Abstract
Penelope-like elements (PLEs) represent a new class of retroelements identified in more than 80 species belonging to at least 10 animal phyla. Penelope isolated from Drosophila virilis is the only known transpositionally active representative of this class. Although the size and structure of the Penelope major transcript has been previously described in both D. virilis and D. melanogaster transgenic strains, the architecture of the Penelope regulatory region remains unknown. In order to determine the localization of presumptive Penelope promoter and enhancer-like elements, segments of the putative Penelope regulatory region were linked to a CAT reporter gene and introduced into D. melanogaster by P-element-mediated transformation. The results obtained using ELISA to measure CAT expression levels and RNA studies, including RT–PCR, suggest that the active Penelope transposon contains an internal promoter similar to the TATA-less promoters of LINEs. The results also suggest that some of the Penelope regulatory sequences control the preferential expression in the ovaries of the adult flies by enhancing expression in the ovary and reducing expression in the carcass. The possible significance of the intron within Penelope for the function and evolution of PLEs, and the effect of Penelope insertions on adjacent genes, are discussed.
INTRODUCTION
Retrotransposons are generally regarded as being of one or other of two classes, either long terminal repeat (LTR) retrotransposons, or non-LTR retrotransposons, also known as LINEs. These classes are distinguished on the basis of their general organization and mode of transposition (1,2). Penelope-like elements (PLEs) do not fit easily into either class, however. Active Penelope elements were originally isolated from Drosophila virilis after hybrid dysgenesis was first described in this species (3,4). This phenomenon is observed when females from strains lacking Penelope are crossed with males carrying multiple active copies (4). Other transposable elements belonging to different classes are mobilized during this form of hybrid dysgenesis but it is the activation of Penelope that is responsible for such cross-mobilization (3,5,6). Penelope elements have been found in all species of the virilis group that have been studied so far. They are probably inactive except in D. virilis itself and have an unusually complex and highly variable organization (3). Database searches and analyses of genomic DNAs have detected PLEs in genomes of crustaceans, echinoderms, fish, amphibians, flatworms, roundworms and rotifers (7–10). These elements code for a protein that represents a fusion between a reverse transcriptase and a GIY-YIG endonuclease (11).
The majority of PLEs in D. virilis and bdelloid rotifers contain introns in different regions of the element, and RT–PCR analysis has confirmed that these can be correctly spliced (8). The ability to retain an intron during transposition, their peculiar structural organization and a distinct placement in the phylogeny of RT-containing elements lead us to conclude that the PLE clade is clearly different from both LTR and non-LTR retrotransposons and constitutes a third, and probably ancient, class of eukaryotic retroelements (8,12), which is supported by analysis of the amino acid sequence of the reverse transcriptase encoded by Penelope. This indicates that PLEs form a clade that is distinct from that of both LTR-retrotransposons and non-LTR retrotransposons (8).
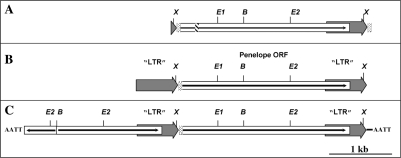
The partial-tandem organization characteristic of active copies of Penelope from D. virilis includes ‘pseudo-LTRs’ (Figure 1B) (3,13). These appear to result from tandem insertions of Penelope and have one or two copies of a 34–37-bp repeat sequence (the 34 bp repeat) at their 3′ end. Copies of Penelope without terminal repeats have been found in the genome of D. virilis. These are flanked by a target site duplication and have therefore transposed without pseudo-LTRs, indicating that these are not generated by reverse transcription. This organization is also observed in several copies of Poseidon/Xena PLEs from Tetraodon nigroviridis (14), and in many other PLEs (I.A., unpublished data). The tandemly repeated PLEs that are often observed may be needed to create a functionally active transcriptional unit, in a way mechanistically similar to that described for Drosophila melanogaster HeT-A elements (15). Indeed, the 2947-nt Penelope transcript initiates within the upstream ‘LTR’ (7).
Figure 1.
Schematic structure of various Penelope copies. (A) Presumptive ancient primitive copy (solo ORF flanked by two 34-bp repeats in direct orientation) isolated from D. texana and D. montana (13). (B) Active copy from the clone ‘p6’ used for inter-specific transformation experiments with ‘pseudo LTRs’ (3). (C) An active form found frequently in both in D. virilis and in D. melanogaster strains transformed with Penelope. It has two tandemly arranged Penelope copies (the 5′ one is truncated at the 5′end) flanked by large inverted terminal repeats which vary in size depending on the copy concerned (3). Restriction sites: E-EcoR1; B-BamH1; X-Xho1; dotted rectangles denote the 34-bp repeat sequences located at 5′ and 3′ ends of Penelope ORF; hatched box means small deletion.
