed for a single peak and the products visualized by agarose gel electrophoresis and ethidium bromide staining to ensure that a single product was generated in the PCR.
Table 1.
Primers used for CHART-PCR, ChIP or mRNA measurements by quantitative PCR
| Primer set | Sense (5′–3′) | Anti-sense (5′–3′) |
|---|---|---|
| 5′ | GAGCTTCTGGAGAGGGAGGT | TCCCAGGCTTAGTCTGTTGC |
| −V | TGGAATGAGCCACCAGAGTA | GGCTCTTGCTTCCATAGCAC |
| −I | GCCTGACAACCTGGGGGAAG | TGATTAATGGTGACCACAGAACTC |
| +I | GAGTTCTGTGGTCACCATTAATCA | CACATCCTCCTCAGGACCTT |
| +II | AAGGTCCTGAGGAGGATGTG | GAGGTTCAGGGCTTCTTTGA |
| 3′ | ATTTGGGCATAGGTGGAGTG | CCTCGATTTCACCTCCCTTT |
| ESP1 | GTCACTCCACCCATCTGGTC | AGGCATTCCTAGATTGCACG |
| GAPDH | AAGTATGATGACATCAAGAAGGTGGT | AGCCCAGGATGCCCTTTAGT |
| RhoM | GCAGCTGGTCTTCACAGTCA | GAAGTCACCCGCATGGTTAT |
CHART-PCR (chromatin accessibility by real-time PCR)
Accessibility of DNA to digestion with restriction enzymes and micrococcal nuclease (MNase) was analysed using CHART-PCR (9). Cell nuclei (5 × 106 nuclei per 100 μl) were treated with 150 U HinfI enzyme (New England BioLabs) at 37°C for 45 min or with 25 U MNase (Roche Boehringer Mannheim, Germany) for 5 min at 20°C. In each case, control samples without enzyme were incubated similarly to monitor for endonuclease activity. Genomic DNA was subsequently isolated using a QIAamp blood kit (Qiagen), and analysed by SYBR Green quantitative PCR in a total volume of 25 μl using the QuantiTect SYBR Green PCR kit (Qiagen). The primer sequences are shown in Table 1. Accessibility was determined by correlating the Ct-values from the amplification plots to a standard curve generated with the AOGM plasmid, and was expressed as a percentage of undigested genomic DNA for each primer set.
Methylation assay
Genomic DNA was isolated from cell lines using the QIAamp Blood Kit (Qiagen) and 1 g was mock digested or digested with AciI (1 U, New England Biolabs) for 16 h at 37°C. DNA (50 ng) was analysed by quantitative PCR using primer sets as shown in Table 1. Percent methylation was determined by correlating the Ct-values from the amplification plots to a standard curve generated with the AOGM plasmid and was expressed as a percentage of the mock digested genomic DNA for each primer set.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as previously described (13). Briefly, chromatin from formaldehyde fixed cells was fragmented to a size range of 100–500 bp. Solubilized chromatin was immunoprecipitated with 4 μg of antibody against acetylH3 and acetylH3K9 (Upstate Biotechnology), 1.5 μg anti-acetylH4 and anti-histone H3 (Abcam) or 1 μg anti-Brg1 antibody (Santa Cruz Biotechnology). Immune complexes were recovered using salmon sperm DNA/protein A-agarose, washed and eluted. Following cross-link reversal and proteinase K treatment, immunoprecipitated DNA was purified by phenol–chloroform extraction and ethanol precipitation. DNA was re-suspended in 50 μl MilliQ water and amplified by real-time PCR. Data were analysed relative to no antibody control immunoprecipitates and normalized to total input samples.
RESULTS
GM-CSF mRNA expression and promoter chromatin remodelling are blocked in B cells
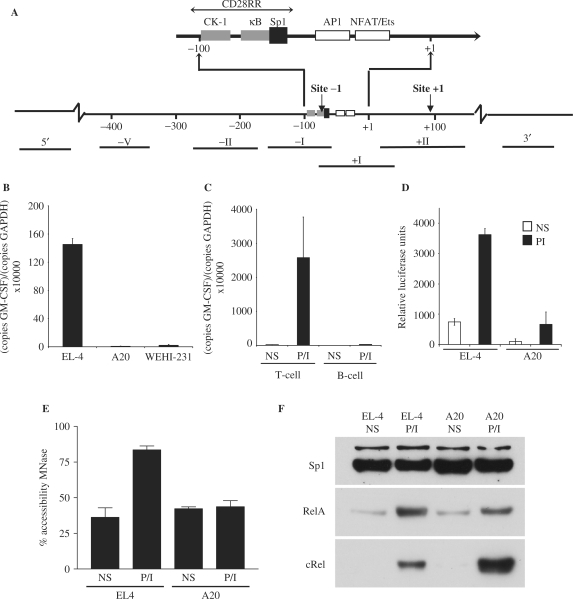
GM-CSF mRNA expression can be rapidly induced in T cells by treatment with the phorbol ester, PMA and calcium ionophore (P/I), which activates the PKC and calcium signalling pathways, respectively. For example, P/I treatment of the murine EL-4 T cell line for 4 h resulted in an increase in GM-CSF mRNA levels, as measured by quantitative PCR analysis (Figure 1B). In contrast, little GM-CSF mRNA expression was detected in the murine A20 and WEHI-231 B-cell lines in response to P/I treatment (Figure 1B). Similarly, GM-CSF mRNA expression was induced in response to P/I treatment in murine splenic CD4+ T cells, but not splenic B cells (Figure 1C).
Figure 1.
GM-CSF expression in T cells versus B cells. (A) Schematic representation of the GM-CSF promoter showing transcription factor-binding sites, CpG sites and the CD28RR. DNA fragments amplified by various primer sets are indicated and numbered with roman numerals. (B and C) mRNA isolated from P/I stimulated EL-4, A20 and WEHI-231 cells (B) or primary CD4+ T and CD19+ B cells (C) was reverse transcribed and amplified by PCR using GM-CSF specific primers. The data are expressed as copies of GM-CSF per copies of GAPDH. The mean and standard error of three (cell lines) or two (primary cells) independent experiments are shown. (D) EL-4 T cells and A20 B cells were transfected with a GM-CSF promoter luciferase reporter construct and stimulated with P/I. Protein was extracted and luciferase activity monitored. The mean and standard error of three independent experiments are shown. (E) Nuclei isolated from non-stimulated and P/I stimulated EL-4 T cells and A20 B cells were incubated with MNase and accessibility to the GM-CSF promoter determined by CHART-PCR. The mean and standard error of three independent experiments are shown. (F) Nuclear extracts prepared from non-stimulated and P/I stimulated EL-4 T cells and A20 B cells were subjected to western analysis using the indicated antibodies.
