Abstract
Mediator is an evolutionary conserved coregulator complex required for transcription of almost all RNA polymerase II-dependent genes. The Schizosaccharomyces pombe Mediator consists of two dissociable components—a core complex organized into a head and middle domain as well as the Cdk8 regulatory subcomplex. In this work we describe a functional characterization of the S. pombe Mediator. We report the identification of the S. pombe Med20 head subunit and the isolation of ts alleles of the core head subunit encoding med17+. Biochemical analysis of med8ts, med17ts, Δmed18, Δmed20 and Δmed27 alleles revealed a stepwise head domain molecular architecture. Phenotypical analysis of Cdk8 and head module alleles including expression profiling classified the Mediator mutant alleles into one of two groups. Cdk8 module mutants flocculate due to overexpression of adhesive cell-surface proteins. Head domain-associated mutants display a hyphal growth phenotype due to defective expression of factors required for cell separation regulated by transcription factor Ace2. Comparison with Saccharomyces cerevisiae Mediator expression data reveals that these functionally distinct modules are conserved between S. pombe and S. cerevisiae.
INTRODUCTION
The Mediator is a multiprotein coregulator complex that is required for the transcription of almost all RNA polymerase II (pol II)-dependent genes in fungi and metazoans (1). Mediator is thought to act as an interface between the general transcription machinery and sequence-specific transcriptional activators. A pure yeast in vitro transcription system consisting of pol II and all the general transcription factors (GTFs) cannot be stimulated by activators in the absence of Mediator (2,3).
Mediator was originally purified in Saccharomyces cerevisiae and has since then been isolated from several other eukaryotic species (1,4,5). These biochemical data revealed a core set of Mediator subunits that are conserved in most, if not all eukaryotes (6–8).
We have previously isolated and characterized the Mediator complex from Schizosaccharomyces pombe (6,9). The S. pombe Mediator exists in at least three states: a smaller core Mediator complex (S-Mediator) consisting of 15 subunits, a larger form (L-Mediator) consisting of core Mediator bound to a four-subunit module known as the Cdk8 module, and finally as a holoenzyme form with the core Mediator bound to pol II (9–11).
The Cdk8 module in both S. cerevisiae and S. pombe consists of four proteins: Med12 and Med13 as well as the cyclin-dependent kinase Cdk8 and its cyclin CycC (10,12). Both S. cerevisiae and S. pombe Cdk8 are able to phosphorylate the C-terminal domain of the largest subunit of pol II in vitro, which is thought to inhibit transcriptional initiation (10,13). Titration of the Cdk8 module-containing L-Mediator into an in vitro S. pombe transcription system has been shown to counteract the stimulatory effect of Mediator on basal transcription (14). Yet there is also evidence of a positive role for the Cdk8 module in activation (15,16).
Electron microscopy (EM) studies of single S. cerevisiae core Mediator particles identified three distinct domains that have been named head, middle and tail (17,18). A similar investigation showed that the S. pombe core Mediator also contained a head and a middle domain, but lacked a visible tail domain (11). In agreement with this, only one of the five S. cerevisiae tail subunits, Med15, is conserved in S. pombe. So far Med15 has not been identified as a stable component of S. pombe Mediator.
Based on work in S. cerevisiae, the S. pombe middle domain is suggested to consist of Med1, Med4, Med7, Med10, Med14, Med19, Med21 and Med31 (19–23). The architecture of the S. cerevisiae head domain has been extensively characterized (20–25). From these data, we predict that the S. pombe head domain consists of Med8, Med17, Med18 and Med22 as well as the proposed Med20 subunit, which has not been identified as a stable Mediator component prior to this work.
Here, we describe a structural and functional characterization of a representative set of subunits of the S. pombe Mediator. We identify the nonessential set of subunits in the head domain including a new subunit homologous to S. cerevisiae Med20. We report the isolation and characterization of conditional alleles of the essential head domain component Med17 and discuss its role in head domain architecture. Phenotypical analysis in combination with expression profiling of a representative set of Mediator mutant alleles allowed us to define two distinct functional classes of subunits within the S. pombe Mediator complex. Finally, the role of these separate classes of Mediator subunits in specific cellular pathways is discussed.
MATERIAL AND METHODS
Schizosaccharomyces pombe strains
All yeast strains used in this study are listed in Table 1. Schizosaccharomyces pombe cells were transformed by the lithium acetate procedure (26). Null mutants of med15+, med18+ and med20+ for expression profiling were generated using the kanMX selectable marker in the haploid h− strain MP9 as described (6) using the primers listed in Supplementary Table S1. The double null mutant of med18+ and med20+ was constructed by crossing and tetrad analysis of the diploid TP396/TP235. Null mutants of med18+, med20+ and med27+ for protein purification were generated with the ura4+ gene as a selectable marker in an Ura− background. A plasmid containing the wild-type ura4+ gene (GenBank accession number X13976), pURA4, was constructed by inserting it into HindIII-digested pBluescript II SK (Stratagene). Segments of 500-bp flanking the med18+, med20+ and med27+ genes were PCR amplified and inserted on either side of the ura4+ marker to generate the plasmids pURA4-spmed18, pURA4-spmed20 and pURA4-spmed27, respectively. The knockout fragments were released from the respective plasmids by digesting with PvuII and transformed into the diploid strain TP219/TP220 carrying a Tandem Affinity Purification (TAP) tag on both copies of the med7+ gene. The obtained Ura+ diploids were sporulated and Ura+/G418-resistant spores were recovered, to generate strains TP416, TP417 and TP306, respectively.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Parent strain(s) | Source |
|---|---|---|---|
| S. pombe | |||
| MP1 | h+ ade6-M210 his7-336 leu1-32 ura4-D18 | ID CHP429 | (6) |
| MP2 | h− ade6-M216 his7-336 leu1-32 ura4-D18 | ID CHP428 | (6) |
| MP9 | h− | L972 | M. Sipiczki |
| MP10 | h90 sep15-598 his3 | 2-506 | (44) |
| MP12 | h− ade6-M210 | IH105 | Ian Hagan |
| MP13 | h+ ade6-M216 | IH107 | Ian Hagan |
| TP21 | h− Δmed1::kanMX4 | MP9 | This study |
| TP26 | h− Δmed18::kanMX4 | MP9 | This study |
| TP27 | h− Δmed12::kanMX4 | MP9 | This study |
| TP28 | h− Δmed12::kanMX4 | MP9 | (10) |
| TP42 | h+ ade6-M210 med7+::TAP-kanMX6 | MP12/MP13a | This study |
| TP47 | h− ade6-M216 med8+::TAP-kanMX6 | MP12/MP13a | This study |
| TP126 | h− Δmed18::kanMX4 | MP9 | This study |
| TP130 | h− Δmed15::kanMX4 | MP9 | This study |
| TP192 | h− cdk8-D158A (dead-kinase mutant) ura4-D18 leu1-32 | (11) | |
| TP207 | h+ cdk8-D158A (dead-kinase mutant) | TP192/MP13a | This study |
| TP216 | h+ ade6-M216 ura4-D18 med7+::TAP-kanMX6 | TP42/MP2b | This study |
| TP234 | h− Δmed20::kanMX4 | MP9 | This study |
| TP235 | h+ Δmed20::kanMX4 ade6-M210 | MP12/MP13a | This study |
| TP274 | h− cdk8-D158A (dead-kinase mutant) | TP207/MP9b | This study |
| TP306 | h+ ade6-M216 ura4-D18 med7+::TAP-kanMX6 Δmed27::ura4+ | TP219/TP220a | This study |
| TP308 | h90 sep15-598 his3 med7+::TAP-kanMX6 | 2-506 | This study |
| TP315 | h+ ade6-M210 his7-336 leu1-32 ura4-D18 med7+::TAP-kanMX med17Δ50 | TP392 | This study |
| TP316 | h− Δmed31::kanMX4 | MP9 | This study |
| TP384 | h+ ade6-M210 his7-336 leu1-32 ura4-D18 med17+::LEU2 | MP1 | This study |
| TP390 | h+ ade6-M210 his7-336 leu1-32 ura4-D18 med17Δ50::ura4+ | TP384/MP2a | This study |
| TP392 | h+ ade6-M210 his7-336 leu1-32 ura4-D18 med17Δ50 | TP390 | This study |
| TP396 | h− Δmed18::hph(hph: Hygromycin B Phosphotransferase | TP26 | This study |
| TP405 | h Δmed18::hph Δmed20::kanMX4 | TP396/TP235b | This study |
| TP416 | h+ ade6-M216 ura4-D18 med7+::TAP-kanMX6 Δmed18::ura4+ | TP216 | This study |
| TP417 | h+ ade6-M216 ura4-D18 med7+::TAP-kanMX6 Δmed20::ura4+ | TP216 | This study |
| ND064 | h− Δace2::kanMX4 | (68) | |
| ND102 | h− Δsep1::kanMX4 | (68) | |
| S. cerevisiae | |||
| BY4741 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF | |
| Y01742 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δmed15::kanMX | BY4741 | EUROSCARF |
| Y04393 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δmed3::kanMX | BY4741 | EUROSCARF |
| Y04494 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δmed31::kanMX | BY4741 | EUROSCARF |
| Y04734 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δmed18::kanMX | BY4741 | EUROSCARF |
| Y05351 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcycc::kanMX | BY4741 | EUROSCARF |
| Y05799 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δmed12::kanMX | BY4741 | EUROSCARF |
| Y06611 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ Δmed20::kanMX | BY4741 | EUROSCARF |
| Y13701 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ Δmed2::kanMX | BY4741 | EUROSCARF |
| TAP-Med8 | MAT a pep4::HIS3/prb1::LEU2 prc1::HISG can1 ade2 trp1 ura3 his3 leu2–3, 112 MED8::TAP-klTRP1 | CB010 | (12) |
| CGC129 | MAT a pep4::HIS3/prb1::LEU2 prc1::HISG can1 ade2 trp1 ura3 his3 leu2–3, 112 MED8::TAP-klTRP1 Δmed18::kanMX6 | TAP-Med8 | This study |
| CGC130 | MAT a pep4::HIS3/prb1::LEU2 prc1::HISG can1 ade2 trp1 ura3 his3 leu2–3, 112 MED8::TAP-klTRP1 Δmed20::kanMX6 | TAP-Med8 | This study |
aSpore obtained after transformation of the diploid and subsequent sporulation of the diploid.