The experiments reported herein have been performed to identify the sequences required for full activity of the promoter, and to identify sequences required for the previously described differential expression of Penelope in the ovaries and carcasses of D. virilis dysgenic hybrids and in D. melanogaster transgenic strains transformed with full-sized Penelope (3,16).
MATERIALS AND METHODS
Fly stocks
The D. melanogaster strain Df(1), y w 67c23(2) was used as recipient in transformation experiments. D. virils strain 160 (with about 35 copies of Penelope) and D. virilis strain 9 (lacking active Penelope) were used in the study to get dysgenic hybrids as described (3). Flies were reared on standard resin-sugar-yeast-agar medium containing propionic acid and methylparaben as mold inhibitors.
Construction of transformation vectors
pWP-CAT vector containing unique XbaI and BamHI sites upstream of the CAT gene was made by Acc65 I and XhoI digestion of p186W8 (17) and subsequent self-ligation. Promoter fragments were amplified by the polymerase chain reaction (PCR), using the following pairs of primers: PenPr1 up–PenPr850 lo; Pen250 up–PenPr850 lo; PenPr570 up–PenPr850 lo; PenPr1 up–PenPr680 lo; PenPr1 up–PenPr611 lo; PenPr1 up–PenPr487 lo; PenPr1 up–PenPr544 lo; PenPr1 up–PenPr416 lo; PenPr1 up–PenPr369 lo; PenPr1 up–PenPr302 lo. The resulting fragments were digested with XbaI, BamHI and ligated into XbaI/BamHI-cut pWP-CAT, yielding plasmids pPenPr A, pPenPr B, pPenPr C, pPenPr D, pPenPr F, pPenPr M, pPenPr N, pPenPr O, pPenPr P and pPenPr R, respectively (Supplementary Table 1). The intronless promoter fragment was synthesized by PCR using primers PenPr1 up–int lo; int up–PenPr850 lo, followed by PCR assembly with primers PenPr1 up–PenPr850 lo. The resulting fragment was digested with XbaI, BamHI and ligated into XbaI/BamHI-cut pWP-CAT to generate plasmid pPenPr G. The construct PB containing 366 bp of the Penelope regulatory region (interval 352–718 in clone p6) was cloned into pCaSpeR-AUG-β-gal as described previously (18).
P-element-mediated transformation
The DNA used for transformation of D. melanogaster was purified by QIAprep Spin Miniprep Kit (Qiagen) and used for embryo injection as described previously (19). Transposase activity was provided by the helper plasmid Turbo Δ2-3 (20) and the recipient embryos were from the D. melanogaster Df(1), y w 67c23(2) strain. Adults emerging from the injected embryos were crossed with Df(1), y w 67c23(2) virgins of the opposite sex, and the eye colour of their progeny was examined. Transformed lines homozygous for the transgene were established by full sibling mating. The presence of homozygous intact transgene copies in each line was confirmed by PCR and Southern blotting. In most cases, the number and exact localization of the inserts were checked by in situ hybridization as described (21). Each transformed line was routinely maintained en masse in 5–6 vials initiated with 20–30 flies per vial.
β-Galactosidase staining
X-gal staining of ovaries was performed essentially as described (22). Ovaries were dissected in PBS and fixed for 10 min in fixing solution (1% gluteraldehyde in PBS). They were washed in PBS for 3 × 10 min, and then incubated for 1 day in staining solution [(10 mM Na/Na2PO4 pH 7.2; 150 mM NaCl; 1 mM MgCl2; 3.1 mM potassium ferricyanide; 3.1 mM potassium ferrocyanide; 0.5 mM Xgal (5-bromo-4-chloro3-indolyl-β-d-galactopyranoside)] at 37°C. The X-gal was added shortly before incubation from a 10% stock solution in DMSO. After staining, the ovaries were washed in PBS and mounted in 50% glycerol solution prior to histological analysis.