To determine whether this may be due to differences in GM-CSF promoter activity, a luciferase reporter construct containing the mouse GM-CSF promoter (31) was transfected into EL-4 T cells or A20 B cells and luciferase activity measured. In both cell types, promoter activity increased ∼6-fold following stimulation of the cells with P/I for 8 h, although basal activity was considerably lower in A20 B cells compared with the EL-4 T cells (Figure 1D). Both cell lines demonstrated approximately equal activity of a control reporter plasmid driven by the SV40 promoter (pGL3-Control) demonstrating equal transfection efficiency (data not shown). Therefore, while expression of mRNA from the endogenous GM-CSF gene is blocked in B cells, the GM-CSF promoter responds to P/I stimulation in a reporter assay with equal efficiency in both cell types.
We have previously shown that induction of GM-CSF mRNA expression in T cells in response to P/I stimulation requires the specific depletion of histones from the gene promoter, resulting in its increased accessibility (11,13). To determine whether these chromatin remodelling events also occur across the GM-CSF promoter in B cells, accessibility of the promoter to MNase was examined using a quantitative PCR-based assay [CHART-PCR, (9)]. EL-4 T cells and A20 B cells were either left unstimulated or stimulated with P/I for 4 h, nuclei isolated, incubated with MNase and genomic DNA isolated. The accessibility of the GM-CSF promoter was then monitored by PCR amplification of the genomic DNA using primer set −I (Figure 1A), with the amount of PCR product generated from digested samples plotted as a percentage of the amount generated from undigested genomic DNA samples. As described previously (11), a relatively high level of basal accessibility was observed at the GM-CSF promoter in unstimulated EL-4 T cells (Figure 1E), which increased following P/I treatment. However, while an equivalent level of inherent accessibility was observed at the promoter in unstimulated A20 B cells (Figure 1E), no change in accessibility was observed following stimulation (Figure 1E). Therefore, the chromatin remodelling events at the promoter that accompany transcriptional activation of the GM-CSF gene in T cells are blocked in B cells.
We have previously shown that the NF-κB family of proteins are important for chromatin remodelling of the GM-CSF promoter in response to T-cell activation (11,35) and it was possible that these transcription factors were not expressed as efficiently in B cells. Therefore, the nuclear level of the NF-κB proteins, cRel and RelA, was determined by western analysis in the B- and T-cell lines. Little or no RelA and cRel was present in the nucleus of unstimulated T and B cells, but nuclear accumulation of the proteins was observed in both cell types following P/I stimulation for 4 h (Figure 1F). Reanalysis of the western blot with an antibody to the constitutively expressed Sp1 protein confirmed equal protein loading (Figure 1F).
Therefore, although the GM-CSF promoter is capable of responding to an activation signal in a reporter assay and NF-κB proteins are equally represented in both B and T cells, chromatin remodelling events at the GM-CSF promoter that precede transcriptional activation in T cells are blocked in B cells. These results raised the possibility that the failure of the endogenous GM-CSF gene to respond to activating signals in B cells was due to differences in chromatin status of the gene in B cells compared to T cells.
CpG methylation status of the GM-CSF promoter does not correlate with gene activity
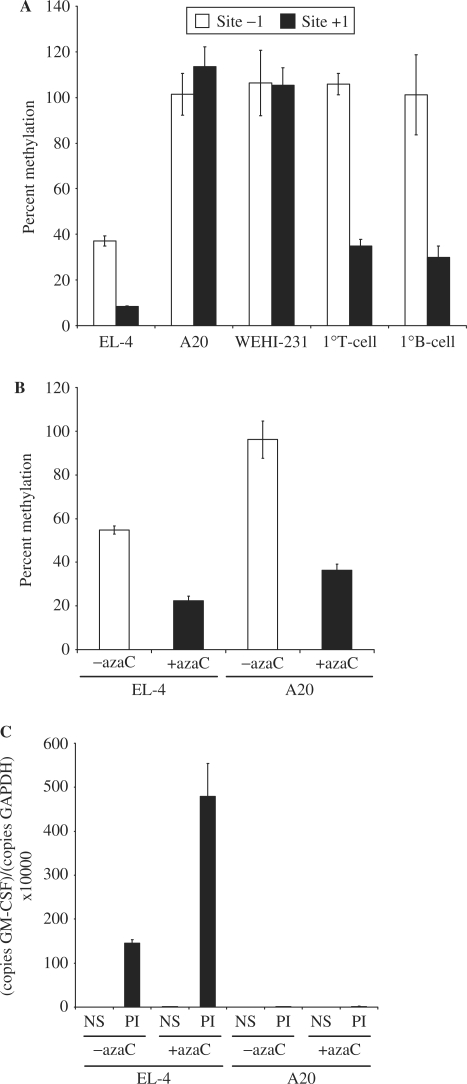
Methylation of CpG dinucleotides within regulatory elements of genes is well established as an epigenetic mechanism that contributes to gene repression (36) and we therefore hypothesized that the GM-CSF promoter may be marked by differential methylation in T and B cells. A single CpG site located within the GM-CSF proximal promoter (Figure 1A) overlapping an Sp1-binding site located within the CD28-responsive region (CD28RR) of the promoter (37) and a second site at +100 are both contained within recognition sites for the methylation sensitive Aci1 restriction enzyme, and therefore methylation of these sites was examined using a restriction enzyme-based assay. Genomic DNA was isolated from EL-4 T cells, A20 and WEHI-231 B cells as well as primary CD4+ T cells and B cells isolated from mouse spleen, digested with Aci1 and analysed by quantitative PCR using primer sets that covered each of the CpG sites (Figure 1A). The amount of PCR product generated from digested samples was plotted as a percentage of the amount generated from undigested genomic DNA samples, generating a measure of methylation at each site. The promoter Site-1 was completely methylated in the two B-cell lines and in both primary T and B cells but was only partially methylated in EL-4 T cells (Figure 1A). The Site + 1 within the transcribed region was completed methylated in the two B-cell lines but only partially methylated in EL-4 cells and both of the primary cell types (Figure 1A). Thus, the methylation status of these sites varies considerably between primary cells and cell lines and there appears to be no correlation between the methylation status of these sites and the ability of the gene to be expressed.