bSpore obtained after mating and sporulation of the diploid.
Isolation of med17ts mutants
A marker switch approach (27) was used for the construction of med17ts mutants. The LEU2 gene was inserted in a PacI site at 159 bp 3′ of med17+ ORF in strain MP1 to generate TP384. The med17+ gene from position 429 bp 5′ to 212 bp 3′ of the med17+ ORF was cloned in pBluescript SK +/− and the 1.8-kb HindIII ura4+ fragment of pREP42X (28,29) was inserted into the PacI site at 159 bp 3′ of the med17+ ORF to generate plasmid pTK1276. Mutagenesis of pTK1276 plasmid DNA was carried out with hydroxylamine (29) as follows: 10 μg of plasmid DNA was added to 500 μl ice-cold hydroxylamine solution (1 M H2NO·HCL, adjusted to pH 7 with NaOH) and incubated at 75°C for 90 min. Reactions were stopped by adding 200 mM NaCl, DNA was collected by EtOH precipitation and transformed in Escherichia coli strain DH5α. The med17::ura4+ fragment was exerted from a pool of mutagenized pTK1276 DNA with ApaLI and PacI and transformed into strain TP384. Ura+ transformants were selected on AA-Ura plates and replica-plated onto YES plates. After 3 days of incubation at 37°C, candidate colonies of temperature-sensitive (ts) mutants were isolated and tested for loss of the LEU2 gene on AA-Leu plates. Screening of 10 000 yeast transformants for a ts phenotype at 37°C yielded four independent mutant strains with a ts phenotype at 37°C on YES plates.
Construction of the med17Δ50 allele
One hundred and fifty nucleotides from position 1601–1751 of the med17+ ORF was deleted from plasmid pTK1276 introducing a NheI site 3′ of the stop codon in the process to generate plasmid pTK1439. The med17Δ50::ura4+ sequence was exerted from pTK1439 with ApaLI and PacI and transformed into diploid strain TP384/MP2. Ura+ transformants were selected on AA-Ura plates and sporulated on MSA plates. After dissection Ura+/Leu− spores were tested for a ts phenotype at 37°C and an ura4+ leu2− med17Δ50 spore called TP390 isolated. The ura4+ gene was removed from the med17Δ50 strain TP390 by transforming with an 812-bp fragment spanning the ura4+ gene but not running into the med17+ ORF followed by selection on 5-FOA plates giving strain TP392.
TAP-tagging of med7+ in strain TP390 (med17Δ50) and strain MP10 (sep15-598/med8ts)
The C-terminal part of the med7+ gene fused to a TAP-tag was exerted from plasmid pFA6a-kanMX6-CTAP2-spmed7 (10) with AvrII and EcoRV and transformed into the med17Δ50 strain TP392 and sep15-598/med8ts strain MP10 giving strains TP315 and TP308, respectively. Correct integration of constructs in the genome was verified by PCR analysis. The MP10 strain was a kind gift from M. Sipiczki (University of Debrecen, Hungary). In all cases, tetrad analysis and/or Southern blot analysis were/was applied to demonstrate that only one copy of the construct had integrated in the genome.
Saccharomyces cerevisiae strains
All S. cerevisiae deletion strains were purchased from Euroscarf. Deletion cassettes of the MED20 and MED18 genes were generated by amplifying the kanMX cassette from Euroscarf deletion strains BY0661 (Δmed20) and BY04734 (Δmed18) with 500-bp flanking sequence on either side of the kanMX marker. Saccharomyces cerevisiae TAP-MED8 cells were transformed with the amplified deletion cassettes using the lithium acetate procedure. Correct genomic integration was assayed by PCR using primers positioned at the very 5′ end of either the upstream or coding region and conversely outside the 3′ flanking regions of the transformation constructs. The TAP-MED8 strain (12) was a kind gift from Roger Kornberg (Stanford University, CA, USA).
Protein purification
Cells were grown at 30°C unless otherwise specified. Saccharomyces cerevisiae cells were grown in YPD medium (10 g/l yeast extract, 20 g/l peptone, 20 g/l glucose) to OD600 3.5–4.0, and S. pombe cells were cultured to OD600 2.0–2.5 in yeast extract supplement (YES) medium (5 g/l yeast extract, 2 g/l casamino acids, 20 g/l glucose) supplemented with 0.2 g/l adenine. For preparation of whole-cell extract, we collected yeast cells from a 10 l culture by centrifugation (Beckman Instruments JLA-10 500 rotor, 9500 r.p.m., 10 min, 4°C), which were washed once with ice-cold water and suspended in 0.5 ml of 3 × lysis buffer (200 mM HEPES-KOH, pH 7.8, 15 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 15% glycerol, 0.5 mM dithiothreitol and protease inhibitors) per gram of cell pellet. For all purifications, we used a 100 × stock of protease inhibitors containing 100 mM phenylmethylsulfonyl fluoride, 200 mM pepstatin, 60 mM leupeptin and 200 mM benzamidine in 95% ethanol. Cells were lyzed by bead beating (Bead-Beater, Stratech, London, UK) for 25 cycles, with each cycle consisting of 30 s of beating and 90 s of rest. We cleared the supernatant by centrifugation (Beckman Instruments JLA-10 500 rotor at 9500 r.p.m., for 10 min, at 4°C), added 1/9 vol of 2 M KCl and stirred for 15 min. Polyethyleneimine was added to a final concentration of 0.02% followed by 15 min of stirring. Insoluble material was removed by ultracentrifugation (Beckman Instruments Ti45 rotor, 42 000 r.p.m., 30 min, 4°C). Mediator was purified according to the original TAP-tag purification protocol (30,31) with the following modifications. Nine hundred microliters of IgG–Sepharose (GE Healthcare) was added to 200 ml of extract and incubated for 2 h at 4°C. The IgG–Sepharose was collected by centrifugation (Beckman Instruments JA-17 rotor, 800 r.p.m., 1 min, 4°C), loaded onto a Poly-Prep column (Bio-Rad) and washed with 100 ml of IgG wash buffer (20 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 0.1% IGEPAL CA-630). Mediator was eluted in two steps: first, by incubation for 2 h at 16°C with 200 U of tobacco etch virus (TEV) NIa protease (Invitrogen) in 2 ml of TEV buffer (10 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% IGEPAL CA-630, 1 mM dithiothreitol), designated IgG eluate 1, and second, by elution of the IgG–Sepharose by incubation with 8 M urea, 100 mM NaH2PO4, 10 mM Tris–HCl, pH 7.7, for 5 min at room temperature, designated IgG eluate 2. For further purification of S. pombe Mediator, 500 µl of IgG eluate 1 was loaded onto a 1-ml heparin HiTrap column (GE Healthcare). The column was washed with 5 ml of buffer A-0.1 (20 mM HEPES-NaOH, pH 7.5, 10% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol, protease inhibitors and the molarity of sodium chloride indicated after the hyphen) and then developed with a linear gradient (12 ml) of A-0.1 to A-1.2.