RNA preparations and analysis
Total RNA was extracted from carcasses and ovaries of 2—5-days-old females of each independent transgenic strain using TriZol Reagent according to protocol provided by Invitrogen. The integrity of each RNA preparation was checked on ethidium bromide-stained 1% agarose/MOPS-formaldehyde gels. A SMART RACE cDNA amplification kit (Clontech) was used for cDNA synthesis and amplification of 5′-ends of cDNAs, as described (23). The sequence of the oligonucleotides used for amplification of 5′-ends of Penelope-containing cDNAs, PenRev1, PenRev2 and PenRev3 and of the SMART PCR, ‘heel-carrier’ and ‘heel-specific primers are given in Supplementary Table 1. Semi-quantitative RT–PCR was used to reveal Penelope-CAT transcripts in total RNA from carcasses ovaries of transformants with constructs N or P, and to study tissue-specific slicing. SMART amplification templates served as the samples for specific amplification using primers IstPenDir10 and PenRev2 (Supplementary Data). Samples were normalized using primers, β-actin D. melanogaster D1 and β-actin D. melanogaster R1, for D. melanogaster β-actin.
Each oligonucleotide was purified through polyacrylamide gel before use. PCR reactions were performed in a 25 µl reaction mixture and contained 1× Advantage KlenTaq Polymerase Mix with provided buffer (Clontech), 200 µM dNTPs, 0.15 µM of gene-specific primers, 0.02 µM of ‘heel-carrier’ oligo and 0.15 µM of ‘heel-specific’ oligo. PCR was carried out using MJ Research PTC-200 DNA Thermal Cycler for 25–30 cycles (95°C for 7 s; 63°C for 20 s; 72°C for 2 min) depending on the transcript's abundance. Reactions without reverse transcriptase have been carried out for all templates to exclude DNA contamination.
Cloning and sequencing
All standard DNA procedures were performed in accordance with published laboratory protocols (24). PCR fragments obtained in the course of 5′-RACE were cloned in the pGEM T-easy vector (Promega) and sequenced. DNA sequence was determined using the Amersham Biosciences Megabase 1000 automated sequencer and FS dye terminator chemistry.
Drosophila CAT assays
One hundred and twenty females of each independent strain (2–5 days old) were used to isolate ovaries and carcasses. The tissues were homogenized in 0.25 M Tris–HCl, pH 7.8 and passed through five freeze–thaw cycles. The extract was then spun in a microcentrifuge to pellet cell debris and denatured proteins, and the supernatant was used as CAT extract (17). The concentration of protein in the supernatant was determined by the BCA Protein Assay Kit (Pierce). CAT assays were performed according to the instruction manual of CAT ELISA kit (Roche). The levels of CAT expression were expressed as pgCAT/μg of total protein estimated.
In our experiments, the original strain Df(1), y w 67c23(2) used for obtaining transgenic strains always exhibited significant and varying background levels of CAT expression. The background was consistently higher in the carcasses than in the ovaries. Therefore, we routinely took the level of CAT expression in the original strain as a unit for each individual ELISA and calculated the CAT levels in the transgenic strains assuming that Df(1), y w 67c23(2) CAT expression equals 1 U. Thus, the formula is: the expression level observed in the transgenic strain divided by the background CAT level (observed in control recipient strain). This approach enabled us to compare the CAT levels measured in ovaries and carcasses and the results obtained in independent ELISA experiments.
Bioinformatics
Promoter prediction was carried out with the BDGP Neural Network Promoter Prediction server with eukaryotic settings (http://www.fruitfly.org/seq_tools/promoter.html; (25), and McPromoter MM:II with Drosophila settings (http://genes.mit.edu/promoterMMII.html; (26). Alignments were generated by ClustalW (http://www.ebi.ac.uk/clustalw/) and presented in the BoxShade format (http://www.ch.embnet.org/software/BOX_form.html).
RESULTS
The promoter region of Penelope
Reporter gene analysis
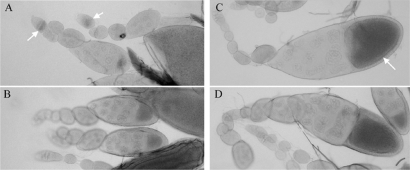
We have previously shown that the p6 copy of D. virilis Penelope (Figure 1B) is functional in D. melanogaster and can actively transpose in transgenic strains (16). Moreover, the full-sized Penelope transcript is restricted to the ovaries of D. melanogaster strains transformed with an active Penelope copy, as is the case with dysgenic females of D. virilis. In order to show that the Penelope 5′ regulatory sequences are able to drive the expression of a reporter gene in the cells of the two species studied, we performed preliminary experiments which have shown that nucleotides 1–850 of this element (Figures 1B and 3B) are sufficient to drive expression of the chloramphenicol acetyl transferase (CAT) reporter gene upon transfection into D. virilis tissue culture cells (data not shown). Furthermore, we observed lacZ expression in the egg cyst and in the cytoplasm of the oocyte at different stages of oogenesis (Figure 2A–D) in the ovaries of transgenic D. melanogaster flies carrying the construct ‘PB’ (see Materials and Methods section) containing nucleotides 352–718 of Penelope linked to a lacZ reporter gene (11).
