Abstract
We have carried out a survey of potential quadruplex structure sequences (PQSS), which occur in the immediate upstream region (500 bp) of human genes. By examining the number and distribution of these we have established that there is a clear link between them and the occurrence of the SP1-binding element ‘GGGCGG’, such that a large number of upstream PQSS incorporate the SP1-binding element.
INTRODUCTION
Certain guanine-rich DNA sequences have the ability to form stable secondary structures, G-quadruplexes, comprising G-tetrad motifs that involve four guanines arranged in a planar array interacting via Hoogsteen hydrogen bonds (1,2). These G-tetrads are stable when a number are stacked on top of one another. There have been numerous topologies observed for G-quadruplexes (3) and a number of studies have mapped out potential quadruplex forming sequences in genomic DNAs (4–11). The search criteria for most of these surveys have been sequences, which contain four or more runs of G-tracts occurring close together on the same strand. Most of the observed topologies follow this pattern (12,13). These potential quadruplex structure sequences (PQSS) have been found to occur with elevated frequency in regions directly upstream of the transcription start site of genes in species as diverse as Escherichia coli (8) and humans (11). There are also a number of specific promoter regions for which there is biophysical and structural evidence for the formation of PQSSs (for example, 11–18), at least in vitro.
The zinc finger protein SP1 acts as a transcription factor, which has been shown to bind to the upstream element sequence ‘GGGCGG’ (19–21). This element has been found in many different promoters, often with a copy number >1 (19,20) and often within the first 100 bp upstream of the transcription start site. The consensus sequence has been shown to be ‘GGGGCGGGGC’ (22,23). The fact that it is guanine-rich, with consecutive guanines, gives it the ability to participate in PQSSs, at least in principle. We show here that many of the PQSSs found in the regions directly upstream of the transcription start site actually contain the SP1 consensus sequence and that there is a correlation between genes, which have the SP1 and PQSS in upstream regions.
Another common upstream promoter element ‘CCAAT’ occurs in a similarly large number of promoters but does not contribute as much guanine-richness as the SP1-binding element and can be employed as a useful comparative group.
METHODS
The mySQL tables for the Ensembl human core database v45 (24) were downloaded from the Ensembl web site (www.ensembl.org) and imported onto a local computer. The human genome was searched for PQSSs in the same way as described earlier (5) and the PQSS data was uploaded into mySQL tables. The search criterion was four runs of guanine Gm Xn Gm Xo Gm Xp Gm where m is between 3 and 5, and X are any combination of bases where n, o and p are between 1 and 7.
Using Perl scripts and the Ensembl Perl API (25), a list of all genes with Ensembl status ‘known’ was compiled and the 500 bp upstream flanking sequences were extracted. These were searched for the sequences ‘GGGCGG’ and ‘CCAAT’ as well as their complementary sequences and the genes were grouped into the following categories:
Genes with ‘GGGCGG’ in the region 500 bps upstream of their transcription start site (500USR).
Genes with ‘CCAAT’ in their 500USR.
Genes with both in their 500USR.
Genes with ‘GGGCGG’ but not ‘CCAAT’ in their 500USR.
Genes with ‘CCAAT’ but not ‘GGGCGG’ in their 500USR.
Genes with neither sequence in their 500USR.
The MySQL table of PQSSs was used to count the PQSSs in the upstream regions of the genes in each category and their distances (−1 bp to −500 bps) from the transcription start sites were noted. In order to represent these graphically the positions of the quadruplex sequences were put into bins of 10 base pairs. When looking at the number of PQSSs we counted each contiguous region of potential quadruplex region as a single sequence element i.e. if there were overlaps between more than one PQSS, then these were counted as one sequence element. For example the sequence: GGGAGGCGGGCGGTGGGGGGGTGGGGGTGGGG, which is in the upstream region of the Ensembl gene ENSG00000204219 (HGNC symbol TCEA3), was counted as one sequence element. Even though it contains up to six runs of guanines, we assume that only one quadruplex structure at a time can be formed in this region.
The number of cytosines and guanines in all known genes was also derived for each of the bins. The cytosine and guanine data were normalized as was the total quadruplex distribution so that a comparison of the relative distributions could be made, since the absolute number of cytosines and guanines was much higher. Our database of PQSSs was also searched for PQSSs, which incorporated the SP1 consensus sequence and the distributions of PQSSs containing the SP1 consensus sequence and those not containing the SP1 consensus sequence could then be determined.