Immunoblotting and production of antibodies
The full-length S. pombe med20+ coding region (GenBank accession NM_001019159) was inserted into the expression plasmid pGEX3X (GE Healthcare). The GST-Med20 fusion-protein was overexpressed in E. coli BL21 (DE3) pLysS cells (Stratagene) and subsequently purified using glutathione–Sepharose 4B (GE Healthcare) according to the recommendations of the manufacturer. The purified GST-Med20 protein was used to immunize rabbits. The antisera used in this study were taken 10 days after the second booster injection (AgriSera, Vännäs, Sweden). Antibodies specific for S. pombe Mediator subunits Med17, Med18, Med27 and Med31 have been described previously (6,9,32). Antibodies specific for S. pombe CycC will be described in a parallel publication (Baraznenok,V. and Gustafsson,C.M., submitted for publication). Schizosaccharomyces pombe TAP-Med7 was detected with the soluble peroxidase anti-peroxidase complex (Sigma). Antibodies specific for S. cerevisiae Mediator subunits Med1, Med2, Med8 and Med11 were kind gifts from Stefan Björklund (Umeå University, Sweden). Antibodies against S. cerevisiae Med18 and Med20 were kind gifts from R.A. Young (Whitehead Institute for Biomedical Research, Cambridge, MA, USA).
Affymetrix GeneChip probe array hybridization
Schizosaccharomyces pombe strains were grown in EMM medium at 30°C until 5 × 106 to 1 × 107 cells/ml. Heat-stressed wild-type and med17Δ50 cells were cultured in EMM at 30°C until 5 × 106 to 1 × 107 cells/ml, spun down briefly and then resuspended in media pre-warmed to 37°C. Cells were harvested after 2 h of incubation at 37°C. Total yeast RNA was isolated using a hot acid phenol extraction protocol (33). Labeling and array hybridization to Affymetrix Yeast Genome 2.0 arrays were performed at the Karolinska Institute Affymetrix core facility (Huddinge, Sweden). Labeling and hybridization protocols are described in the Affymetrix users’ manual (34). Washing and staining of arrays were performed using the GeneChip Fluidics Station 450. Arrays were scanned with the Affymetrix GeneArray scanner 3000 7G. Acquisition and quantification of array images as well as primary data analysis including normalization was performed using Affymetrix GeneChip Operating Software (GCOS). Data were processed with the MAS5 algorithm (scaling: all probe sets with target signal 100, normalization: user defined with normalization value 1). The expression profiling was done on two sets of samples. The first set contained transcript level data from three wild-type (MP9) cultures and three cultures of each of the cdk8mut, Δmed12, Δmed15, Δmed18 and Δmed20 strains. The second set contained transcript level data from two med17Δ50 mutant cultures and two corresponding wild-type (MP1) cultures. To rule out the possibility of global changes to the transcriptome in the Mediator mutants, all datasets were normalized to an internal housekeeping control mRNA. Through quantitative real-time PCR (see below) we established that the atp1+ gene displayed <1.5-fold change in all mutants when normalized to 28 S rRNA levels and was therefore chosen as the internal mRNA control. Within each set of samples the transcript signal intensities was compared to all the corresponding wild-type baseline controls, generating a total of nine comparisons for each mutant in the first series. The Affymetrix change-call algorithm designated every transcript in each individual comparison as increased, decreased or invariant (P-value threshold 0.0025) with reference to the wild-type baseline intensity. An average ratio change was calculated for each transcript in each strain by first adjusting the ratio change to one of all comparisons designated invariant and then calculating the geometric mean for the total number of comparisons. The normalized and filtered dataset is available as Supplementary Data in the online version of this article.
Clustering and statistical analysis of expression data
Hierarchical clustering was carried out with the TIGR Multiexperiment Viewer version 3.0.3 (35). Ortholog tables for S. pombe and S. cerevisiae were kindly provided by Valerie Wood at the Wellcome Trust Sanger Institute S. pombe Genome Project. Genomic coordinates for S. pombe genes were retrieved from the S. pombe Genome Database. Expression data for the S. pombe med8ts/sep15-598 and med31ts/sep10-412 mutants (36) (ArrayExpress accession E-MEXP-193) as well as the Δace2 and Δsep1 mutants (37) (ArrayExpress accession E-MEXP-62) were downloaded from the Bähler lab homepage (http://www.sanger.ac.uk/PostGenomics/S_pombe/projects/). Missing values were set to ratio 1. The list of S. pombe core environmental stress response (CESR) genes is from ref. 38.
Quantitative real-time PCR
Total RNA was isolated as described above. RNA quality and concentration was determined using a BioAnalyser 2100 (Agilent). cDNA was synthesized with the AMV 1st Strand Synthesis Kit (Roche) using random hexamers. Relative quantification of cDNA was carried out in triplicate on a LightCycler 2.0 (Roche) using SYBR green technology and the FastStart DNA Master SYBR Green 1 mix (Roche). Standard curves were generated by at least four 5-fold serial dilutions of a single control sample and values within the linear exponential phase were used to calculate relative concentrations after normalization. Primers used for quantitative PCR are listed in Supplementary Table S2.
Northern blotting
Northern blotting was done as previously described (39) using 2 μg of heat-denatured total RNA (65°C for 3 min) in each lane. The 32P-labeled poly(dT) probe was obtained by reverse transcription: a 10 μl reaction containing 50 ng poly(rA), 100 pmol oligo(dT)18 primer, 1 μl 10 × M-MuLV reverse transcriptase reaction buffer [50 mM Tris–HCl (pH 8,3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT], 100 pmol dTTP, 10 μCi [α-32P] dTTP (400 Ci/mmol), 100 U Moloney murine leukemia virus (M-MuLV) reverse transcriptase (NEBiolabs) was carried out at 37°C for 1 h. The poly(rA) template was removed by RNase H treatment where after RNase H was heat inactivated.
RESULTS
Biochemical characterization of the S. pombe Mediator head domain
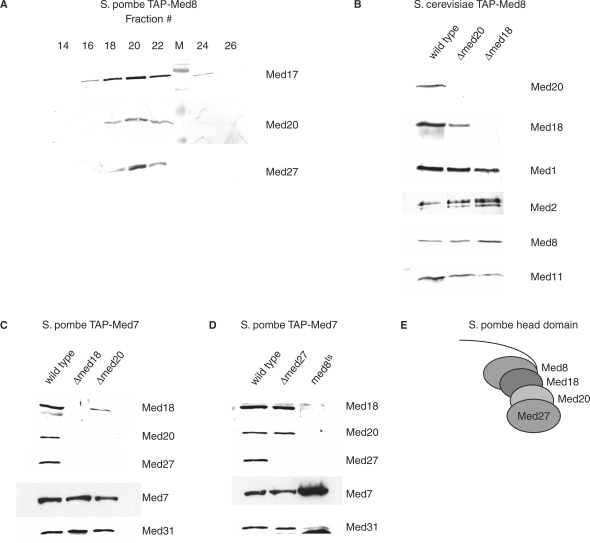
Mass spectrometric analysis of partially purified S. pombe Mediator identified the hypothetical protein SPAC17G8.05 (GenBank accession NP_593728) as a potential Mediator subunit with an apparent mass of around 20 kDa (data not shown). SPAC17G8.05 has previously been predicted to be the S. pombe homolog of S. cerevisiae and mammalian Med20 (7). To demonstrate its stable Mediator association, S. pombe Mediator was purified with TAP-tag on the Med8 subunit and resolved by Heparin–Sepharose chromatography. Western analysis showed co-elution of Med20 with Med17 and Med27 (Figure 1A). Thus, Med20 is a stable subunit of S. pombe Mediator.
Figure 1.
Structural organization of the S. pombe Mediator head domain. (A) Schizosaccharomyces pombe TAP-Med8 Mediator was eluted from IgG–Sepharose by TEV cleavage and further resolved over Heparin–Sepharose as described in ‘Material and methods’ section. Western analysis of different fractions. (B) Western analysis of S. cerevisiae Mediator purified through a TAP-tag on the Med8 subunit from wild-type, Δmed20 and Δmed18 cells. (C) Western analysis of S. pombe Mediator purified through a TAP-tag on the Med7 subunit from wild-type, Δmed18 and Δmed20 cells. (D) Western analysis of S. pombe Mediator purified through a TAP-tag on the Med7 subunit from wild-type, Δmed27 and med8ts/sep15-598 cells. (E) Proposed subunit organization of the S. pombe head domain based on the results in (C) and (D).
In S. cerevisiae, Med20 and Med18 form a distinct submodule within the head domain (24,25,40). Med20 is completely lost from S. cerevisiae Mediator isolated from a Δmed18 strain (Figure 1B, right lane). However, we could still detect reduced amounts of Med18 associated with Mediator isolated from a Δmed20 strain (Figure 1B, middle lane).