Figure 3.
(A) Histogram illustrating CAT levels in the ovaries and in carcasses of the transgenic strains. The schematic structure of all constructs used is depicted at the bottom. (B) Presumptive structure of the Penelope regulatory region with positions of promoter and other regulatory sequences indicated. Letters denote the constructs used, and CAT expression in ovaries (ov) and carcasses (ca) is illustrated by + or −. Figures at the bottom indicate the boundaries of constructs within the Penelope 5′ region (interval 1–850 bp). The dotted grey line shows promoter prediction scores from the NNPP program (from 0 to 1, indicated on the right). McPromoter scores yielded a qualitatively similar picture with different ratios of peak intensities (data not shown). The shaded region at the beginning of the ORF marks the 34-bp repeat.
Figure 2.
Detection by X-gal staining of β-galactosidase expression in the D. melanogaster transgenic strain with construct PB containing nucleotides 352–718 of the Penelope 5′ region driving expression of the reporter. (A) Histochemical detection of β-galactosidase in the germarium. A band of stained follicle cells enveloping the egg cyst can be seen (indicated by the arrows). Five-day-old females; (B) Control Df(1), y w 67c23(2) strain. Five-day-old females. (C) Strong staining in the cytoplasm of the oocyte at stage 9 in 1–2-days-old females (indicated by the arrow). (D) Control Df(1), y w 67c23(2) strain. 1–2-days-old females.
We have previously identified the first nucleotide of Penelope transcripts as being predominantly at position 371 (Figure 3B) (8). Since other non-LTR retrotransposons in Drosophila are transcribed from internal promoters (17,27,28) it is likely that the same is true of Penelope. We have analysed the first 850 nucleotides of the p6 Penelope element (Figure 1B) using two programs previously applied to Drosophila promoters. ‘McPromoter’ (26), which combines a generalized hidden Markov model for sequence features and Gaussian distributions for the predicted structural features of DNA, and ‘BDGP Neural Network Promoter Prediction’ (25), which utilizes time-delay neural networks. Both programs predicted basal promoter elements between nucleotides 302 and 680 (Figure 3B) with peaks at the start of transcription and in the vicinity of the 34-bp repeat sequence.
In order to determine experimentally the boundaries of the presumptive promoter and other regulatory sequences, we generated constructs containing various fragments of the 5′sequence (1–850) of Penelope linked to the CAT reporter gene as was done for the I factor (17). These constructs were verified by sequencing, and introduced into the genome of D. melanogaster by P-element-mediated transformation (19). Figure 3A shows the level of CAT protein in ovary and carcass, as determined by ELISA assays, in each transformed line, while Figure 3B summarizes the expression of CAT protein for lines carrying each construct.
These data indicate that promoter and regulatory sequences of Penelope lie between nucleotides 369 and 680 (lines with constructs R and D) while the CAT levels in transgenic lines carrying constructs R and P suggest that sequences required for expression lie between nucleotides 369 and 416. This region appears to be sufficient for transcription as the lacZ reporter construct PB expresses β-galactosidase in ovaries. This contains nucleotides 352–718, and it is unlikely that the 18 nt between positions 352 and 369 contain a significant transcription signal. We conclude that it is likely that Penelope contains an internal promoter. This is consistent with the computer prediction indicating that the Penelope promoter is within the transcribed region. Alignment of this presumptive Penelope promoter with several TATA-less internal promoters of other Drosophila non-LTR retrotransposons (Figure 4) shows that Penelope contains the downstream promoter element RGACGTGY, the only promoter sequence conserved in other Drosophila non-LTR retrotransposons (29).
Figure 4.