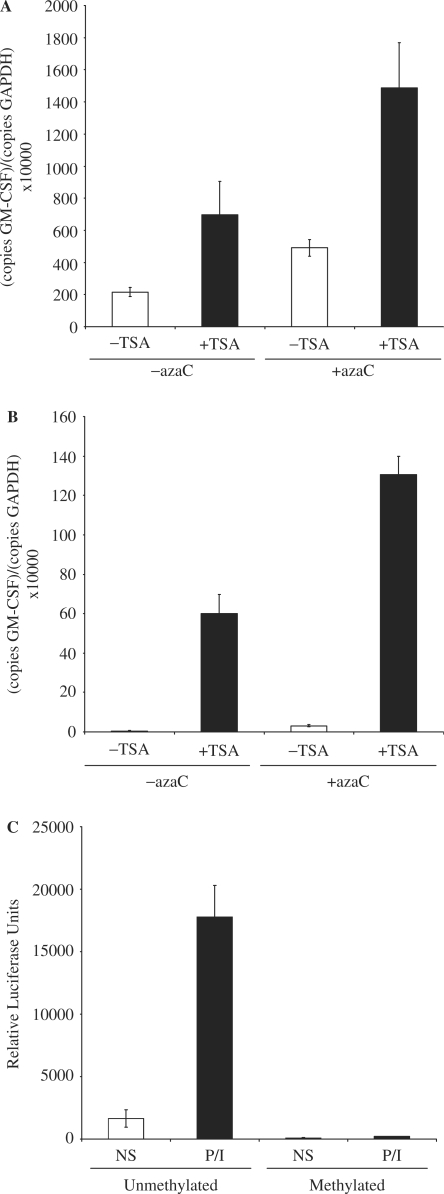
The effect of demethylating these sites in T- and B-cell lines was next examined. EL-4 T cells and A20 B cells were treated with 2-deoxy-5-azacytidine (azaC) to reduce CpG methylation. While azaC treatment decreased methylation of the promoter CpG site in both EL-4 T cells and A20 B cells (to 20 and 30% methylation, respectively, Figure 2B), it augmented P/I stimulation in EL-4 T cells only (∼4-fold, Figure 2C) and had no effect on GM-CSF expression in A20 B cells, either alone or following P/I stimulation (Figure 2C). Therefore, demethylation of the promoter in B cells is not sufficient to relieve the block in transcriptional activation of the GM-CSF gene in these cells.
Figure 2.
Methylation status of the GM-CSF promoter and enhancer in T cells and B cells. (A) Genomic DNA was isolated from EL-4 T cells, A20 and WEHI-231 B cells and primary CD4+ T and CD19+ B cells, and digested with the methylation sensitive AciI enzyme. Digested and undigested DNA was analysed by quantitative PCR using the indicated primer sets. The mean and standard error of three independent experiments are shown. (B) Genomic DNA was isolated from EL-4 T cells and A20 B cells either left untreated or pre-treated with azaC. Promoter methylation was determined using primer set −I. The mean and standard error of three independent experiments are shown. (C) mRNA was isolated from EL-4 T cells and A20 B cells treated as in (B) prior to stimulation with P/I. cDNA was prepared and GM-CSF expression levels monitored by PCR using primer set +II. The mean and standard error of three independent experiments are shown.
Put together, these data suggest that the methylation status of the GM-CSF promoter does not influence the ability of the GM-CSF promoter to undergo chromatin remodelling and gene transcription in response to activating signals and that demethylation of the promoter in B cells is not sufficient to relieve the block in transcription activation.
GM-CSF promoter is hyperacetylated in T cells compared to B cells
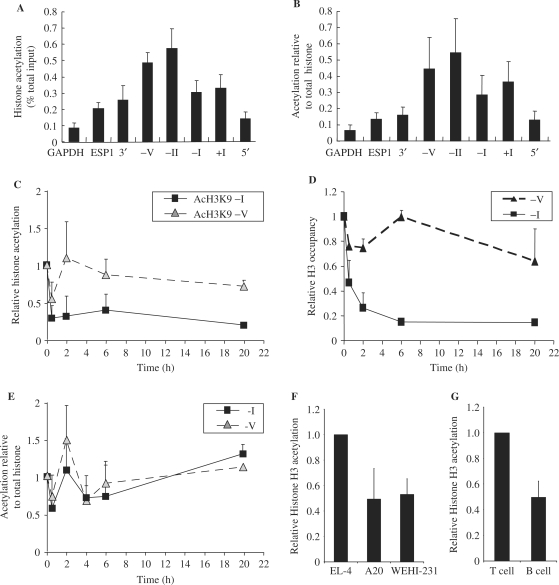
The association between histone acetylation and gene transcription is well established, both from studies of individual genes, as well as global gene analysis studies (17–21). We, therefore, speculated that differences in histone acetylation at the GM-CSF promoter may account for differences in the transcriptional competence of the promoter in T and B cells. To investigate this possibility, ChIP analysis was used to map histone acetylation levels across different regions of the GM-CSF gene in unstimulated EL-4 T cells. DNA immunoprecipitated with acetylated-H3K9 antibodies was amplified by quantitative PCR using primer sets that covered various regions of the GM-CSF gene (Figure1A). H3K9 acetylation was enriched at regions surrounding and including the GM-CSF promoter (primer sets −V, −II, −I, +I) compared to regions >2-kb upstream or downstream from the promoter (primer sets 5′ and 3′), a region of the GM-CSF enhancer (ESP1) and the constitutively expressed GAPDH gene (Figure 3A). The acetylation level of GAPDH is still at least 5-fold higher than a non-expressed gene (rhodopsin) where there is barely detectable acetylation (data not shown). The enrichment of histone acetylation at the promoter was highlighted when the level of acetylation was plotted relative to histone occupancy levels (Figure 3B). A similar pattern was observed for H4 acetylation (data not shown).
Figure 3.
Histone acetylation is enriched at the GM-CSF promoter in T cells. (A) ChIP assays were performed with anti-acH3K9 antibodies on unstimulated EL-4 T cells. Immunoprecipitated DNA was amplified using the indicated primer sets (described in Figure 1A). The data are presented as the ratios of immunoprecipitated DNA to total input. The mean and standard error of three independent experiments are shown. (B) Histone acetylation levels [from (A)] presented as a ratio of total H3 levels. (C, D) ChIP assays were performed with anti-acetylated H3K9 (C) or core H3 (D) antibodies on EL-4 T cells stimulated with P/I for the indicated times. Immunoprecipitated DNA was amplified with primer sets as indicated. The data are presented as the ratios of immunoprecipitated DNA to total input and normalized to the unstimulated cell samples, which was set at 1. The mean and standard error of three independent experiments are shown. (E) AcH3K9 levels [from (C)] presented as a ratio of total H3 levels. (F) EL-4 T cells, A20 and WEHI-231 B cells were subjected to ChIP analysis using an anti-acH3 antibody. Immunoprecipitated DNA was amplified using primer set -I. Data are plotted as acetylation level relative to EL-4 T cells. The mean and standard error of three independent experiments are shown. (G) ChIP analysis as above was also performed using primary CD4+ T and CD19+ B cells. Data are plotted as acetylation relative to primary T cell. The mean and standard error of two independent experiments are shown.
To determine whether there is a further increase in acetylation at the promoter following activation, histone H3 and Ac-H3K9 levels were measured in EL-4 cells at various times following activation with P/I. As previously described (13), the level of Ac-H3K9 and as well as total H3 decreased at the promoter following activation (Figure 3C and D). While there was some fluctuation at early time points, when AcH3K9 was plotted relative to total H3 there was no evidence for either a significant increase or decrease in acetylation across the GM-CSF promoter (Figure 3E). Therefore, reduced acetylation observed at the promoter in response to stimulation reflects nucleosome depletion rather than histone deacetylation.
