Abstract
The mechanism and role of RNA structure elements in the replication and translation of Caliciviridae remains poorly understood. Several algorithmically independent methods were used to predict secondary structures within the Norovirus, Sapovirus, Vesivirus and Lagovirus genera. All showed profound suppression of synonymous site variability (SSSV) at genomic 5′ ends and the start of the sub-genomic (sg) transcript, consistent with evolutionary constraints from underlying RNA structure. A newly developed thermodynamic scanning method predicted RNA folding mapping precisely to regions of SSSV and at the genomic 3′ end. These regions contained several evolutionarily conserved RNA secondary structures, of variable size and positions. However, all caliciviruses contained 3′ terminal hairpins, and stem–loops in the anti-genomic strand invariably six bases upstream of the sg transcript, indicating putative roles as sg promoters. Using the murine norovirus (MNV) reverse-genetics system, disruption of 5′ end stem–loops produced ∼15- to 20-fold infectivity reductions, while disruption of the RNA structure in the sg promoter region and at the 3′ end entirely destroyed replication ability. Restoration of infectivity by repair mutations in the sg promoter region confirmed a functional role for the RNA secondary structure, not the sequence. This study provides comprehensive bioinformatic resources for future functional studies of MNV and other caliciviruses.
INTRODUCTION
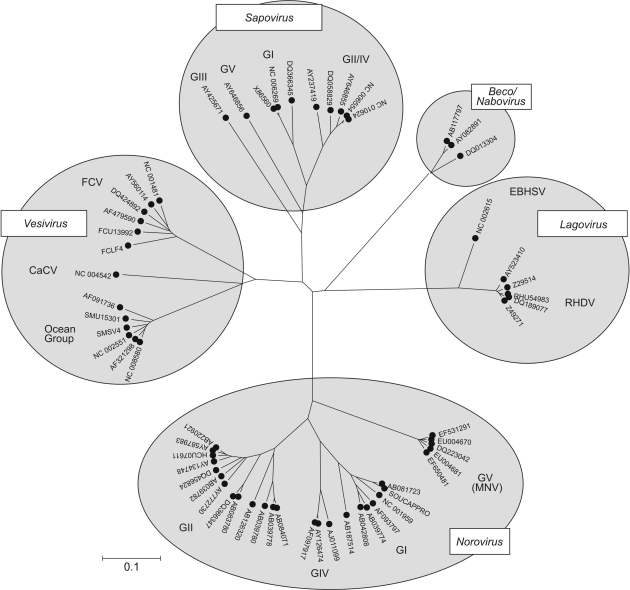
The virus family Caliciviridae is a heterogeneous group of non-enveloped viruses with positive-stranded RNA genomes. Caliciviruses are currently classified into five genera based on differences in genome organization, coding strategies, host range and persistence (Figure 1) (1–3). Caliciviruses infecting humans are found in two of the genera, the noroviruses and sapoviruses, both responsible for highly infectious, acute gastroenteritis. Viruses in the Vesivirus genus are widely distributed in a variety of mammalian species, where they cause systemic and frequently persistent infections, often with high morbidity and mortality. Vesiviruses fall into two main groups, feline calicivirus (FCV) infecting domestic and wild cats, and a second ‘ocean virus’ group, including San Miguel sea lion virus, which are widely distributed in marine and terrestrial mammals (4). Lagoviruses [rabbit haemorrhagic fever virus (RHDV) and the related European brown hare syndrome virus (EBHSV)] are restricted to rabbits and hares, in which they cause rapidly progressive and frequently fatal haemorrhagic disease and necrotizing hepatitis (1). Finally, two groups of caliciviruses that cause diarrhoea and acute intestinal diseases in calves have been recently characterized (3,5,6). One group is related to human noroviruses and has been classified as genogroup III in the Norovirus genus; the second group is genetically distinct from other genera and its classification as members of a fifth genus in the Caliciviridae has been proposed [Becovirus or Nabovirus; (3)].
Figure 1.