To establish whether the S. pombe head domain is organized as that of S. cerevisiae diploid strains heterozygous for either Δmed18 or Δmed20 were made. After tetrad analysis, viability segregated 4:0 showing that both med18+ and med20+ are nonessential genes in S. pombe like their S. cerevisiae homologs. Both Δmed18 and Δmed20 S. pombe cells have a slow growth phenotype and fail to sporulate (data not shown) (41). We purified Mediator from strains lacking either med18+ or med20+, respectively, utilizing a TAP-tag on Med7 (Figure 1C). In agreement with our observations in budding yeast, Δmed20 S. pombe Mediator contained reduced amounts of the Med18 subunit, whereas mutant Δmed18 Mediator completely lacked Med20 after purification over IgG–Sepharose. In addition, both the Δmed18 and Δmed20 mutant Mediator lacked the Med27 subunit. No homolog of Med27 has been identified in the S. cerevisiae Mediator. Cells lacking med27+ (systematic name SPAC17C9.05c) are viable without any apparent phenotype at 30°C (data not shown) (6). Mediator purified from Δmed27 TAP-med7+ cells still contained both Med18 and Med20 (Figure 1D, middle lane) consistent with the wild-type growth phenotype at 30°C of Δmed27 cells. This suggests that Med27 is located proximally to the Med18/Med20 dimer on the periphery of the head domain.
A ts allele of the essential head domain subunit Med8 called sep15-598 (or med8ts for clarity) has been isolated (42–44). Schizosaccharomyces pombe cells carrying the med8ts allele display a similar growth phenotype as Δmed18 cells suggesting that this allele could affect subunit interactions within the head domain (42). Western blot analysis revealed that Mediator purified from a med8ts TAP-med7+ strain had lost Med18, Med20 and Med27 (Figure 1D, right lane). Furthermore, Med8 interacts with Med20 in a yeast two-hybrid interaction analysis (data not shown). Thus, our observations suggest a step-wise structural organization of the nonessential S. pombe head domain subunits where Med27 is found on the very exterior connected to Med20, which in turn contacts Med18 that binds Med8 (Figure 1E). This model is in agreement with recent structural data of a Med8-Med18-Med20 trimer in S. cerevisiae (40).
Isolation and characterization of S. pombe med17 ts alleles
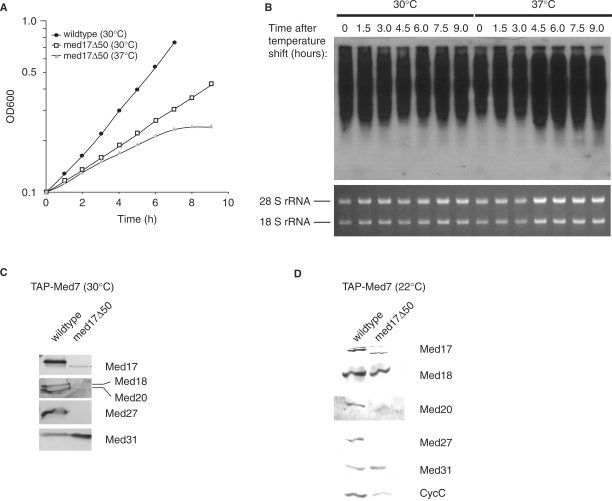
The S. cerevisiae Med17 subunit is located in the head domain and essential for cell viability (20,24,45) reflecting its importance as a scaffold for the remaining head domain subunits (22,25). The ts allele of S. cerevisiae MED17 termed srb4-138 was instrumental in demonstrating the universal requirement for Mediator in pol II-dependent transcription in vivo (45,46). The med17+ gene is also essential in S. pombe (6). We constructed a conditional med17 allele in S. pombe to understand Mediator function in fission yeast. We employed a marker switch approach (27) to establish a ts allele by targeted hydroxylamine mutagenesis at the med17+ locus and isolated four independent mutant strains with a ts phenotype at 37°C on YES plates. All four med17 mutants had acquired base changes in the C-terminal part of the med17+ coding sequence resulting in premature stop codons within the last 50 C-terminal amino acids of the Med17 protein. We considered the possibility that low levels of read-through of the premature stop codon could still produce full-length Med17 protein and that a de facto C-terminal truncation would be lethal. Thus, we constructed a strain lacking the last 50 C-terminal amino acids and termed this med17 allele med17Δ50 allele, which has a ts phenotype indistinguishable from the four previously isolated med17ts alleles.
Saccharomyces cerevisiae cells with the srb4-138 allele immediately stop growing after a shift to 37°C (45). Schizosaccharomyces pombe med17Δ50 cells displayed a slightly more gradual response when heat stressed. After shifting a mid log culture of S. pombe med17Δ50 cells growing at 30°C to 37°C an initial decrease of growth rate after 2.5 h was observed with growth ceasing altogether after 7.5 h (Figure 2A). The same pattern was obtained with the isolated med17ts alleles (data not shown). As the S. pombe med17Δ50 allele appeared subtly different from the S. cerevisiae srb4-138 allele, we isolated total RNA from med17Δ50 cells and wild-type controls cultured at 30°C as well as 2 h after a shift to 37°C. In S. cerevisiae srb4-138 cells, transcription ceases abruptly with most mRNA transcripts gone after 1 h (45). In contrast, northern blot analysis of heat-shocked S. pombe med17Δ50 cells show no detectable decrease in mRNA levels even after 9 h at 37°C (Figure 2B).
Figure 2.
Isolation and characterization of temperature-sensitive alleles of the S. pombe med17+ gene. (A) med17Δ50 cells and wild type (MP1) were grown to OD600 = 0.1 (106 cells/ml) in YES medium at 30°C. Cultures were then divided in two, spun down and resuspended in medium preheated to either 30°C or 37°C. Cell density was continually measured until growth of the med17Δ50 culture at 37°C had ceased. Due to septation defects in the med17Δ50 cells, samples were sonicated briefly prior to absorbance measurement. (B) Northern blot analysis of global mRNA levels in med17Δ50 cells following resuspension in medium preheated at either 30°C or 37°C. The upper panel shows a membrane probed with radioactively labeled poly-T as described in ‘Material and Methods’ section. Lower panel indicates rRNA loading control as observed in the ethidium bromide-stained agarose gel. (C) Western analysis of Tap-purified S. pombe Mediator from med17Δ50 TAP-med7+ cells grown at 30°C. (D) The same purification as in (C) but cells were cultured at 22°C.
Mediator was purified from med17Δ50 TAP-med7+ strain grown at 30°C in order to characterize its subunit composition. Western blotting of the TEV eluate revealed that the head module proteins Med17 and Med27 were completely lost while the amounts of Med18 and Med20 were severely reduced (Figure 2C). This drastic destabilization of S. pombe med17Δ50 Mediator was reminiscent of S. cerevisiae srb4-138 Mediator, which fractures at the head/middle domain boundary irrespective of growth temperature (19). We repeated the purification, culturing the med17Δ50 cells at 22°C and in this case Med17 was present in the med17Δ50 Mediator (Figure 2D). The head module subunit Med18 was still present whereas both Med20 and Med27 were lost from the mutant complex. This suggested that Med17 could be interacting with Med20 and Med27 independently of Med18. In support of this notion, yeast two-hybrid interaction analysis revealed that Med17 has physical contacts with Med20 and Med27 (data not shown). We have no two-hybrid data regarding interactions between Med17 and Med18. Direct interactions between Med17 and Med20 have also been reported previously in S. cerevisiae Mediator (22,24). Thus, we suggest the structural organization of the S. pombe head domain subunits depicted in Figure 1E with Med27 situated on the exterior connected to Med20, which contacts Med18 that ultimately binds Med8. Med17 could serve as a scaffold interacting with Med8, Med20 and Med27.