To determine whether the higher level of acetylation at the promoter in non-stimulated EL-4 cells correlates with the transcriptional competence of the GM-CSF gene, histone H3 acetylation levels at the GM-CSF promoter were examined in EL-4 T cells compared to A20 and WEHI-231 B cells. We found that histone H3 acetylation at the GM-CSF promoter (primer set −I) was almost 50% lower in both of the B-cell lines in which GM-CSF expression is not inducible compared to EL-4 T cells (Figure 3F). This result was confirmed in primary mouse cells, with splenic B cells also displaying ∼50% lower histone H3 acetylation levels compared to CD4+ T cells (Figure 3G).
While there is no evidence for an increase in acetylation following activation, enriched histone acetylation marks the GM-CSF promoter relative to flanking regions in a T-cell line. Promoter acetylation is also higher in primary T cells and a T-cell line compared to B cells and may play a role in the ability to remodel promoter chromatin.
Hyperacetylation of the GM-CSF promoter facilitates chromatin remodelling and transcription in both T and B cells
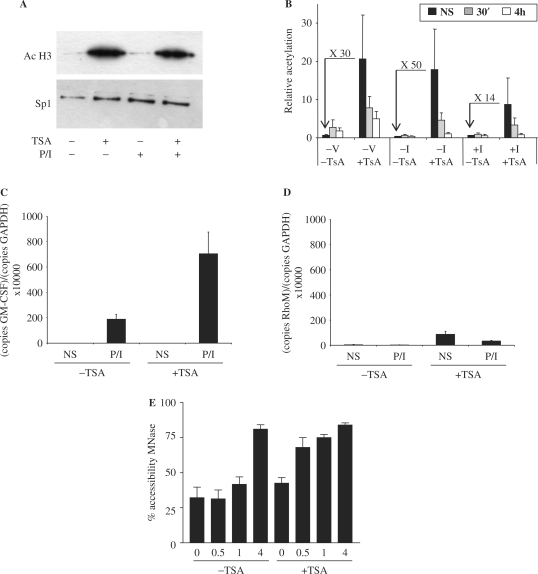
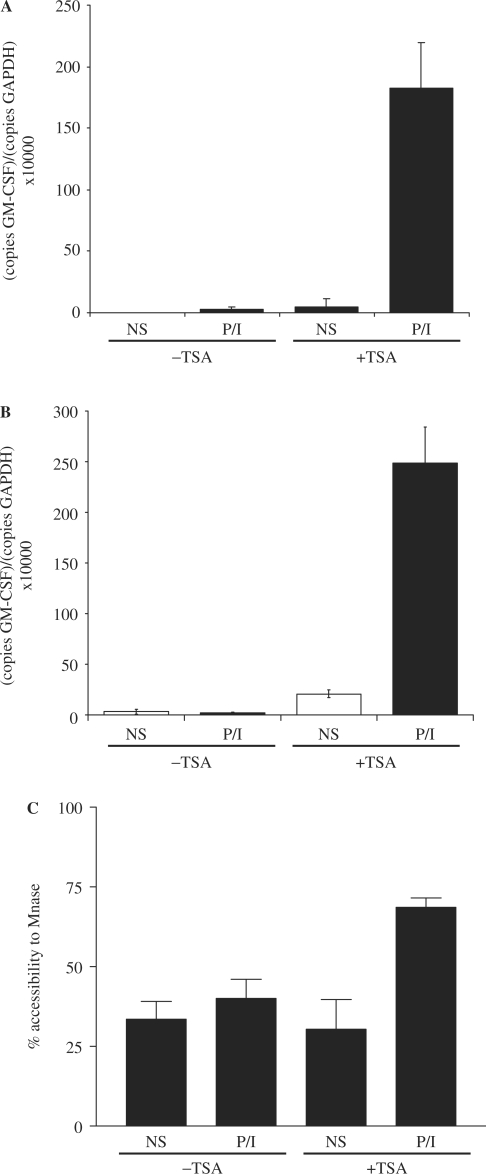
To determine whether histone acetylation facilitates transcriptional activation of the GM-CSF gene, we examined GM-CSF expression in EL-4 T cells treated with the histone deacetylase inhibitor, TSA. As expected TSA treatment of EL-4 T cells caused a dramatic increase in global acetylated H3 levels, as measured by western blotting of nuclear extracts (Figure 4A) but global acetylation levels were not affected by P/I stimulation (Figure 4A). This increase in global histone acetylation in response to TSA treatment was reflected by a similarly dramatic increase in acetylated H3K9 levels across the GM-CSF gene, as determined by ChIP assay (Figure 4B), both at the promoter (primer set −I and +I) and upstream regions (set −V). We have previously demonstrated that acetylated histone levels decrease across the GM-CSF promoter in T cells following stimulation due to the specific depletion of histones from this region [(13) and Figure 3C]. Consistent with this, a decrease in acetylated histones was observed at the promoter region (primer set −I and +I) in TSA-treated cells following P/I stimulation (Figure 4B). Although not observed normally (13), a decrease in acetylated histones was also observed to some extent at the upstream region (primer set −V, Figure 4B) in the TSA-treated cells suggesting that acetylation may facilitate histone removal following activation.
Figure 4.
Histone hyperacetylation facilitates GM-CSF gene activation in T cells. (A) Nuclear extracts from EL-4 T cells pre-treated with TSA for 4 h followed by P/I treatment for 4 h as indicated, were subjected to SDS–PAGE and analysed by western blotting with the indicated antibodies. (B) ChIP assays were performed with an antibody against acetylated H3K9 on EL-4 T cells treated with or without TSA and then either unstimulated (NS) or stimulated with P/I for the indicated times. Immunoprecipitated DNA was analysed by PCR using primer sets as indicated. Data are graphed as ratios of the immunoprecipitated sample to total input sample. The mean and standard error of three independent assays are shown. (C) GM-CSF mRNA levels were determined by PCR analysis of cDNA prepared from EL-4 T cells treated with or without TSA for 4 h and then either unstimulated (NS) or stimulated with P/I for 4 h. (D) Rhodopsin mRNA levels were determined by PCR analysis of EL-4 cells treated with or without TSA for 4 h, and then either unstimulated (NS) or stimulated with P/I for 4 h. The mean and standard error of three independent experiments are shown. (E) Nuclei were isolated from EL-4 T cells pre-treated with or without TSA for 4 h, then stimulated with P/I for the indicated times. Nuclei were incubated with MNase and genomic DNA was analysed by PCR using primer set −I. The mean and standard error of three replicate assays are shown.