Phylogenetic analysis of the RNA-dependent RNA polymerase (RdRp)-encoding region of representative variants (up to five per genus or genogroup) of each genus of caliciviruses, shown as an unrooted tree. Sequences were translated and aligned using ClustalW with default settings, and a neighbour-joining phylogenetic tree constructed from pair wise translated amino acid distances. The scale bar indicates a distance of 0.1 (10% amino sequence divergence).
Recently, a new calicivirus was characterized as the causative agent of a highly transmissible virus among immunosuppressed laboratory mice in which it caused a fatal hepatitis and encephalitis (7). Subsequently it was shown that the virus is widely distributed in laboratory mice worldwide, shows genetic diversity and altered patterns of disease among different laboratory strains of immunocompetent mice, generally establishing persistent infections and little or no disease (8,9). Because of its genetic relatedness to human noroviruses, it has been named murine norovirus (MNV), and is classified as genogroup V within the Norovirus genus.
Caliciviruses are thought to replicate in a manner typical of positive-stranded RNA viruses, through the synthesis of a full-length anti-genomic strand (reverse complement copy) using the virally encoded RNA-dependent RNA polymerase (RdRp) translated initially from the RNA genome entering the cell (10). The minus strand then acts as a template for the synthesis of full-length genomic RNA from which non-structural proteins are translated, including the RdRp, helicase and protease. In a manner comparable to the replication of Togaviridae and hepatitis E virus, caliciviruses express a downstream sub-genomic (sg) transcript encoding structural genes.
Studies of the replication of caliciviruses have been hampered in the past through the difficulty in culturing infectious virus in mammalian cell culture. Particularly intractable are human noroviruses and sapoviruses for which no efficient cell culture system exists. However, recent reports have demonstrated limited genome replication and encapsidation following transfection of cDNA constructs and subsequent infection with recombinant Vaccinia virus expressing T7 RNA polymerase (11,12). In these systems, replication occurs despite the initial lack of a covalently attached, virally encoded VPg protein at the 5′ end of the genomic RNA that is required for translation initiation (13). In addition, studies suggest that a differentiated intestinal cell culture may support Norwalk virus replication (14). Although these systems represent a significant advance in the study of norovirus biology, they lack the efficiency with which to study the basic mechanisms of calicivirus translation and replication.
In contrast, FCV and MNV can be efficiently grown in tissue culture (7). Infectious virus can be artificially generated through transfection of in vitro transcribed 5′ capped RNA transcripts in the case of FCV (15) or cDNA clones under control of T7 RNA polymerase for both FCV (16) and MNV (17). These advances provide the means for much more detailed characterization of the calicivirus replication cycle and its mechanisms of transcription and translation.
In the current study, we have applied a number of algorithmically independent bioinformatic methods to identify, locate and characterize RNA secondary structure potentially implicated in the replication cycle of different calicivirus genera. This approach was prompted by the success of such methods in the definition of RNA secondary structures subsequently shown to play essential roles in the replication and translation of other positive-stranded RNA viruses, such as members of the Picornaviridae (18–22) and hepatitis C virus (HCV) (23–25). Bioinformatic methods for RNA structure prediction in caliciviruses are becoming increasingly feasible through the growing availability of nucleotide sequence data in each of the genera that enables evolutionary constraints, phylogenetic conservation of RNA structures and co-variant substitutions to be identified. We subsequently used our predictions as the basis for a preliminary reverse genetic analysis of MNV replication. These functional studies strongly support our bioinformatic predictions and define RNA secondary structures involved in virus replication. The catalogue of evolutionarily conserved RNA secondary structures provides a valuable resource for future investigations of the replication of MNV, FCV and members of other calicivirus genera.
MATERIALS AND METHODS
Nucleotide sequences
RNA structure analysis was based on sets of available complete genome sequences of epidemiologically unlinked variants from Norovirus, Sapovirus, Vesivirus and Lagovirus genera (Genbank in May, 2007; Table 1; individual sequences listed in Supplementary Table S1).
Table 1.