RESULTS AND DISCUSSION
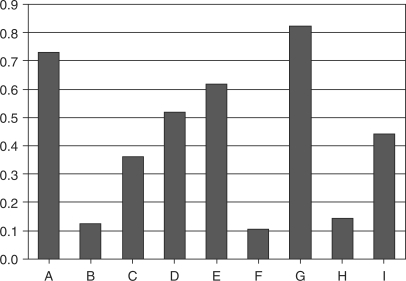
Figure 1 shows the number of PQSS elements per gene for a number of different grouping of genes. Bars A and B show that PQSSs in upstream regions of genes that contain SP1 elements are much more common than PQSSs in upstream regions of genes without SP1 elements. In bars C and D we see that there is no such relationship in genes whose upstream regions contain CCAAT elements. If anything the reverse is true and genes without CCAAT elements contain more potential PQSSs. This trend is repeated in bars E, F, G and H where E and G contain SP1 elements and contain many more PQSSs per gene than bars F and H. The trend of fewer PQSSs in the upstream regions with CCAAT is also apparent here. In bar I the number of PQSS elements per gene in the upstream region of all genes is much lower than those which contain SP1 sequence elements and higher than those which do not.
Figure 1.
PQSS sequence elements per gene for potential quadruplex sequences in upstream regions (A) containing SP1 elements, (B) not containing SP1 elements, (C) containing CCAAT elements, (D) without CCAAT elements, (E) containing CCAAT and SP1 elements, (F) containing CCAAT and no SP1 elements, (G) containing SP1 and no CCAAT elements, (H) containing neither SP1 nor CCAAT elements, (I) of all known Ensembl genes.
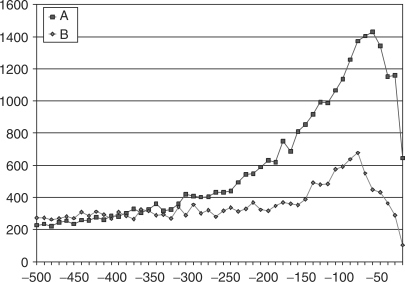
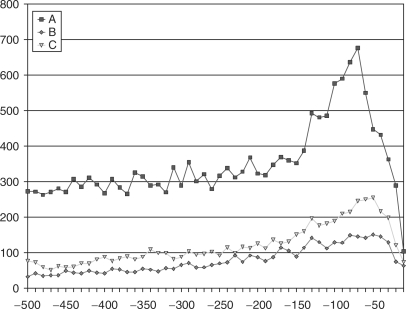
Figure 2 shows the distribution of the promoter-binding elements SP1 (GGGCGG) and CCAAT with distance from the transcription start site. The shape of this graph shows that the frequency of sequence elements rises steeply, reaching a peak in the −50 to −41 range for SP1 and in the −70 to −61 range for CCAAT before falling off gradually.
Figure 2.
(A) SP1 sequence elements, which occur in upstream regions, (B) CCAAT sequence elements, which occur in upstream regions.
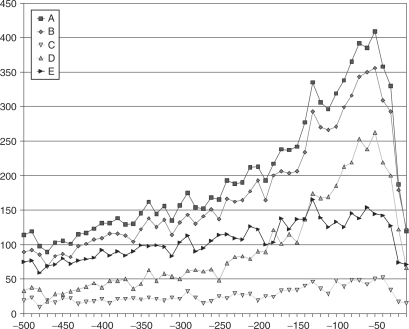
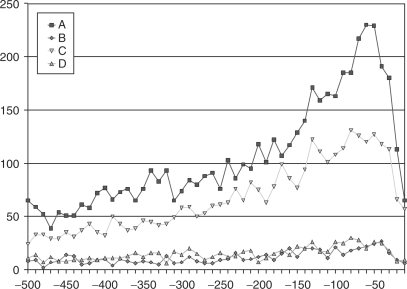
The distribution of PQSSs in the 500 bp upstream region of transcription start sites is very similar to that of the regulatory motifs in Figure 2. Figure 3A shows the distribution for PQSSs in the upstream region of all Ensembl genes with status ‘known’; the maximum peak is in the same region as the peak for the distribution of SP1-binding elements, −50 to −41 bases. A search for the SP1-binding site motif revealed that of the 22 633 known genes, just over half (52.5%) contained the motif in their upstream region (Table 1). However in Figure 3B we can see that this set of genes accounts for the vast majority of PQSSs in upstream regions (86.6%). Not only are the absolute number of PQSSs different, but the distribution of PQSSs which are in the upstream regions of genes with and without the SP1 consensus sequence differ markedly, as seen in Figure 3B and C, respectively. The PQSSs in non-SP1 upstream regions have a much flatter distribution than that of the SP1 motif genes.