The effect of increased histone acetylation on activation of the GM-CSF gene was then examined. As seen earlier, GM-CSF mRNA levels increased ∼450-fold following P/I stimulation (Figure 4C) and while TSA pre-treatment did not affect basal GM-CSF mRNA levels, expression in response to P/I was increased >3-fold by the presence of TSA (Figure 4C). The P/I response of a control gene, rhodopsin, which is not normally expressed in T cells was not facilitated by TSA treatment although TSA alone led to a small increase in expression (Figure 4D). CHART-PCR was then used to determine how the increased histone acetylation levels across the GM-CSF gene influenced chromatin remodelling events at the promoter. As we have observed previously (11), following P/I stimulation little change in accessibility of the promoter to MNase was seen at 30 min or 1 h, but an increase in accessibility occurred by 4-h post-stimulation (Figure 4E). However, in TSA pre-treated cells an increase in chromatin accessibility was detected as early as 30 min post-stimulation (Figure 4E).
Therefore, increasing histone acetylation across the GM-CSF promoter resulted in faster kinetics of chromatin remodelling at the promoter in response to P/I stimulation, which translated into increased mRNA levels. This suggests that histone hyperacetylation at the GM-CSF promoter facilitates chromatin remodelling and subsequent transcriptional activation of the GM-CSF gene in T cells.
The effect of TSA treatment on the GM-CSF gene in B cells in which the promoter is normally resistant to P/I-induced chromatin remodelling and subsequent transcriptional activation was next examined. As seen previously, P/I treatment could not induce GM-CSF expression in either A20 or WEHI-231 B cells (Figure 5A and B, respectively). While TSA treatment alone had a small effect on GM-CSF mRNA levels, an ∼200-fold increase in GM-CSF mRNA levels was observed in both cells lines in response to P/I stimulation following pre-treatment with TSA (Figure 5A and B). Furthermore, analysis by CHART-PCR revealed that following TSA treatment, P/I stimulation was able to induce chromatin remodelling across the GM-CSF promoter in A20 B cells (Figure 5C).
Figure 5.
Histone hyperacetylation relieves the block in GM-CSF gene activation in B cells. (A) GM-CSF mRNA levels were determined by PCR analysis of cDNA prepared from A20 B cells treated with or without TSA for 4 h and then either left unstimulated (NS) or stimulated with P/I for 4 h. The mean and standard error of three replicate assays are shown. (B) GM-CSF mRNA levels were determined in WEHI-231 B cells treated as in (A). (C) Nuclei were isolated from A20 B cells treated as in (A). Nuclei were incubated with MNase and genomic DNA was analysed by PCR using primer set −I. The mean and standard error of three replicate assays are shown.
Therefore, increasing histone acetylation levels in B cells removes the block in chromatin remodelling at the GM-CSF promoter in response to P/I stimulation.
Hyperacetylation and demethylation cooperate to facilitate activation of the GM-CSF gene in both T and B cells
Although demethylation alone had no effect on GM-CSF expression in A20 B cells, we next examined whether demethylation of the promoter had any effect in conjunction with histone hyperacetylation. As seen earlier, in EL-4 T cells both TSA-induced histone hyperacetylation and azaC-induced DNA demethylation increased GM-CSF mRNA expression in response to P/I stimulation, and GM-CSF mRNA expression was increased even further by azaC and TSA treatment in combination (Figure 6A). While azaC treatment had only minimal effect on GM-CSF gene expression in A20 B cells, TSA treatment led to increased GM-CSF mRNA levels in response to P/I (Figure 6B) and azaC treatment was again able to augment P/I-induced GM-CSF mRNA levels in combination with TSA (Figure 6B).
Figure 6.
DNA demethylation facilitates increased transcription of the GM-CSF gene. (A) EL-4 T cells were either left untreated or pre-treated with azaC then treated with or without TSA prior to stimulation with PI for 4 h. GM-CSF mRNA levels relative to GAPDH were then determined by quantitative PCR analysis. The mean and standard error of three independent experiments are shown. (B) GM-CSF mRNA levels were determined in A20 B cells treated as in (A). (C) EL-4 T cells were transfected with a GM-CSF promoter luciferase reporter construct either mock methylated or methylated with the CpG methylase M.SssI. Cells were either left unstimulated or stimulated with P/I for 8 h; protein extracted and luciferase activity monitored. The mean and standard error of two independent experiments are shown.
Put together, these data suggest that demethylation of the GM-CSF promoter only influences GM-CSF mRNA levels in cells in which the promoter has undergone chromatin remodelling implying that promoter methylation may affect transcriptional events subsequent to chromatin remodelling. To test this possibility the GM-CSF promoter reporter construct was methylated with the CpG methylase M.SssI prior to transfection into EL-4 T cells. As before basal expression of the unmethylated GM-CSF reporter was detected, which increased dramatically upon P/I stimulation (Figure 6C). In contrast, little basal or induced expression of the methylated GM-CSF reporter was detected, suggesting that CpG methylation inhibits transcriptional activation of the GM-CSF promoter.
Put together, these data suggest that methylation of the GM-CSF promoter does not affect chromatin remodelling events at the promoter, but influences transcriptional activation of the GM-CSF gene subsequent to chromatin remodelling.
Brg1 is enriched at the GM-CSF promoter in T cells, and is required for efficient gene activation
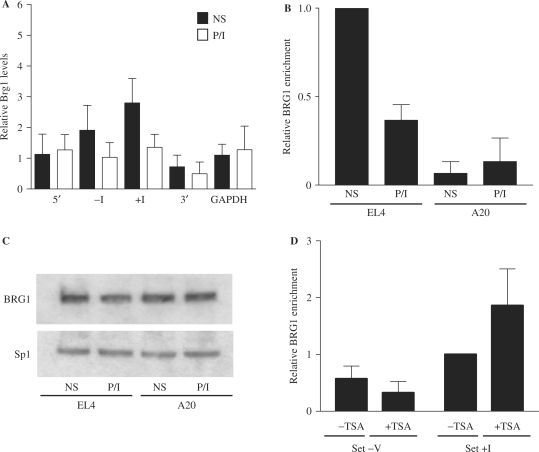
The chromatin remodelling events at the GM-CSF promoter involving the specific depletion of histones (13) facilitated by histone acetylation is reminiscent of the chromatin remodelling events observed at the Pho5 promoter in yeast (29,38). At the Pho5 promoter histone depletion is facilitated by the SWI/SNF chromatin remodelling complex recruited to the promoter via hyperacetylated histones (25,26). We, therefore, asked whether the mammalian SWI/SNF remodelling complex was associated with the GM-CSF promoter in T cells. ChIP analysis with antibodies to the SWI/SNF component, BRG1 was performed on EL-4 T cells before and after stimulation with P/I for 4 h. Immunoprecipitated DNA was quantified using PCR primers designed to specific regions of the GM-CSF gene (Figure 1A). We found that in unstimulated cells, relative levels of BRG1 were 2–3-fold higher at the GM-CSF promoter (primer sets −I and +I) than at regions 2 kb distal from the promoter (primer set 3′ and 5′) or the GAPDH gene (Figure 7A). Surprisingly, BRG1 levels decreased at the promoter following T-cell activation (Figure 7A, set −I P = 0.037 and set +I P = 0.040, paired t-test). In contrast, there was little change in BRG1 levels associated with the 3′ and 5′ regions of the gene, or with the constitutively expressed GAPDH gene following stimulation (Figure 7A).