Sequence divergence in different calicivirus genera/groups
| Genus/Group | Total | Non-structurala (%) | structurala (%) | ORF2/3a (%) |
|---|---|---|---|---|
| Norovirus GII | 20 | 13.1 (5.8) | 26.1 (23.7) | 28.6 (29.7) |
| MNV | 9 | 10.8 (3.0) | 9.5 (4.0) | 7.8 (4.8) |
| Lagovirus | 13 | 11.4 (5.8) | 9.7 (6.2) | 8.9 (7.1) |
| Vesivirus | 14 | 19.9 (8.8) | 21.7 (11.6) | 13.4 (3.7) |
| Sapovirus | 15 | 31.0 (25.8) | 43.5 (48.0) | NA.b |
| Beco/Nabovirus | 3 | 16.3 (6.1) | 10.5 (4.2) | 12.3 (5.9) |
aMean pairwise nucleotide sequence divergence between group members (amino acid sequence divergence in parentheses).
bRegion could not be aligned.
Because of the high sequence divergence of variants within the Norovirus and Vesivirus genera, subsets corresponding to previously assigned genogroups were used. For noroviruses, sequences from MNV and human norovirus genogroup II were separately analysed, the GII/4 subset of human noroviruses used for RNA structure analysis of the 3′ end of the genome, while in the Vesivirus genus, analysis was restricted to FCV (there were too few sequences from the ocean virus group for separate analysis). However, the limited availability of complete genome sequences of sapoviruses precluded a separate analysis of individual genogroups, and analysis was carried out on the genus collectively. All nucleotide (non-coding) and codon-based (coding region) alignments were created using ClustalW with default settings.
Comparative analysis was carried out with sequences from human enteroviruses species A, B and C. To maximize the amount of phylogenetic information, single example of each classified serotype within each species was used (Table S1).
Suppression of synonymous site variation
Synonymous sequence variability was determined by measurement of mean pair wise distances at each codon position in the two or three open reading frames (ORFs) of caliciviruses and the ORF of enteroviruses. Variability at each codon was calculated using the program Sequence Scan in the Simmonic sequence editor (26). Mean pair wise synonymous variability was restricted to aligned codons, where the translated amino acid was the same. Each pair wise value was normalized by dividing by the degeneracy of the codon, with normalization factors for 2-fold degenerate sites of 0.5, 3-fold: 0.6666; 4-fold: 0.75 and 6-fold: 0.8333. This takes into account the different sequence distances achievable at maximally diverged sites. Variability at each codon position was averaged over a sliding window of 21 or 41 codons.
RNA secondary structure prediction
Secondary structure predictions with PFOLD (27) and Alifold (28) used web interfaces provided by the authors at: http://www.daimi.au.dk/~compbio/rnafold/ and http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi, respectively. Both programs were run with default settings.
High-resolution thermodynamic scanning was carried out using a locally implemented compiled version of the program UNAFold (29) downloaded from http://www.bioinfo.rpi.edu/%7Ezukerm/export/mfold-3.html. Each sequence in alignments of calicivirus genera/groups and enterovirus species was split into sets of 200 base fragments overlapping by 152 bases (∼130–170 fragments per virus genome). Fifty copies of each fragment were also generated and the sequence order scrambled using the algorithm NDR that preserves the dinucleotide frequencies of the native sequence (30), implemented in the Simmonic Sequence Editor package (30). Minimum free energies (MFEs) of each native sequence fragment were compared with the mean MFE of the NDR-scrambled controls to produce a MFE difference (MFED):
where MFENATIVE and MFESCRAMBLED are MFEs of native sequences and mean MFEs of 50 sequence order randomized controls respectively.
Mean MFEDs for each fragment were calculated for each alignment group, and plotted against the mid-point of each fragment to localize areas of sequence order-dependent RNA secondary structure. MFEDs were also similarly calculated for the reverse complement of each genome sequence. All results were expressed as a mean of five adjacent fragments.
The position of the native sequence MFE in the distribution of MFE values for controls can also be used to generate Z-scores (31,32). However, for the current study, the use of MFEDs to express folding energy differences is more appropriate since it provides a biological scale with which to quantify the extent of sequence order-dependent RNA secondary structure formation in different genome regions. MFEDs have therefore been used for presentation of results.