Multiple sequence alignment of the putative Penelope promoter from D. virilis (Dv) and D. willistoni (Dw) with internal promoters of Drosophila LINE elements (I-factor, Jockey, F, G and Doc) listed in refs. 17 and 19). Transcription start site is indicated by an arrow. Inr, initiator, DPE, downstream promoter element; SD, splice donor site conserved in D. virilis and D. willistoni Penelope elements. ‘de1 and ‘de2’ represent motifs which stimulates transcription in various Drosophila LINEs (27).
RNA analysis
We have complemented our measurements of CAT proteins levels by RNA analysis of transcripts from some of the transgenic lines in Figure 3A to distinguish effects on CAT expression due to changes in transcription or RNA stability from those due to changes in translation and to check that transcription originated from the Penelope promoter rather than from adjacent chromosomal sequences. RT–PCR and 5′ RACE, with the primers indicated in the Materials and Methods section and Supplementary Data, was used to analyse RNA from ovaries and carcass of strains carrying constructs N or P, each of which contains sequences from the Penelope promoter region. Canonical Penelope transcripts are normally detected in ovaries in both D. virilis dysgenic hybrids and in D. melanogaster strains transfected with active copies of Penelope (30,31). We could not detect such transcripts in ovaries of strains with constructs N or P, although they were present in carcass RNA. (Figure 5), suggesting that the lack of CAT expression in ovaries was the result of an effect on transcription.
Figure 5.
Semi-quantitative RT–PCR analysis using SMART-amplified templates and primers for detection real transcripts of Penelope-CAT initiated at the usual start site in strains with constructs N and P. Lane 1–100 bp size markers; Results of amplification of cDNA: lane 2, carcasses of P-33 strain; lanes 3, ovaries of strain P-33; lane 4, ovaries of strain P-46; lane 5, ovaries of strain N-10; lane 6, ovaries of strain N-39; lane 7, carcasses of strain P-33 without reverse transcriptase; lane 8, ovaries of strain P-33 without reverse transcriptase.
In the case of strain P-25, which is exceptional in showing CAT expression in ovaries (Figure 3A), 5′ RACE revealed a long ovarian transcript that had initiated within the Cct1 (phosphocholine cytidylyltransferase 1) gene (Supplementary Figure 1), which we have confirmed, by in situ hybridization, is the site of insertion of the P construct in this strain (data not shown). This gene is active in ovaries and we conclude that expression of the P construct in this line results from read-through transcription from the Cct1 gene, and have excluded it from further analysis.
We have previously shown that all active copies of Penelope isolated from D. virilis contain a copy of the 34-bp repeat at the beginning of the ORF and there is often a second copy at the 3′ end. Moreover, structurally similar copies with 34-bp repeats flanking the Penelope ORF in direct orientation have been found in two other species of the virilis group (13). In D. virilis, the most frequently observed structure of a Penelope element contains a partial-tandem repeat of two Penelope ORFs flanked with inverted terminal repeats (Figure 1C) and may have evolved from a single Penelope ORF flanked by 34-bp repeats (Figure 1A). Our computer analysis of the promoter regions suggested that this sequence may be part of a strong promoter. We have tested this using transgenic lines carrying construct C which has nucleotides 570–850 linked to the CAT gene. This lacks both the promoter that we have identified as being between nucleotides 369 and 416 and the transcriptional start site, and yet strains carrying it express CAT in ovaries and weakly in carcass. This appears to be due to transcription that has initiated at cryptic transcription start sites within the transformation vector, presumably under the influence of Penelope regulatory sequences (Supplementary Figure 1).
Tissue-specific regulation of Penelope expression
The data in Figure 3A and B indicate that the sequence in between positions 611 and 680 controls tissue-specific expression from the presumptive Penelope promoter. Strains containing constructs A, B, C or D each express CAT in the ovary whereas strains carrying constructs F, N, O and P that lack this sequence do not. This suggests that the sequence between 611 and 680 enhances expression in the ovary. Strains carrying constructs F, N, O, P that contain the Penelope promoter and sequences up to position 611, show high CAT expression in the carcass but none in the ovary, whereas strains with constructs containing the promoter and nucleotides 611 to 680 show reduced CAT protein in the carcass. This suggests that the sequence between positions 611 and 680 also down-regulate expression in the carcass.