Figure 7.
BRG1 is enriched at the GM-CSF promoter in T cells. (A) ChIP assays were performed with an antibody against BRG1 on EL-4 T cells either unstimulated (NS) or stimulated with P/I for 4 h (PI). Immunoprecipitated DNA was amplified using the indicated primer sets. The data are plotted as the ratio of immunoprecipitated DNA to total input DNA. The mean and standard error of at least three independent assays are shown. (B) ChIP assays were performed with a BRG1 antibody on EL-4 T cells and A20 B cells either non-stimulated (NS) or stimulated with P/I for 4 h. Immunoprecipitated DNA was analysed using primer set +I. The data are plotted as the ratio of immunoprecipitated DNA to total input DNA and normalized to the EL-4 NS value, which was set at 1. The mean and standard error of three replicate assays are shown. (C) Nuclear extracts from EL-4 T cells and A20 B cells treated as indicated were subjected to SDS–PAGE and analysed by western blotting with the indicated antibodies. (D) ChIP assays were performed using a BRG1 antibody on EL-4 T cells pre-treated with or without TSA for 4 h, and analysed with primer sets −V and +I. The data are plotted as the ratio of immunoprecipitated DNA to total input DNA and normalized to the untreated value using primer set +I, which was set at 1. The mean and standard error of three replicate assays are shown.
Since BRG1 is enriched at the GM-CSF promoter in unstimulated EL-4 T cells we speculated that this may be a requirement for GM-CSF gene activation. Therefore, association of BRG1 with the GM-CSF promoter was examined in different cell types. As shown above, BRG1 was associated with the GM-CSF promoter in unstimulated T cells, with levels decreasing upon stimulation (Figure 7B). In contrast, little BRG1 was associated with the GM-CSF promoter in A20 B cells, with relative levels significantly lower than observed in T cells even after stimulation (Figure 7B). Furthermore, there was no decrease in BRG1 levels at the GM-CSF promoter in A20 B cells following stimulation. The difference in BRG1 levels at the GM-CSF promoter in B and T cells is not due to differences in BRG1 expression in these cell types, as western analysis of nuclear extracts from A20 B cells and EL-4 T cells confirmed equal levels of BRG1 protein in these cells (Figure 7C). In addition, BRG1 protein levels did not change in either cell type upon P/I stimulation (Figure 7C).
The cell-type specific enrichment of BRG1 at the GM-CSF promoter correlates with histone acetylation levels at the promoter in these cell types. We therefore asked whether BRG1 enrichment at the GM-CSF promoter is facilitated by histone acetylation. EL-4 T cells were treated with TSA to increase histone acetylation, and BRG1 levels monitored by ChIP assay. TSA treatment resulted in increased binding of BRG1 at the GM-CSF promoter (Figure 7D, set +I), although no increased binding of BRG1 was detected at an adjacent region (Figure 7D, set −V), where increased acetylation was also observed (Figure 4B). These data suggest that while increasing histone acetylation may facilitate BRG1 recruitment to the GM-CSF promoter, acetylation alone is not sufficient to recruit BRG1.
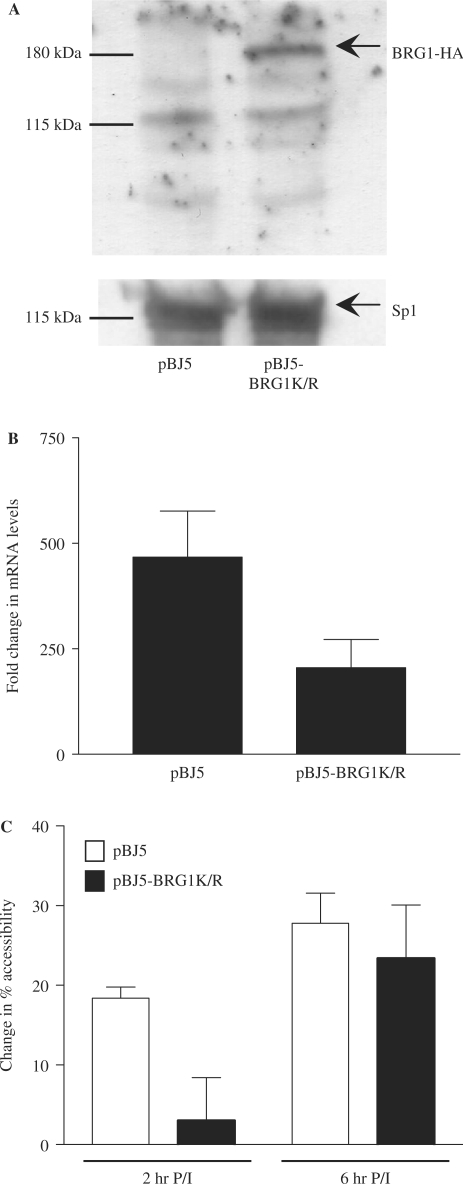
Enrichment of BRG1 at the GM-CSF promoter correlates with the transcriptional competence of the GM-CSF gene and the ability of the promoter to undergo remodelling in response to activating signals. To determine whether BRG1 is required for GM-CSF promoter remodelling and gene activation, GM-CSF expression was monitored in EL-4 T cells transfected with an ATPase BRG1 mutant construct, BRG1K/R (32). This BRG1 mutant protein has been found to assemble into the SWI/SNF complex, but is unable to remodel nucleosomes (39). Cells were transfected with the HA tagged BRG1 mutant construct along with a truncated Kk.II receptor protein that enabled sorting of transfected cells. Expression of the mutant BRG1 protein in EL-4 T cells was confirmed by western blotting with an HA antibody (Figure 8A). Sorted cells were either left unstimulated or stimulated with P/I for 6 h and GM-CSF expression monitored by quantitative PCR. GM-CSF mRNA levels increased ∼500-fold following P/I stimulation in cells transfected with vector alone (Figure 8B). In contrast, accumulation of GM-CSF mRNA was reduced by 50% in cells expressing the BRG1 ATPase mutant, suggesting that BRG1 contributes to activation of the GM-CSF gene. Basal levels of GM-CSF expression were unaffected in the mutant expressing cells (data not shown).
Figure 8.