Phenotypical classification of head and Cdk8 module Mediator mutant alleles
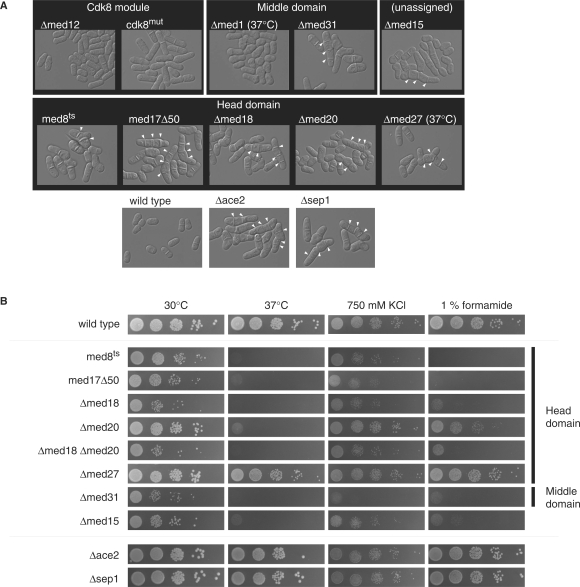
To discern functional subdivisions among Mediator subunits, we investigated the phenotypes of null alleles of med1+, med12+, med18+, med20+, med27+, med31+ as well as the SPBC146.01 gene encoding the proposed MED15 homolog. Deletion of med15+ in a diploid followed by tetrad analysis showed that med15+ is nonessential in S. pombe. We included the dead-kinase mutant allele of cdk8+ called ckd8mut (11) as well as med8ts (42) and med17Δ50. All mutants displayed one of two specific cellular morphologies—flocculation or hyphal growth (Figure 3A). Flocculation signifies the aggregation of yeast cells into larger clumps (47) and has previously been observed in Δcdk8, Δmed12 and Δmed13 cells (10,48). Hyphal growth occurs when cells are unable to cleave the primary septum that separates two daughter cells following medial septation at the end of mitosis (49). This phenotype had previously been observed in ts alleles of med8+, med18+ and med31+ (41,44). The cellular morphology of the head domain mutants med17Δ50, Δmed18 and Δmed20 corresponded to the hyphal growth seen for the med8+ ts allele sep15-598. The Δmed27 mutant did not have a visible phenotype at 30°C, but showed some septation defects at 37°C (Figure 3A). A complete deletion of middle domain subunit med31+ also displayed a hyphal growth phenotype in agreement with previous report (41). Interestingly, the Δmed15 mutant also showed this phenotype. We included the two null mutants of G2/M transcription factors Ace2 and Sep1 for comparison, both of which display a severe septation defect (50–52). The Cdk8 module mutants flocculated, but showed no septation defects. Mutants lacking the med1+ gene did not have a visible phenotype at 30°C, but flocculated at 37°C. In summary, we observed a clear distinction in cellular morphology between Cdk8 module mutants (and the associated middle domain subunit Med1) and those of the head domain as well as of middle domain subunit Med31 and the potential Mediator subunit Med15.
Figure 3.
Phenotypes of S. pombe Mediator mutants. (A) Light microscopy of wild-type and mutant cells cultured in YES medium grown at 30°C unless otherwise indicated. Mediator mutants are organized according to their location within the Mediator. Unresolved septa are indicated by white arrows. The two well-characterized hyphal growth mutants Δace2 and Δsep1 are included for comparison. (B) Ten-fold serial dilutions of wild-type and mutant cells under different forms of stress.
We observed varying degrees of severity in the hyphal growth phenotype in the head domain mutants, Δmed15 and Δmed31. A spot assay was carried out to further discern the magnitude of growth inhibition in the septating mutants (Figure 3B). We included Δace2 and Δsep1 for comparison. Cells were challenged with elevated temperatures (37°C), high salt (750 mM KCl) and 1% formamide, respectively. The Δmed27 mutant appeared nearly indistinguishable from wild-type cells at all conditions tested. The Δmed20 mutant was slightly more sensitive than wild type at high salt, but was very sensitive to heat and formamide. The remaining head domain mutants med8ts, med17Δ50, Δmed18 as well as a Δmed18 Δmed20 double mutant grow very poorly at all conditions tested. Thus, in these assays Δmed18 is epistatic to Δmed20. Equal degree of sensitivity was observed for the Δmed31 and Δmed15 mutants. The two mutants Δace2 and Δsep1 were indistinguishable from wild type indicating that it was not the hyphal growth defect in itself that caused slow growth and stress sensitivity in the Mediator mutants. The Cdk8 module mutants were indistinguishable from wild type under all conditions tested (data not shown).
Expression profiling of Mediator mutants
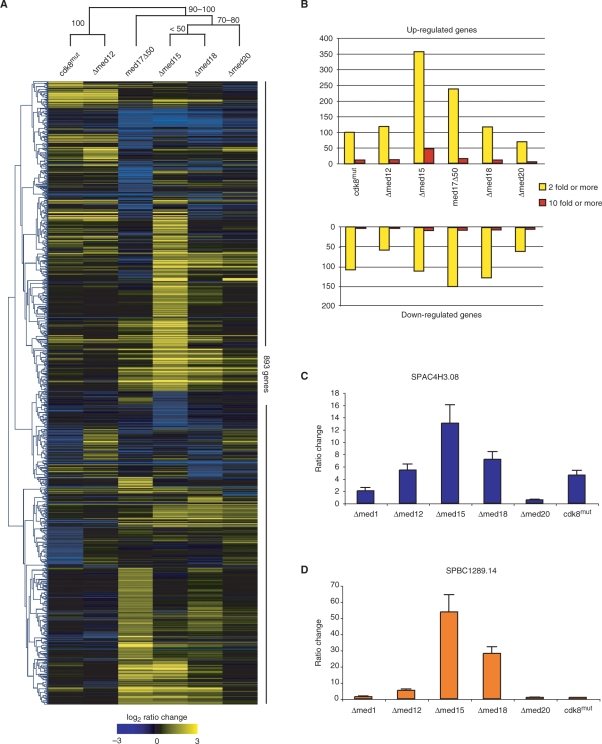
We asked whether the distinct morphological phenotypes of Mediator mutants reflected distinct effects on global gene transcription. Hence, we performed expression profiling on six mutant alleles that we considered a representative cross section of the S. pombe Mediator. Global expression profiles of mutant alleles in conjunction with hierarchal clustering analysis can be used in an analogous way to phenotypes as a way to discern functional relationships (53).
We made two sets of comparisons. The first set of comparisons consisted of three independent cultures of each of the mutant strains cdk8mut (TP274), Δmed12 (TP27), Δmed15 (TP130), Δmed18 (TP126) and Δmed20 (TP234) as well as a wild-type (MP9) control. The second set of comparisons consisted of two independent med17Δ50 cultures (TP392) with two wild-type (MP1) controls. We employed the Affymetrix platform and the number of genes changing in each mutant by a defined ratio threshold is shown in Figure 4B.
Figure 4.
Genome-wide expression analysis of nonlethal S. pombe Mediator alleles. (A) Heatmap cluster diagram of the 893 transcripts that change 2-fold or more in at least one of the mutants. Experiments and genes were hierarchically clustered according to average Pearson correlation linkage. The robustness of the experimental dataset clusters were analyzed by bootstrap analysis in MeV using 10 000 replicates. Numbers on branches indicate the range of bootstrap values expressed as percentages. (B) Bar diagram showing the number of genes changing 2-fold or more (yellow bars) and 10-fold or more (red bars) in each mutant strain. (C) Quantitative real-time PCR analysis of SPAC4H3.08 transcript levels of the mutant strains indicated as compared to wild-type cells. SPAC4H3.08 levels were normalized to 28 S rRNA. Error bars indicate 1 SD. (D) Quantitative real-time PCR analysis of SPBC1289.14 levels as described in (C).
We performed hierarchal clustering (54) on all genes changing 2-fold or more in the mutant datasets. The robustness of the clustering was tested by bootstrap analysis of 10 000 replicates with re-sampling of genes. The Cdk8 module mutants were clearly separated from the head domain subunits Med17, Med18 and Med20 (Figure 4A) as illustrated by the 100% bootstrap support of the Cdk8 module clade. The Δmed15 mutant clustered within the head domain clade (Figure 4A), which is in agreement with previous results from S. cerevisiae Mediator (53). The genes that did change 2-fold or more belonged to diverse set of functional groups with no particular group of genes dominating any one expression profile—a reflection of the central role of Mediator in all pol II-dependent transcription. In addition, despite the significant correlation between expression profiles shown Figure 4A, each profile contained a set of genes particular to each mutant that showed a 2-fold or more change in transcript levels—in total 531 genes.
Mediator mutations elicit a partial stress response
Expression profiling of S. cerevisiae Mediator mutants have reported the increase of stress response gene transcript levels (46,53,55). We investigated whether this is also the case for S. pombe Mediator mutants. The central enviromental stress response (CESR) in S. pombe consists of a set of genes whose transcript levels consistently increase or decrease during various forms of stress (38). The Affymetrix array platform used in our study contained 134 of the induced CESR genes. We studied the occurrence of CESR genes above a defined fold change threshold and then calculated P-values using the hypergeometric probability distribution. We applied a 2-fold ratio change threshold for the cdk8mut, Δmed12, Δmed18 and Δmed20 datasets, while the large number of genes changing in the Δmed15 and med17Δ50 datasets required us to apply a 3-fold threshold in order to allow computation. The induced set of CESR genes were significantly overrepresented in the genes increasing their transcript levels above the threshold in all six datasets (Table 2, P < 0.001). In some mutants we also observed a significant overlap between downregulated genes and the genes belonging to the repressed set of CESR genes (data not shown). We confirmed the elevated transcription levels of the two induced CESR genes SPAC4H3.08 and SPBC1289.14 by quantitative real-time PCR analysis (Figure 4C and D). These genes are known to become induced 10-fold or more following heat shock and hydrogen peroxide treatment, respectively (38). Although all strains tested showed at least a 2-fold increase in transcript levels, a substantially higher degree of upregulation of these two genes were observed in the Δmed15 mutant closely followed by the Δmed18 mutant. These observations taken together suggest a central role for Mediator in the regulation of some CESR genes. The actual molecular mechanism of this regulation remains to be elucidated.