Figure 3.
Potential quadruplex sequences (A) occurring in upstream regions, (B) in upstream regions which contain SP1 sequence elements (C) in upstream regions which do not contain SP1 sequence elements, (D) which incorporate SP1 sequence elements and are within upstream regions, (E) which do not incorporate SP1 sequence elements and are within upstream regions.
Table 1.
Summary of quadruplex occurrences in upstream regions of the human genome
| Quadruplex occurences | Number of genes | Number of sequence elements |
|---|---|---|
| SP1 sequence elements which occur in upstream regions | 11 872 | 29 991 |
| PQSSs in upstream regions which contain SP1 sequence elements | 5415 | 8660 |
| Genes without SP1 consensus sequence in their upstream regions | 10 761 | |
| PQSSs in upstream regions which do not contain SP1 sequence elements | 1096 | 1335 |
| PQSSs which incorporate SP1 sequence elements and are within upstream regions | 3596 | 4721 |
| PQSSs which do not incorporate SP1 sequence elements and are within upstream regions | 4150 | 5274 |
| CCAAT sequence elements which occur in upstream regions | 10 963 | 17 574 |
| PQSSs in upstream regions containing CCAAT sequence elements | 2718 | 3948 |
| Genes whose upstream regions contain no CCAAT sequence elements | 11 670 | |
| PQSSs in upstream regions containing no CCAAT sequence elements | 3794 | 6047 |
| Genes whose upstream regions contain CCAAT and SP1 | 5451 | |
| PQSSs in upstream regions containing SP1 and CCAAT sequence elements | 2236 | 3368 |
| Genes whose upstream regions contain CCAAT and no SP1 | 5512 | |
| PQSSs in upstream regions containing CCAAT and no SP1 sequence elements | 482 | 580 |
| Genes whose upstream regions contain SP1 and no CCAAT | 6421 | |
| PQSSs in upstream regions containing SP1 and no CCAAT elements | 3180 | 5292 |
| Genes whose upstream regions contain neither SP1 nor CCAAT | 5249 | |
| PQSSs in upstream regions containing neither SP1 nor CCAAT elements | 614 | 755 |
| All genes | 22 633 | |
| PQSSs in all upstream regions | 6512 | 9995 |
Since the SP1 consensus sequence is guanine-rich and can be incorporated into PQSSs, we examined the number of PQSSs, which contained the SP1 consensus sequence. These account for just under half the total PQSSs (47.2%). The distribution of PQSSs incorporating the SP1 sequence (Figure 3D) and PQSSs without the SP1 consensus sequence (Figure 3E) is very different. Figure 3D shows a distribution similar to the SP1 motif while that in Figure 3E is much flatter.
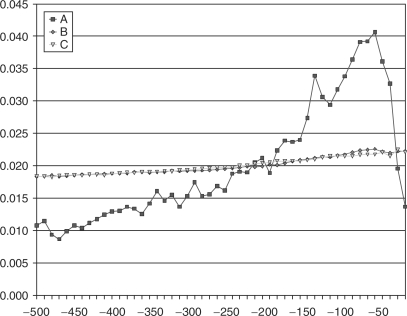
For a random sequence the probability of finding a PQSS is related to its guanine content so we looked at the guanine density within upstream regions. In Figure 4 we have the normalized distributions of PQSSs (A), guanine bases (B) and cytosine bases (C). Both guanine and cytosine do indeed get more frequent closer to the transcription start site although it is hard to say whether this is related to PQSS distribution.
Figure 4.
Normalized distributions in the 500 bp upstream regions of Ensembl genes with status ‘known’ of (A) PQSSs, (B) guanines, (C) cytosines.
Figure 5 shows the distributions of PQSSs within genes that contain the regulatory element CCAAT in their upstream region (Figure 5B) and PQSSs within genes, which do not contain CCAAT in their upstream region (Figure 5C). The distribution of PQSSs in CCAAT, although having a maximum at the same place as the CCAAT elements themselves (151 elements in the −70 to −61 region) has virtually the same number (149) in the region of the peak of the SP1 consensus sequence distribution (−70 to −61 bases). The PQSSs that occur in genes without upstream CCAAT elements have a peak in the same regions as the SP1 sequence elements.
Figure 5.
(A) CCAAT sequence elements occurring in upstream regions. (B) PQSSs in upstream regions containing CCAAT sequence elements. (C) PQSSs in upstream regions containing no CCAAT sequence elements.