Prediction of specific RNA secondary structures
The positions of RNA secondary structures in the genomes of different calicivirus groups and genera were first determined by scanning genomes for regions of suppression of variability at synonymous sites, and high MFED values. Regions with evidence for consistent structure between virus groups were then investigated using algorithms to characterize the RNA secondary structures in detail (PFOLD, AliFold).
MNV reverse genetics
Synonymous mutations were introduced into predicted RNA structures in MNV using established molecular approaches (see Supplementary Methods) to investigate the effect of their disruption and restoration on virus replication ability. Mutated MNV cDNA sequences were transfected into baby hamster kidney (BHK) cells engineered to express T7 RNA polymerase [BSRT7; (33)] and infected with a fowlpox virus expressing T7 RNA polymerase to initiate transcription of MNV RNA (17). Further details of the virus recovery method and quantitation of infectivity (34) are provided in Supplementary Methods.
RESULTS
Screening calicivirus genomes for RNA secondary structure
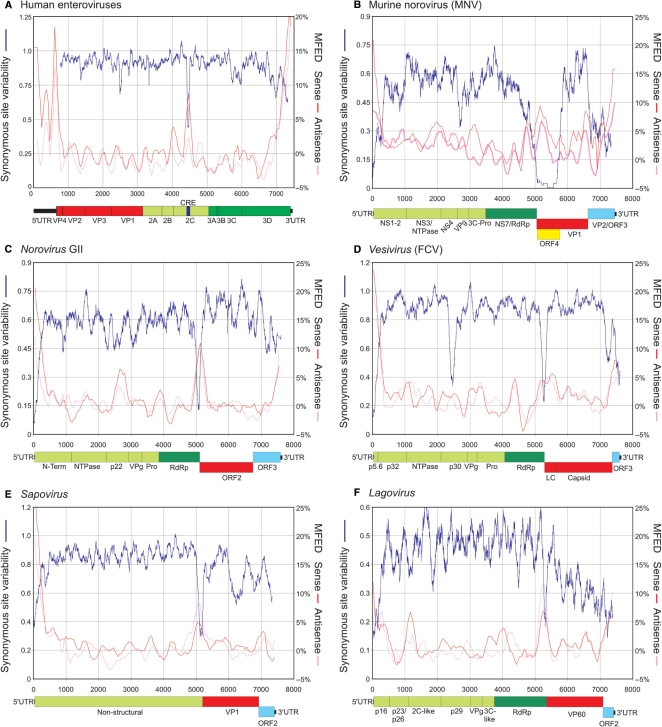
Functional RNA secondary structures in coding regions place constraints on codon usage absent in unstructured regions of the genome (26). Sequence changes in base-paired regions are restricted predominantly to compensated (co-variant) substitutions that maintain base pairing, which typically occur at a lower frequency than in unstructured regions. Suppression of synonymous site variability (SSSV) was observed among each species of human enterovirus sequences in the 2C region coinciding with the cis-replicating element (CRE, Figure 2A), a small stem–loop (between positions 4442 and 4493) with a known role in enterovirus replication (20,19).
Figure 2.
Scanning alignments of complete genome sequences of enteroviruses and caliciviruses for RNA secondary structure. Variability at synonymous sites (left y-axis) was computed at each codon position in alignments, plotted with a window size of 41 codons. MFED values (right y-axis) for sense and anti-sense RNA sequences were calculated for 200 base fragments, incrementing by 48 bases; values plotted represent mean values of five consecutive fragments. All nucleotide positions were calculated relative to reference sequences listed in methods; gene names, boundaries and other structural features followed the annotation provided for the reference sequences used for numbering. A higher resolution vector diagram of the figure is available from Supplementary Data (Figure S1).
Regions of marked suppression of synonymous variability were observed in each of the genera or groups of caliciviruses (Figure 2B–F), irrespective of the degree of overall nucleotide sequence divergence within each group (analysis of the Beco/Nabovirus genus was precluded by the limited number of complete genome sequences). There was some consistency in regions of observed SSSV; variability was markedly suppressed at the start of the non-structural (NS) protein coding sequence in each group and a second region of generally very localized SSSV in the region between the NS and structural (S) gene regions, irrespective of whether their reading frames were offset (Norovirus, Vesivirus) or in-frame (Sapovirus, Lagovirus). In MNV, more extensive suppression of variability was observed, starting at the NS/S junction, and extending for the length of the alternative ORF [Figure 2B; (35)]. Finally, there was a lesser degree of SSSV at the 3′ end of the coding sequence, particularly among vesivirus and lagovirus sequences.