BRG1 is required for GM-CSF gene activation. (A) Nuclear extracts from EL-4 T cells transfected with the pBJ5 plasmid or pBJ5-BRG1K/R were subjected to SDS–PAGE and analysed by western blotting with the indicated antibodies. Positions of protein molecular weight markers are indicated. (B) EL-4 T cells transfected with the indicated plasmids and the K.kII plasmid were purified using magnetic beads labelled with antibodies to the K.kII receptor, then either left untreated or stimulated with P/I for 6 h. GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from these cells using primer set +II. Data were graphed as fold change in mRNA levels following stimulation. The mean and standard error of three replicate assays are shown. (C) Nuclei from EL-4 T cells transfected and purified as in (B), then either left unstimulated or stimulated with P/I for 2 h and 6 h were incubated with MNase and genomic DNA was analysed by PCR using primer set −I. Data were graphed as change in accessibility relative to the unstimulated sample. The mean and standard error of at least three replicate assays are shown.
To determine whether BRG1 is also required for chromatin remodelling events at the promoter, transfected cells were analysed by CHART-PCR. Cells were either left unstimulated or stimulated with P/I, nuclei isolated and incubated with MNase. Genomic DNA was then isolated, and analysed using primer set −I that spans the GM-CSF promoter. In cells transfected with the control plasmid, an increase in accessibility (∼20%) was observed following stimulation for 2 h with a further increase at 6 h (Figure 8C). However, while a similar change in accessibility was observed in cells containing the mutant BRG1 protein at 6 h, a considerably smaller change in accessibility was observed at 2 h (Figure 8C), suggesting that chromatin remodelling at the promoter was delayed in these cells.
Put together, these data suggest that BRG1 is enriched at the GM-CSF promoter in unstimulated EL-4 T cells and plays a direct role in chromatin remodelling events associated with GM-CSF gene activation. Loss of BRG1 from the promoter following stimulation correlates with depletion of histones from the promoter and the associated increased accessibility.
DISCUSSION
Activation of the GM-CSF gene in response to T-cell stimulatory signals involves highly targeted changes in chromatin structure, involving the depletion of histones and resulting in increased accessibility across the promoter and enhancer regions of the gene (11–14). These chromatin changes are a prerequisite for efficient transcriptional activation of the GM-CSF gene in T cells, and we have found that these remodelling events are blocked in B cells, which do not express GM-CSF in response to immune signals. The chromatin environment of the GM-CSF promoter is differentially marked in T cells compared to B cells, and this appears to at least partially dictate the ability of the gene promoter to respond to immune stimuli. The GM-CSF promoter is specifically marked in uninduced T cells by increased histone acetylation and enrichment of the chromatin remodeller BRG1, and these marks contribute to the ability of the GM-CSF promoter to undergo chromatin remodelling in response to immune signalling.
We found that the methylation status of two CpG sites located up and downstream from the transcription start site did not correlate with the response of the gene. Furthermore, demethylation of the CpG site in the GM-CSF promoter in B cells by azaC treatment was not sufficient to render the gene responsive to activating signals and this is likely to be due to the fact that demethylation of the promoter is not sufficient to relieve the block in chromatin remodelling of the promoter (data not shown). However, azaC treatment augmented GM-CSF mRNA expression once the block in chromatin remodelling had been relieved by TSA treatment. While it is possible that azaC is indirectly affecting GM-CSF gene expression, the data presented are also consistent with a model in which methylation of the promoter CpG site inhibits binding of transcription factors to the gene promoter, and therefore inhibits transcriptional activation of the GM-CSF gene, subsequent to promoter remodelling. This is supported by data demonstrating that a methylated reporter construct cannot be activated, but further studies are required to investigate whether methylation of the CpG site in the promoter affects transcription factor binding, particularly at the Sp1 site which contains the methylated CpG. It should also be noted, however, that the luciferase gene has a 5-fold higher density of CpG sequences than the GM-CSF gene, and this density of methylation may itself be sufficient to block activation of a methylated reporter gene plasmid.
Recent studies have demonstrated that chromatin remodelling events that occur at the GM-CSF promoter in response to immune signals involve the depletion of histones from the promoter region. Similar histone eviction has also been described at the IL-2 gene promoter following T-cell activation (13) and at the yeast Pho5 and Pho8 promoters (29,30,38,40). In the case of both Pho5 and Pho8, histone eviction was preceded by a transient increase in histone acetylation and stimulation-driven increases in histone acetylation have been found to accompany activation of a number of other inducible genes (25,28,29). However, such increases in acetylation were not observed at the GM-CSF promoter in response to activation. Instead, the GM-CSF promoter already has a relatively high degree of histone acetylation in uninduced T cells compared to regions upstream or downstream to the promoter or compared to the GM-CSF promoter in B cells. This basal enrichment of histone acetylation in uninduced T cells may generate an activation competent environment at the GM-CSF promoter and obviate the need for stimulation-induced increases in acetylation. In support of this, artificially increasing histone acetylation levels at the GM-CSF promoter in T cells resulted in more rapid chromatin remodelling of the promoter and enhanced GM-CSF mRNA expression. More importantly, treatment of B cells with the histone deacetylase inhibitor TSA relieved the block in chromatin remodelling at the GM-CSF promoter, enabling promoter activation in response to stimulatory signals. Thus, we would speculate that unlike many other inducible genes the GM-CSF promoter does not become hyperacetylated following activation, but rather needs to be constitutively acetylated to respond to P/I activation signals.
The SWI/SNF complex has been implicated in displacement of nucleosomes at the yeast Pho5 and Pho8 genes. Nucleosome remodelling is inhibited at the Pho8 promoter and significantly delayed at the Pho5 promoter in yeast strains in which the SWI/SNF ATPase snf2 is mutated (26,27,41). Such a role for the SWI/SNF complex is also supported by in vitro experiments demonstrating that the SWI/SNF complex can displace histone octamers in trans, although this is less efficient than histone sliding (42). Our finding that chromatin remodelling at the GM-CSF promoter is dependent on BRG1 also implicates the SWI/SNF remodelling complex in histone loss at the GM-CSF promoter. We found that chromatin remodelling events across the promoter were impaired, but not completely inhibited in the presence of a BRG1 ATPase mutant. This may be because wild-type BRG1 protein is still present in these cells, and not completely competed by the BRG1 mutant, or may reflect the fact that remodelling can still occur, although less efficiently in the absence of BRG1. A similar situation exists at the Pho5 promoter where the SWI/SNF complex is required for efficient histone eviction, but remodelling can still occur in the absence of the core SNF2 component (43) and has led to the suggestion that an alternative, although less efficient, eviction mechanism is operating in these circumstances.