Table 2.
Occurrence of CESR transcripts in Mediator mutant expression profiles
| Mutant | Fold-change cut-off | Increased transcripts | Induced CESR transcripts in the increased set |
|---|---|---|---|
| cdk8mut | 2 | 100 | 14 (p = 2.7 × 10−7) |
| Δmed12 | 2 | 119 | 12 (p = 5.5 × 10−5) |
| Δmed15 | 3 | 153 | 41 (p = 1.6 × 10−31) |
| med17Δ50 | 3 | 68 | 8 (p = 3.5 × 10−4) |
| Δmed18 | 2 | 125 | 41 (p = 1.5 × 10−35) |
| Δmed20 | 2 | 68 | 24 (p = 1.4 × 10−21) |
Effects on genes involved in cell wall organization and metabolism
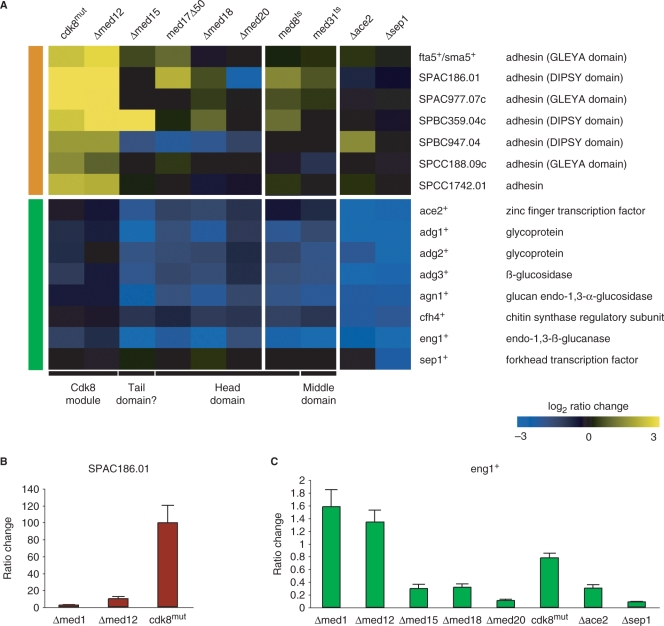
We looked specifically at genes annotated in the S. pombe genome database as being involved in cell wall metabolism to explain the clear morphological differences between mutants of the Cdk8 module and those of the core Mediator. Specifically, we investigated genes known or proposed to be involved in the process of cell-to-cell adhesion as well as postmitotic septum dissolution. We could discern two major groups of genes with distinct expression patterns. The first group consisted of a family of genes that code for putative cell surface adhesins (56). These adhesins displayed increased transcript levels in the cdk8mut and Δmed12 strains, but were largely unchanged in the head domain mutants (Figure 5A, orange bar). The increased transcript levels of these genes agree with the flocculation phenotype observed in S. pombe Cdk8 module mutants (Figure 3A) (10,48). The majority of the adhesin genes displayed only moderate increases in transcript levels (between 2- and 5-fold). One adhesin however, the DIPSY domain-containing SPAC186.01 gene, was upregulated nearly a 100-fold in the cdk8mut strain as assayed by quantitative real-time PCR (Figure 5B). This corresponded well with the stronger flocculation phenotype in this strain as compared to the somewhat milder phenotype observed for the Δmed12 mutant (data not shown). Some of the other members of the S. pombe adhesin family that displayed more moderate increases in transcript levels are related to the FLO1, FLO5 and FLO9 genes of S. cerevisiae (56). The FLO genes are known to be upregulated in S. cerevisiae Cdk8 module mutants as well (46,53,55) and we conclude that the role of the Cdk8 module as a repressor of Flo-like adhesins appears conserved between S. pombe and S. cerevisiae.
Figure 5.
Mediator mutations affect genes involved in cell wall organization and metabolism. (A) Expression profiles of genes involved in cell wall metabolism, cell-to-cell adhesion and degradation of the post-mitotic primary septum. The first group (orange bar) includes cell surface proteins proposed to be involved in cell-to-cell adhesion during stress. The second group (green bar) includes genes previously known to cause hyphal growth when deleted. The corresponding datasets from previously described studies of other Mediator mutants (16) as well as null alleles of ace2+ and sep1+ (37) are included for comparison. (B) Quantitative real-time PCR analysis of SPAC186.01 transcript levels of the indicated mutant strains as compared to wild-type cells. SPAC186.01 levels were normalized to 28 S rRNA. Error bars indicate 1 SD. (C) Quantitative real-time PCR analysis of eng1+ levels as described in (B).
The second group (Figure 5A, green bar) consisted of genes whose transcript levels were significantly decreased in the Δmed15 and head domain mutants, but with no significant change in the Cdk8 module mutants. This group included genes regulated by the transcription factor Ace2 as well as ace2+ itself (37,51,57). These genes are expressed late in the cell cycle and involved the dissolution of the primary septum separating the newly formed daughter cells (51,57). Cells lacking these genes display septation defects. The Sep1 forkhead transcription factor regulates ace2+ expression (37,57). Sep1 precedes Ace2 in the regulatory pathway and the sep1+ gene was not affected by any of the mutants tested, suggesting that Sep1 requires the head domain subunits for ace2+ transcription.
We also compared our profiles with published datasets of ts alleles of med8+ and med31+ (36). In agreement with their hyphal phenotypes only the Ace2-regulated genes were affected in these two mutants. Likewise, Δace2 and Δsep1 expression data (37) did not exhibit any significant changes in expression of surface adhesins indicating that the Cdk8 module does not play a role in the expression of Ace2-dependent genes. The ace2+-regulated eng1+ gene encodes an endoglucanase that is required for the dissolution of the primary division septum following mitosis, and its deletion leads to a septation phenotype similar to that of Δace2 and Δsep1 cells (51). We performed quantitative real-time PCR analysis of eng1+ transcript levels in the Mediator mutants and the Δace2 and Δsep1 strains to directly compare the magnitude of decrease in this particular gene. We observed that the decrease in the head domain mutants and Δmed15 was equivalent to that of the Δace2 and Δsep1 mutants (Figure 5C). We therefore conclude that the S. pombe Mediator head domain and Med15 subunit play a crucial role in the transcription of ace2+/sep1+-dependent genes. We would like to point out that this analysis was done on asynchronous cells and therefore does not reveal any potential effect on the timing of cell cycle-dependent expression patterns.
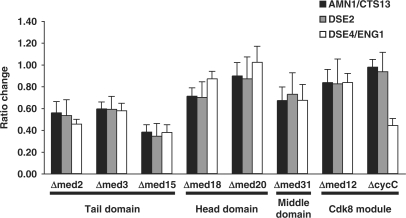
Degradation of the primary septum in S. cerevisiae is controlled in part by the ACE2 gene, the homolog of S. pombe ace2+ (58,59). However, primary septum dissolution and cell separation is fundamentally different and most of the required factors lack S. pombe homologs (60,61). Nevertheless, quantitative real-time PCR analysis of three genes wholly or partially regulated by S. cerevisiae ACE2-AMN1/CTS13, DSE2 and DSE4/ENG1 (58,59) all displayed decreased transcript levels in head and tail domain mutants compared to wild-type cells when grown in rich medium (Figure 6). Analysis of previously published expression data for S. cerevisiae Mediator mutants grown in minimal media revealed that this effect can be observed in other ACE2-regulated genes as well (53). We therefore conclude that the requirement of the head and tail domain for Ace2-dependent expression appears conserved between S. pombe and S. cerevisiae.
Figure 6.
The requirement of the Mediator head and tail domain for Ace2-dependent transcription in S. cerevisiae. Quantitative real-time PCR analysis of transcript levels of the indicated mutant strains as compared to wild-type cells. Transcript levels for all genes were normalized to TUB1 in accordance with previous studies of S. cerevisiae Mediator mutants (53).
DISCUSSION
In this study, we use a combined genetic and biochemical approach to perform a functional characterization of the S. pombe Mediator complex. Our data suggest a head module architecture with a stepwise structural organization: Med27 located on the very exterior connected to Med20, which in turn contacts Med18 which binds Med8 (Figure 1E). Biochemical characterization of the med17Δ50 allele suggested that Med17 may act as scaffold for the head domain subunits and this function requires an intact C-terminus of the Med17 subunit (Figure 2C and D).