Previous studies have shown that the minority of Penelope transcripts in both D. virilis dysgenic hybrids and transgenic D. melanogaster contain the intron that lies between nucleotides 416 and 491 while a majority of transcripts lack the intron (7), but as yet there is no indication as to whether the distribution of these RNAs differs between tissues. We have addressed this question by RT–PCR using primers that allow detection of both the spliced and unspliced forms, and RNA extracted from carcass or ovary. Both spliced and unspliced forms were detected in RNA from each tissue of both dysgenic D. virilis females and females of the Penelope containing D. virilis strain 160, and from each tissue of flies containing construct A or G (Figure 6A, B and Supplementary Figure 2).
Figure 6.
Semi-quantitative RT–PCR analysis using SMART-amplified templates performed to detect spliced and unspliced transcripts. (A) Lane 1–100 bp size markers; Results of amplification of RNA: lane 2, carcasses of strain A-6 (33cycles); lane 3, carcasses of strain G-27 (30 cycles); lane 4, ovaries of strain G-27 (32 cycles); lane 5, carcasses of strain G-7 (30 cycles). (B) Lane 1, 100 bp size markers; results of amplification of RNA: lane 2, carcasses of D. virilis dysgenic hybrids, (30 cycles); lane 3, ovaries of D. virilis dysgenic hybrids, (25 cycles); lane 4, carcasses of D. virilis strain 160, (30 cycles); lane 5, ovaries of D. virilis strain 160, (27 cycles). The sequences of PCR fragments depicted in this figure are presented in the Supplementary Data.
Transgenic flies containing construct G show virtually no CAT activity in either carcass or ovary extracts whereas flies with construct A show significant CAT expression in the ovaries and weak expression in carcasses (Figure 3A). Constructs A and G differ in that A contains Penelope intron but G does not. The lack of CAT expression from construct G could result from transcriptional or post-transcriptional effects, or both. We have investigated this using RT–PCR to assay the presence of RNA from construct G in carcasses and ovaries of transgenic flies. The results (Figure 6A and Supplementary Figure 2) show that the transcripts are present in both ovary and carcass ruling out the possibility that sequences essential for Penelope transcription are contained within the intron. The lack of CAT activity in flies with construct G presumably reflects an effect of the intron on normal RNA processing or translation.
DISCUSSION
The active Penelope retrotransposon from D. virilis was the first PLE retrotransposon to be described, and its structural and functional organization can be expected to display characteristics that are typical of other members of this widespread but relatively little-studied class of elements. The present study demonstrates that sequences necessary and sufficient for expression of Penelope in vivo reside within the element itself, in the region previously characterized as the pseudo-LTR. This region results from a partial-tandem arrangement of two elements, one 5′-truncated and one full-length, and consists primarily of the 3′ UTR of the upstream copy which, because of the partial tandem duplication, is also part of the 5′ UTR of the downstream copy. This is reminiscent of the composite promoter previously described in HeT-A retrotransposons in D. melanogaster for which the sequences in the 3′ UTR of the upstream copy drive transcription of the downstream copy in a tandem repeat (15). In the case of Penelope, all the sequences necessary for transcription in the carcass are present in the 3′ pseudo-LTR. This gives Penelope elements the ability to change patterns of gene expression by driving transcription of downstream genes, or downstream exons of genes into which they insert.
Our data show that sequences controlling the tissue-specific transcription directed by the Penelope promoter are also downstream of the RNA start site and in the region that includes the 34-bp repeat and part of the ORF. Sequences directing tissue-specific expression have been identified in a similar position in the I factor (32) and F element (33), two other non-LTR retrotransposons of Drosophila. These regulatory elements apparently confer tissue specificity on the Penelope promoter, and may be essential for the expression in ovaries that results in germ-line transposition and gonadal sterility during hybrid dysgenesis in D. virilis. This suggests that these signals may have been conserved for over 60 million years, the time since the divergence of the lines that lead to D. virilis and D. melanogaster.
LTR retrotransposons are transcribed from a promoter within the U3 region of the 5′ LTR while all non-LTR retrotransposons studied so far, with the possible exception of R2Bm from Bombyx mori which is thought to be transcribed from an adjacent rDNA promoter (34), are transcribed from a promoter that lies within the transcription unit. The fact that most PLEs appear to require a partial-tandem repeat, rather like an LTR, for transcription sets them apart from both LTR and non-LTR retrotransposons. There is no obvious mechanism for pseudo-LTR regeneration other than the propensity to form tandem repeats. In fact, as many as 30% of all D. virilis genomic Penelope copies, as well as most Penelope copies amplified in D. melanogaster strains transformed by full-sized Penelope (Figure 1B), possess a tandem structure, with a full-length downstream copy and a variably 5′-truncated upstream copy (30). In future studies, we plan to investigate the basis for frequent tandem formation in PLEs.