The SWI/SNF complex has been implicated in chromatin remodelling events required for activation of a range of genes (23,24,44), however in such studies, BRG1 has generally been found to be recruited to the gene promoters in response to stimulatory signals, consistent with a model in which the SWI/SNF complex is recruited to gene promoters through DNA bound transcription factors. In contrast, we observed that BRG1 was constitutively associated with the GM-CSF promoter in uninduced T cells and then lost from the promoter concomitant with histone loss in response to T-cell activation. This raises the question of how the BRG1 protein is specifically enriched at the GM-CSF promoter in uninduced EL-4 T cells. BRG1 contains a bromodomain which can interact with acetylated histones (45) and its recruitment to the IFNβ enhancer as well as the Pho5 and Pho8 promoters is facilitated by histone acetylation (25,29,46). It is possible, therefore, that BRG1 is constitutively recruited to the GM-CSF promoter via histone acetylation. In support of this model, enrichment of both acetylated histones and BRG1 was observed at the GM-CSF promoter in uninduced T cells but not B cells. Furthermore, increasing GM-CSF promoter acetylation in T cells by TSA treatment resulted in an associated increase in BRG1 levels at the promoter. However, it is likely, that BRG1 requires the presence of a constitutive transcription factor(s) acting together with higher levels of acetylation, for recruitment to the promoter in uninduced T cells because increasing histone acetylation levels was not sufficient to increase BRG1 levels at a distal region of the gene. The nature of such factors remains to be determined. It should be noted that these findings are in contrast to our previous in vitro studies, which demonstrated stimulation-induced recruitment of BRG1 to a GM-CSF template (11), however this previous study used naked DNA templates and therefore was not able to take into account interactions between chromatin and BRG1.
The data presented here are consistent with a model in which the SWI/SNF complex is poised at the GM-CSF promoter in uninduced T cells and this together with histone acetylation generates a chromatin environment which is conducive to rapid remodelling and gene activation in response to the appropriate cellular signals. Along this line, analysis of the yeast GAL1 gene demonstrated that more rapid gene induction was achieved in situations when the SWI/SNF complex is already poised at the gene promoter (47).
It is becoming apparent that activation of inducible genes in response to stimulatory signals involves a series of precisely orchestrated events aimed at remodelling nucleosomes positioned over the promoter so that transcription can occur. Activation of the GM-CSF gene occurs relatively rapidly in response to immune signals, but cannot be induced in B cells by these same signals. We propose that this is because the gene promoter is specifically poised for remodelling in T cells by high basal levels of histone acetylation and the constitutive recruitment of the SWI/SNF complex, which create an environment conducive to gene activation. These data demonstrate that the epigenetic status of the GM-CSF promoter varies between cell types and dictates its potential for activation.
ACKNOWLEDGEMENTS
We thank Dr P Cockerill for GM-CSF plasmids, Dr G Crabtree for BRG1 plasmids and Shannon Ray and Lina Ma for technical assistance. This work was funded by National Health and Medical Research Council, Australia (302202 to A.F.H. and M.F.S); Royal Hobart Hospital Research Foundation (to A.F.H). Funding to pay the Open Access publication charges for this article was provided by the NHMRC, Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gasson JC, Baldwin GC, Sakamoto KM, DiPersio JF. The biology of human granulocyte-macrophage colony-stimulating factor (GM-CSF) Prog. Clin. Biol. Res. 1990;352:375–384. [PubMed] [Google Scholar]
- 2.Holloway AF, Rao S, Shannon MF. Regulation of cytokine gene transcription in the immune system. Mol. Immunol. 2002;38:567–580. doi: 10.1016/s0161-5890(01)00094-3. [DOI] [PubMed] [Google Scholar]
- 3.Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Cockerill PN, Shannon MF, Bert AG, Ryan GR, Vadas MA. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc. Natl Acad. Sci. USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siebenlist U, Durand DB, Bressler P, Holbrook NJ, Norris CA, Kamoun M, Kant JA, Crabtree GR. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol. Cell. Biol. 1986;6:3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward SB, Hernandez-Hoyos G, Chen F, Waterman M, Reeves R, Rothenberg EV. Chromatin remodeling of the interleukin-2 gene: distinct alterations in the proximal versus distal enhancer regions. Nucleic Acids Res. 1998;26:2923–2934. doi: 10.1093/nar/26.12.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockerill PN, Bert AG, Roberts D, Vadas MA. The human granulocyte-macrophage colony-stimulating factor gene is autonomously regulated in vivo by an inducible tissue-specific enhancer. Proc. Natl Acad. Sci. USA. 1999;96:15097–15102. doi: 10.1073/pnas.96.26.15097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 10.Attema JL, Reeves R, Murray V, Levichkin I, Temple MD, Tremethick DJ, Shannon MF. The human IL-2 gene promoter can assemble a positioned nucleosome that becomes remodeled upon T cell activation. J. Immunol. 2002;169:2466–2476. doi: 10.4049/jimmunol.169.5.2466. [DOI] [PubMed] [Google Scholar]
- 11.Holloway AF, Rao S, Chen X, Shannon MF. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor kappaB proteins. J. Exp. Med. 2003;197:413–423. doi: 10.1084/jem.20021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson BV, Bert AG, Ryan GR, Condina A, Cockerill PN. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol. Cell. Biol. 2004;24:7914–7930. doi: 10.1128/MCB.24.18.7914-7930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell. Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bert AG, Johnson BV, Baxter EW, Cockerill PN. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol. Cell. Biol. 2007;27:2870–2885. doi: 10.1128/MCB.02323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 17.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 18.Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Dynamic assembly of silent chromatin during thymocyte maturation. Nat. Genet. 2004;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- 19.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell. Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 22.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 23.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinke H, Gregory PD, Horz W. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell. 2001;7:529–538. doi: 10.1016/s1097-2765(01)00200-3. [DOI] [PubMed] [Google Scholar]
- 26.Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory PD, Schmid A, Zavari M, Lui L, Berger SL, Horz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell. 1998;1:495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 28.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 29.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 30.Korber P, Luckenbach T, Blaschke D, Horz W. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 2004;24:10965–10974. doi: 10.1128/MCB.24.24.10965-10974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborne CS, Vadas MA, Cockerill PN. Transcriptional regulation of mouse granulocyte-macrophage colony-stimulating factor/IL-3 locus. J. Immunol. 1995;155:226–235. [PubMed] [Google Scholar]
- 32.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 33.Holloway AF, Occhiodoro F, Mittler G, Meisterernst M, Shannon MF. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. J. Biol. Chem. 2000;275:21668–21677. doi: 10.1074/jbc.M909058199. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brettingham-Moore KH, Rao S, Juelich T, Shannon MF, Holloway AF. GM-CSF promoter chromatin remodelling and gene transcription display distinct signal and transcription factor requirements. Nucleic Acids Res. 2005;33:225–234. doi: 10.1093/nar/gki161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 37.Cakouros D, Cockerill PN, Bert AG, Mital R, Roberts DC, Shannon MF. A NF-kappa B/Sp1 region is essential for chromatin remodeling and correct transcription of a human granulocyte-macrophage colony-stimulating factor transgene. J. Immunol. 2001;167:302–310. doi: 10.4049/jimmunol.167.1.302. [DOI] [PubMed] [Google Scholar]
- 38.Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- 39.de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 42.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 43.Barbaric S, Reinke H, Horz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 2003;23:3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 45.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 46.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 47.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]