Figure 6 shows the effect of the presence of SP1 and CCAAT sequence elements on the PQSS distribution, focusing on the distribution in additional groupings of genes. PQSSs upstream of genes with upstream SP1 elements but no upstream CCAAT elements have a distribution very similar to the SP1 element and the PQSS distribution of all known Ensembl genes (Figure 6A). There are very few PQSSs in genes, which contain upstream CCAAT sequences but no SP1 elements, and their distribution is very flat (Figure 6B). Genes with both upstream SP1 and CCAAT elements have many more PQSS however not such a distinct maximum (Figure 6C) and genes with neither upstream SP1 nor CCAAT elements have very few PQSSs and a rather flat distribution (Figure 6D).
Figure 6.
(A) PQSSs in upstream regions containing SP1 but no CCAAT sequence elements. (B) PQSSs in upstream regions containing CCAAT but no SP1 sequence elements. (C) PQSSs in upstream regions containing SP1 and CCAAT sequence elements. (D) PQSSs in upstream regions containing neither SP1 nor CCAAT sequence elements.
The distribution of PQSSs resembles that of regulatory motifs such as SP1 and CCAAT, although it would appear that upstream SP1 elements have a positive effect on the number of upstream PQSSs while the presence of CCAAT has a deleterious effect. Almost half of the total upstream PQSSs have the SP1 consensus sequence incorporated. Thus we can demonstrate that PQSSs linked to SP1 sequence motifs in the upstream regions is perhaps unsurprising but what is not necessarily so obvious is how dominant this effect is. It has been proposed that induction of quadruplex formation in promoter sequences by quadruplex-selective small molecules can be a viable therapeutics strategy (26). The present study provides support for this approach, and suggests that effort could be focused on those genes in which PQSSs are linked to SP1 sites, as is the case, for example, of the c-kit gene implicated in gastrointestinal cancers (15,27). A very recent report (28), in contrast, finds G-rich sequences in the first intron of many human genes, and considers that these are more likely to be PQSSs suitable for therapeutic intervention, in part because of the potential for structural polymorphism in the upstream sites. It is not clear that this would be a problem since it is likely that small molecule binding would tend to drive the equilibrium towards discrete quadruplex species. In addition, some PQSS sites such as those in the c-kit promoter (15,27), comprise isolated runs of just four G-tracts each, and are much less likely to participate in quadruplex polymorphism.
We also note that the presence of the zinc finger motif in SP1 may be significant in view of findings that the motif has been selected out from phage libraries to bind to quadruplex DNAs (29–31), and that transcription factors containing zinc fingers have been reported to bind to G-tract promoter sequences, notably the insulin promoter factor Pur-1/MAZ (32).
ACKNOWLEDGEMENTS
This work has been support by Cancer Research UK (programme grant C129/A4489). We are grateful to an anonymous reviewer for constructive comments. Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Burge S, Parkinson GP, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Antonio L, Bagga PS. CSB Proceedings of the 2004 IEEE Computational Systems Bioinformatics Conference (CSB 2004) CA: Stanford University; 2004. Computational methods for predicting intramolecular G-Quadruplexes in nucleotide sequences; pp. 561–562. [Google Scholar]
- 5.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawal P, Kummarasetti VBR, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2006;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 15.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 18.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett RD, Baty D, Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983;11:2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 21.Dynan WS, Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. Nature. 1985;316:774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- 22.Kadonaga JT, Jones KA, Tjian R. Promoter-specific activation of RNA polymerase II transcription by Sp1. Trends Biochem. Sci. 1986;11:20–23. [Google Scholar]
- 23.Thiesen HJ, Bach C. Target Detection Assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 1990;18:3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkm1138. doi:10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isalan M, Patel SD, Balasubramanian S, Choo Y. Selection of zinc fingers that bind single-stranded telomeric DNA in the G-quadruplex conformation. Biochemistry. 2001;40:830–836. doi: 10.1021/bi001728v. [DOI] [PubMed] [Google Scholar]
- 30.Patel SD, Isalan M, Gavory G, Ladame S, Choo Y, Balasubramanian S. Inhibition of human telomerase activity by an engineered zinc finger protein that binds G-quadruplexes. Biochemistry. 2004;43:13452–13458. doi: 10.1021/bi048892t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladame S, Schouten JA, Roldan J, Redman JE, Neidle S, Balasubramanian S. Exploring the recognition of quadruplex DNA by an engineered Cys2-His2 zinc finger protein. Biochemistry. 2006;45:1393–1399. doi: 10.1021/bi050229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]