The 34-bp repeat sequence, which is found at both ends of the ORF in ancient copies of Penelope from other species of the virilis group (13), generates relatively high scores in computational analyses of basal promoter elements but is not required for transcription from the normal initiation site. The 34-bp repeat may be a simple basal promoter of ancestral nature, and more complex structures, including upstream promoter elements as seen in LTR retrotransposons and retroviruses, may have arisen later.
The 75-bp intron located 46-bp downstream from the Penelope transcription start site is within the promoter region. The splice donor and acceptor sites of the intron are well conserved in two Penelope elements from D. willistoni, that display 67% overall nucleotide sequence identity to Penelope from D. virilis (I.A., unpublished data). The intron appears to have a role in post-transcriptional regulation of Penelope as deleting it virtually abolishes CAT expression in transgenic strains with construct G, without apparently affecting RNA levels in ovary or carcass. The difference in CAT activity from constructs A and G could reflect differences in the secondary structure of the corresponding RNAs in the context of CAT reporter, as has been described for CAT reporter constructs of the Doc retrotransposon (27). It is also possible that nuclear export and/or translation of spliced Penelope RNA is facilitated by a component of the intron junction complex, indeed introns have been shown to affect RNA and protein levels in S. cerevisiae (35). RNA lacking the intron, such as transcripts from construct G, would not have this complex and might therefore be exported or translated inefficiently, or not at all. If this is the case, then Penelope elements without the intron would be at a selective disadvantage, providing an explanation for retention of the intron.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr Boris Andrianov (Institute of General Genetics RAS, Moscow) for performing transient expression experiments exploring D. virilis cell culture. We thank Dr Olga Zatsepina for critically reading the manuscript and fruitful discussion of the results. This work was supported by grants from Russian Academy of Sciences (Cell and Molecular Biology to M.E.), and Welcome Trust Grant (075698) to M.E and D.J.F. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Finnegan DJ. Transposable elements. Curr. Opin. Genet. Dev. 1992;2:861–868. doi: 10.1016/s0959-437x(05)80108-x. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 3.Evgen’ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, Lankenau DH, Corces VG. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc. Natl Acad. Sci. USA. 1997;94:196–201. doi: 10.1073/pnas.94.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozovskaya ER, Scheinker VS, Evgen’ev MB. A hybrid dysgenesis syndrome in Drosophila virilis. Genetics. 1990;126:619–623. doi: 10.1093/genetics/126.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc. Natl Acad. Sci. USA. 1995;92:8050–8054. doi: 10.1073/pnas.92.17.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira J, Vieira CP, Hartl DL, Lozovskaya ER. Factors contributing to the hybrid dysgenesis syndrome in Drosophila virilis. Genet. Res. 1998;71:109–117. doi: 10.1017/s001667239800322x. [DOI] [PubMed] [Google Scholar]
- 7.Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat. Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 8.Evgen’ev MB, Arkhipova IR. Penelope-like elements–a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet. Genome Res. 2005;110:510–521. doi: 10.1159/000084984. [DOI] [PubMed] [Google Scholar]
- 9.Volff JN, Hornung U, Schartl M. Fish retroposons related to the Penelope element of Drosophila virilis define a new group of retrotransposable elements. Mol. Genet. Genomics. 2001;265:711–720. doi: 10.1007/s004380100468. [DOI] [PubMed] [Google Scholar]
- 10.Zelentsova ES, Pyatkov KI, Shostak NG, Evgen’ev MB. The unusual moile element Penelope and its behaviour in distant Drosophila species. Genetika (Russ.) 2003;39:269–279. [PubMed] [Google Scholar]
- 11.Pyatkov KI, Arkhipova IR, Malkova NV, Finnegan DJ, Evgen’ev MB. Reverse transcriptase and endonuclease activities encoded by Penelope-like retroelements. Proc. Natl Acad. Sci. USA. 2004;101:14719–14724. doi: 10.1073/pnas.0406281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arkhipova IR. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst. Biol. 2006;55:875–885. doi: 10.1080/10635150601077683. [DOI] [PubMed] [Google Scholar]