Removal of Med27 does not affect the association of Med18 or Med20 with Mediator to any detectable extent (Figure 1D). In addition, Δmed27 cells do not display visible phenotypes unless heat stressed (Figure 3A) and appear only marginally less stress tolerant than wild-type cells (Figure 3B). Removal of Med20 also caused the complete loss of Med27 as well as an apparent decrease in the amount of Med18 (Figure 1C). A corresponding increase in severity is observed both in the cellular morphology of Δmed20 cells (Figure 3A) as well as their stress tolerance (Figure 3B). The strongest effect was seen in Δmed18 Mediator where both Med20 and Med27 were lost (Figure 1C). Similarly, the Δmed18 phenotype was more severe than that of Δmed20 cells. A Δmed18 Δmed20 double mutant appeared indistinguishable from a Δmed18 strain at 30°C demonstrating that med18 is epistatic to med20. This is what we would expect as Med20 is unable to associate with Δmed18 Mediator (Figure 1C). Consolidation of the biochemical data with the observed phenotypes indicated that the loss of Med18, which occurred in Δmed18 (Figure 1C) and med8ts cells (Figure 1D) as well as med17Δ50 cells cultured above room temperature (Figure 2C), appeared sufficient to cause the septation defect and stress hypersensitivity. The somewhat milder phenotype in Δmed20 cell may well be caused by a decrease in the number of Med18 containing Mediator in the Δmed20 cells (Figure 1C) rather than the loss of the Med20 subunit in itself.
The Med17 subunit has played a key role in establishing the almost absolute requirement for Mediator in pol II-dependent transcription in S. cerevisiae (45,46). The conditional S. pombe med17Δ50 allele isolated in this study is different to the S. cerevisiae srb4-138 allele in several respects. First, the S. pombe med17Δ50 allele is a C-terminally truncated form while the srb4-138 allele has a number of single nucleotide substitutions along the entire coding region (62). Second, we do not see an equally rapid growth arrest at the nonpermissive temperature in S. pombe (Figure 2A) as reported in the S. cerevisiae mutant (45). The growth arrest in S. cerevisiae appears directly linked to the immediate cessation of pol II-dependent transcription as the Mediator complex comes apart (19). The effect on Mediator stability in S. pombe med17Δ50 is less clear-cut with an increase in severity at higher temperatures (Figure 2C and D). We also observed that transcription does not shut down globally at the nonpermissive temperature in S. pombe med17Δ50 cells (Figure 2B). In addition, global chromatin immunoprecipitation of med17Δ50 Mediator showed a clear pattern of genomic reorganization after heat shock rather than a global reduction of Mediator binding (63).
In this work, we addressed the function of the potential S. pombe Mediator subunit Med15. Our data clearly demonstrate a functional link between Med15 and Mediator. Specifically, the phenotype and transcript profile of Δmed15 cells links Med15 to the head domain and to the middle domain subunit Med31 (Figures 3A and 4A). Presently, we do not know whether Med15 is able to stably associate with Mediator.
A relatively large number of core environmental stress-related (CESR) transcripts changed significantly in the expression profiles of all six Mediator mutants analyzed in this study. We interpret these data as the result of either one of two scenarios. First, the mutation might perturb cellular homeostasis to such a degree that the stress response is triggered as a secondary effect of the respective mutation. Alternatively, Mediator components could act as direct regulators of the stress response and so by removal of the regulator, the transcriptional program is triggered inadvertently. Further work will be required to shed light on this observation. However, Cdk8 in S. cerevisiae is known to specifically phosphorylate transcription factors directly involved in stress gene regulation (64), which therefore also would implicate Mediator as a direct regulator of the cellular stress response.
Expression profiles of genes known to be involved in cell wall metabolism correlated very well with the aberrant cellular morphologies observed in S. pombe Mediator mutants (Figures 3A and 5A–C). The flocculation phenotype seen in Cdk8 module mutants was likely the result of overproduction of surface adhesins (Figure 5A and B), a typical stress response in yeast and regulated by stress-related pathways such as MAP kinase cascades (47). Deletion of Cdk8 module genes in S. cerevisiae also leads to flocculation (46,65,66), suggesting that regulation of adhesins is conserved between S. pombe and S. cerevisiae. Indeed, transcript levels of the S. cerevisiae adhesin-coding genes FLO1, FLO5 and FLO9 are significantly elevated in Cdk8 module mutants (53,67).
Conversely, the decreased expression of genes involved in the degradation of primary septum would explain the hyphal growth in head domain mutants and the Δmed15 strain. Although transcription was not completely abolished the cumulative effect of several downregulated genes might gives rise to the hyphal growth. The process of septum dissolution to achieve complete cell separation is fundamentally different between S. pombe and S. cerevisiae (49,60). In S. pombe the primary septum consists of 1,3-β-glucan which is primarily degraded by the Eng1 endoglucanse (51) while in S. cerevisiae the primary septum is made up of chitin and is subsequently degraded by the Cts1 endochitinase (51). The cell wall material surrounding the septum, the so-called septum edging, also differs in its composition between the two species. In S. pombe it is mainly composed of 1,3-α-glucan that is degraded by the Agn1 endoglucanse (68). In S. cerevisiae, the septum edging is 1,3-β-glucan and degraded by the Eng1 endo-glucanse (61,69). The process is regulated by the conserved Ace2 transcription factor in both species but most of the target genes are not conserved between S. pombe and S. cerevisiae. We were therefore quite struck by the fact that the Mediator head domain and tail domain (only the Med15 subunit in S. pombe) were required for proper expression of Ace2-dependent genes in both S. pombe and S. cerevisiae despite the many differences in the process (Figures 5A and 6) (53). Even though the tail domain is not conserved in S. pombe, the Med15 protein still has an essential role to play in the Ace2-regulated septation cascade.
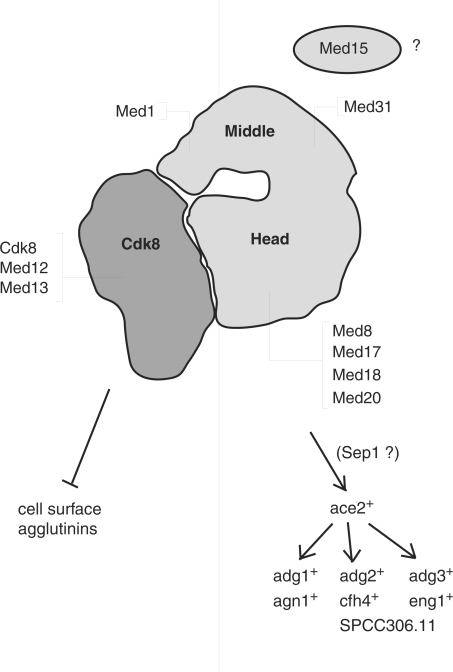
In summary, the data presented in this article suggest a structural model for the S. pombe head domain and clearly defines two distinct functional classes of subunits within the S. pombe Mediator complex (Figure 7). The first class consists of all four components of the Cdk8 module as well as middle domain subunit Med1. If deleted, this group all flocculate due to de-repression of cell surface adhesions, some of which may be involved in mating, stress response or adaptation to starvation conditions. The second class is composed of the head domain as well as middle domain subunit Med31 and the as yet unassigned Med15 protein. When mutated these genes cause septation defects leading to hyphal growth due to impaired expression of the Ace2-dependent genes involved in cell separation. Our results and previously published expression data indicate that the head domain, Med31 and Med15 are required for the proper expression of the ace2+ gene, possibly by the Sep1 transcription factor. Thus, our work identifies two distinct functional submodules of the S. pombe Mediator that have conserved roles in the regulation of specific cellular pathways.
Figure 7.