- 13.Lyozin GT, Makarova KI, Velikodvorskaya VV, Zelentsova H, Khechumian RR, Kidwell MG, Koonin E, Evgen’ev M. The structure and evolution of Penelope in the Drosophila virilis species group: an ancient lineage of retrotransposons. J. Mol. Evol. 2001;52:445–456. doi: 10.1007/s002390010174. [DOI] [PubMed] [Google Scholar]
- 14.Dalle Nogare DE, Clark MS, Elgar G, Frame IG, Poulter RT. Xena, a full-length basal retroelement from tetraodontid fish. Mol. Biol. Evol. 2002;19:247–255. doi: 10.1093/oxfordjournals.molbev.a004078. [DOI] [PubMed] [Google Scholar]
- 15.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 16.Pyatkov KI, Shostak NG, Zelentsova ES, Lyozin GT, Melekhin MI, Finnegan DJ, Kidwell MG, Evgen’ev MB. Penelope retroelements from Drosophila virilis are active after transformation of Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2002;99:16150–16155. doi: 10.1073/pnas.252641799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean C, Bucheton A, Finnegan DJ. The 5′ end of the I factor, a LINE-like Retrotransposon of Drosophila, contains an Internal promoter and Sequences that Regulate Expression. Mol. Cell. Biol. 1993;13:1042–1050. doi: 10.1128/mcb.13.2.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyatkov KI, Zelentsova ES, Evgen’ev MB. The determination of nuclease activity of Penelope retrotransposon. Mol. Biol. (Russ.) 2004;38:609–616. [PubMed] [Google Scholar]
- 19.Spradling AC. In: Drosophila: a Practical Approach. Roberts DB, editor. ORL Press: Oxford; 1986. pp. 175–197. [Google Scholar]
- 20.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim JK. In situ hybridisation with biotinylated DNA. Dros. Inf. Serv. 1993;72:73–77. [Google Scholar]
- 22.Lachaume P, Bouhidel K, Mesure M, Pinon H. Spatial and temporal expression of the I factor during oogenesis in Drosophila melanogaster. Development. 1992;115:729–735. doi: 10.1242/dev.115.3.729. [DOI] [PubMed] [Google Scholar]
- 23.Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 26.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Contursi C, Minchiotti G, Di Nocera PP. Identification of sequences which regulate the expression of Drosophila melanogaster Doc elements. J. Biol. Chem. 1995;270:26570–26576. doi: 10.1074/jbc.270.44.26570. [DOI] [PubMed] [Google Scholar]
- 28.Minchiotti G, Di Nocera PP. Convergent transcription initiates from oppositely oriented promoters within the 5′ end regions of Drosophila melanogaster F elements. Mol. Cell. Biol. 1991;11:5171–5180. doi: 10.1128/mcb.11.10.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minchiotti G, Contursi C, Di Nocera PP. Multiple downstream promoter modules regulate the transcription of the Drosophila melanogaster I, Doc and F Elements. J. Mol. Biol. 1997;267:37–46. doi: 10.1006/jmbi.1996.0860. [DOI] [PubMed] [Google Scholar]
- 30.Pyatkov KI, Shostak NG, Evgen’ev MB. Amplification of retrotransposon Penelope after interspecific transformation. Dokladi Russ. Acad. Sci. 2002;381:268–270. doi: 10.1023/a:1013347026102. [DOI] [PubMed] [Google Scholar]
- 31.Zelentsova H, Poluectova H, Mnjoian L, Lyozin G, Veleikodvorskaja V, Zhivotovsky L, Kidwell MG, Evgen’ev MB. Distribution and evolution of mobile elements in the virilis species group of Drosophila. Chromosoma. 1999;108:443–456. doi: 10.1007/s004120050396. [DOI] [PubMed] [Google Scholar]
- 32.Udomkit A, Forbes S, Arkhipova I, McLean C, Finnegan DJ. Control of expression of the I Factor, a LINE-like transposable element in Drosophila melanogaster. EMBO J. 1996;15:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 33.Kerber B, Fellert S, Taubert H, Hoch M. Germ line and embryonic expression of Fex, a member of the Drosophila F-element retrotransposon family, is mediated by an internal cis-regulatory control region. Mol. Cell. Biol. 1996;16:2998–3007. doi: 10.1128/mcb.16.6.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George JA, Eickbush TH. Conserved features at the 5 end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol. Biol. 1999;8:3–10. doi: 10.1046/j.1365-2583.1999.810003.x. [DOI] [PubMed] [Google Scholar]
- 35.Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW. Introns regulate RNA and protein abundance in yeast. Genetics. 2006;174:511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.