Two classes of nonessential mutant alleles in fission yeast Mediator. A diagram of S. pombe L-Mediator based on single-particle EM structures (11). The functional data presented in this work allow us to draw a virtual line through the Mediator. To the left are components of the Cdk8 module and the middle domain subunit Med1. When deleted these cells aggregate due to the overexpression of adhesive surface proteins as outlined in the text. To the right are mutant alleles of the head domain as well as the nonessential middle domain subunit Med31 and the Med15 factor. These mutants have a hyphal growth defect due to the faulty expression of genes required for efficient cell separation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are very grateful to the labs of Stefan Björklund, Frans Hochstenbach, Roger Kornberg, Matthias Sipiczki and Richard A. Young for the generous gifts of yeast strains and antibodies. Valerie Wood of the Sanger Centre Schizosaccharomyces pombe sequence database was very helpful in providing functional annotations and tables of S. cerevisiae orthologs for S. pombe genes as well as answering any questions we had. We are also very grateful to David Brodin of the Huddinge Affymetrix Facility for assistance with the expression analysis. C.M.G. was supported by grants from the Swedish Research Council, the Swedish Cancer Society and the Swedish Foundation for Strategic Research. S.H. was supported by grants from the Danish Research Councils and Manufacturer Vilhelm Pedersen and Wife Memorial Legacy (this support was granted on recommendation from the Novo Nordisk Foundation). Funding to pay the Open Access publication charges for this article was provided by the Danish Research Councils.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Kelleher RJ, III, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 3.Flanagan PM, Kelleher RJ, III, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 4.Conaway JW, Florens L, Sato S, Tomomori-Sato C, Parmely TJ, Yao T, Swanson SK, Banks CA, Washburn MP, Conaway RC. The mammalian Mediator complex. FEBS Lett. 2005;579:904–908. doi: 10.1016/j.febslet.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 5.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Spahr H, Samuelsen CO, Baraznenok V, Ernest I, Huylebroeck D, Remacle JE, Samuelsson T, Kieselbach T, Holmberg S, Gustafsson CM. Analysis of Schizosaccharomyces pombe Mediator reveals a set of essential subunits conserved between yeast and metazoan cells. PNAS. 2001;98:11985–11990. doi: 10.1073/pnas.211253898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 8.Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Spahr H, Beve J, Larsson T, Bergstrom J, Karlsson KA, Gustafsson CM. Purification and characterization of RNA polymerase II holoenzyme from Schizosaccharomyces pombe. J. Biol. Chem. 2000;275:1351–1356. doi: 10.1074/jbc.275.2.1351. [DOI] [PubMed] [Google Scholar]
- 10.Samuelsen CO, Baraznenok V, Khorosjutina O, Spahr H, Kieselbach T, Holmberg S, Gustafsson CM. TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution. PNAS. 2003;100:6422–6427. doi: 10.1073/pnas.1030497100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl Acad. Sci. USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. A complex of the srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 13.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 14.Spahr H, Khorosjutina O, Baraznenok V, Linder T, Samuelsen CO, Hermand D, Makela TP, Holmberg S, Gustafsson CM. Mediator Influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro. J. Biol. Chem. 2003;278:51301–51306. doi: 10.1074/jbc.M306750200. [DOI] [PubMed] [Google Scholar]
- 15.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 16.Larschan E, Winston F. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 2005;25:114–123. doi: 10.1128/MCB.25.1.114-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 18.Davis JA, Takagi Y, Kornberg RD, Asturias FJ. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 19.Linder T, Zhu X, Baraznenok V, Gustafsson CM. The classical srb4-138 mutant allele causes dissociation of yeast Mediator. Biochem. Biophys. Res. Commun. 2006;349:948–953. doi: 10.1016/j.bbrc.2006.08.099. [DOI] [PubMed] [Google Scholar]
- 20.Lee YC, Kim Y-J. Requirement for a functional interaction between Mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YC, Park JM, Min S, Han SJ, Kim YJ. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 23.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon H-M, Holstege FCP, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol. Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 25.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J. Biol. Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 26.Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacIver FH, Glover DM, Hagan IM. A ‘marker switch’ approach for targeted mutagenesis of genes in Schizosaccharomyces pombe. Yeast. 2003;20:587–594. doi: 10.1002/yea.983. [DOI] [PubMed] [Google Scholar]
- 28.Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto T, Sekiguchi M. Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine. J. Bacteriol. 1976;127:1561–1563. doi: 10.1128/jb.127.3.1561-1563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigaut G. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 31.Puig O. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 32.Linder T, Gustafsson CM. The Soh1/MED31 Protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator. J. Biol. Chem. 2004;279:49455–49459. doi: 10.1074/jbc.M409046200. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GeneChip Expression Analysis - Technical Manual. 2004. Affymetrix.
- 35.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 36.Miklos I, Szilagyi Z, Watt S, Zilahi E, Batta G, Antunovics Z, Enczi K, Bahler J, Sipiczki M. Genomic expression patterns in cell separation mutants of Schizosaccharomyces pombe defective in the genes sep10 (+) and sep15 (+) coding for the Mediator subunits Med31 and Med8. Mol. Genet. Genomics. 2008;279:225–238. doi: 10.1007/s00438-007-0296-z. [DOI] [PubMed] [Google Scholar]
- 37.Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 38.Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira JM, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lariviere L, Geiger S, Hoeppner S, Rother S, Straszer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 41.Szilagyi Z, Grallert A, Nemeth N, Sipiczki M. The Schizosaccharomyces pombe genes sep10 and sep11 encode putative general transcriptional regulators involved in multiple cellular processes. Mol. Genet. Genomics. 2002;268:553–562. doi: 10.1007/s00438-002-0773-3. [DOI] [PubMed] [Google Scholar]
- 42.Zilahi E, Miklos I, Sipiczki M. The Schizosaccharomyces pombe sep15+ gene encodes a protein homologous to the Med8 subunit of the Saccharomyces cerevisiae transcriptional mediator complex. Curr. Genet. 2000;38:227–232. doi: 10.1007/s002940000158. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Gimeno MA, Munoz I, Arino J, Sanz P. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J. Biol. Chem. 2003;278:47744–47752. doi: 10.1074/jbc.M306157200. [DOI] [PubMed] [Google Scholar]
- 44.Grallert A, Grallert B, Zilahi E, Szilagyi Z, Sipiczki M. Eleven novel sep genes of Schizosaccharomyces pombe required for efficient cell separation and sexual differentiation. Yeast. 1999;15:669–686. doi: 10.1002/(SICI)1097-0061(19990615)15:8<669::AID-YEA411>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl Acad. Sci. USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 47.Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- 48.Watson P, Davey J. Characterization of the Prk1 protein kinase from Schizosaccharomyces pombe. Yeast. 1998;14:485–492. doi: 10.1002/(SICI)1097-0061(19980330)14:5<485::AID-YEA239>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 49.Feierbach B, Chang F. Cytokinesis and the contractile ring in fission yeast. Curr. Opin. Microbiol. 2001;4:713–719. doi: 10.1016/s1369-5274(01)00273-9. [DOI] [PubMed] [Google Scholar]
- 50.Sipiczki M, Grallert B, Miklos I. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J. Cell Sci. 1993;104(Pt 2):485–493. doi: 10.1242/jcs.104.2.485. [DOI] [PubMed] [Google Scholar]
- 51.Martin-Cuadrado AB, Duenas E, Sipiczki M, Vazquez de Aldana CR, del Rey F. The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 2003;116:1689–1698. doi: 10.1242/jcs.00377. [DOI] [PubMed] [Google Scholar]
- 52.Ribar B, Banrevi A, Sipiczki M. sep1+ encodes a transcription-factor homologue of the HNF-3/forkhead DNA-binding-domain family in Schizosaccharomyces pombe. Gene. 1997;202:1–5. doi: 10.1016/s0378-1119(97)00390-9. [DOI] [PubMed] [Google Scholar]
- 53.van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, Holstege FC. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 54.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang Y-W, Howard SC, Herman PK. The Ras/PKA signaling pathway directly targets the srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell. 2004;15:107–116. doi: 10.1016/j.molcel.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 56.Linder T, Gustafsson CM. Molecular phylogenetics of ascomycotal adhesins - A novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 2007 doi: 10.1016/j.fgb.2007.08.002. doi:1016/j.fgb.2007.08.002 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57.Alonso-Nunez ML, An H, Martin-Cuadrado AB, Mehta S, Petit C, Sipiczki M, del Rey F, Gould KL, de Aldana CR. Ace2p controls the expression of genes required for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell. 2005;16:2003–2017. doi: 10.1091/mbc.E04-06-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 59.Doolin MT, Johnson AL, Johnston LH, Butler G. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 2001;40:422–432. doi: 10.1046/j.1365-2958.2001.02388.x. [DOI] [PubMed] [Google Scholar]
- 60.Yeong FM. Severing all ties between mother and daughter: cell separation in budding yeast. Mol. Microbiol. 2005;55:1325–1331. doi: 10.1111/j.1365-2958.2005.04507.x. [DOI] [PubMed] [Google Scholar]
- 61.Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 2001;276:19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- 62.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of mediator interactions. Mol. Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 64.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 66.Chang YW, Howard SC, Budovskaya YV, Rine J, Herman PK. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase II holoenzyme during stationary phase entry in Saccharomyces cerevisiae. Genetics. 2001;157:17–26. doi: 10.1093/genetics/157.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang YW, Howard SC, Herman PK. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell. 2004;15:107–116. doi: 10.1016/j.molcel.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 68.Dekker N, Speijer D, Grun CH, van den Berg M, de Haan A, Hochstenbach F. Role of the alpha-glucanase Agn1p in fission-yeast cell separation. Mol. Biol. Cell. 2004;15:3903–3914. doi: 10.1091/mbc.E04-04-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baladron V, Ufano S, Duenas E, Martin-Cuadrado AB, del Rey F, Vazquez de Aldana CR. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell. 2002;1:774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.