For enteroviruses, constraints imposed by RNA secondary structure created the regions of SSSV in the CRE. To scan caliciviruses for sequence order-dependent RNA secondary structure (i.e. secondary structures dependent on order of bases in the nucleotide sequence, rather than compositional variables such as G + C content), consecutive, free energies on folding closely spaced fragments were determined for sequences in each alignment and compared to those of the same sequences in which sequence order was randomized. For human enteroviruses, substantial differences in folding energies between native and scrambled sequences were detected in the 5′UTR, in the terminal region of the 3Dpol gene and the 3′UTRs, consistent with previous studies. For the rest of the coding region, folding energy differences approximated to zero (indicating an absence of sequence order-dependent RNA structure), except for a restricted area of predicted RNA secondary structure in the 2C-encoding region, superimposable with the known location of the CRE (Figure 2A; MFED values on right-hand y-axis). For the 5′UTR and 2C CRE region, MFEDs for sequences in their sense orientation were consistently higher than those calculated for anti-sense sequences, a difference that reflects the preferential distribution of G:U pairings in the sense sequence and asymmetries in folding energies between 5′G:C3′ and 5′C:G3′ Watson–Crick pairings (29), and consistent with the function of these sequences in the positive-sense genome.
High-resolution thermodynamic scanning similarly identified regions of RNA secondary structure in the four calicivirus genera (Figure 2B–F). For each, the 5′ end of the genome, including the short 5′UTR and 200–300 bases of coding sequence showed evidence for substantial RNA structure formation. Folding energies for sequences in the genomic orientation were more energetically favoured than anti-sense sequences. All five groups additionally showed localized regions of high-folding energy differences around the NS/S junction, particularly marked in the Norovirus, Sapovirus and Lagovirus genera that coincided precisely with regions of suppression of synonymous variability. Folding energies for anti-genomic sequences were frequently greater (sapoviruses, lagoviruses; Figure 2E and F) or similar (noroviruses, vesiviruses; Figure 2B–D) to those of the genomic strand. There was frequent evidence for RNA secondary structure at the 3′ end of the genomic strand, frequently overlapping the coding region of ORF2/3, and associated with the observed SSSV. Vesiviruses showed a further region of suppression of synonymous variability in the NS encoding region between positions 2350 and 2600, although this was not associated with higher MFEDs (Figure 2D). Similar findings were made for the area of SSSV in MNV between positions 2600 and 2800 (Figure 2B).
Identification of specific RNA structures in 5′, NS/S junction and 3′ regions of calicivirus genomes
Paired, co-variant substitutions provide independent evidence supporting RNA secondary structure models, and can be used to refine predictions from energy-minimizing methods. To identify and characterize RNA secondary structures in caliciviruses, we used the programs Alifold and PFOLD, an algorithmically independent, non-energy-minimizing algorithm that also exploits comparative sequence information (27,36). Outputs from both programs provide probabilities for specific pairings, from which specific structures can be visualized in the form of a half-diagonal matrix (Figures 3–5). For both programs, probabilities of pairings are reflected in the size of the points plotted, while Alifold output shows additional information on the variability (degree of co-variance) of each pairing through different colours (red: invariant; green: variable). To assist comparison of the predictions from each program, output from Alifold was plotted in the lower right quadrant, and PFOLD in the upper left (Figures 3 and 4).
Figure 3.
RNA structure prediction for the first 400 bases of genome sequence alignments of (A) the first 800 bases of genome sequence alignments of human enteroviruses species A, and (B–F) the first 400 bases of genome sequence alignments of different calicivirus genera/groups. Pairing predicted by Alifold shown in lower right quadrant by PFOLD in the upper right. For both predictive algorithms, the size of the dots is proportional to the probability of pairing; predicted paired sites containing co-variant or semi-co-variant substitutions shown in green. Synonymous variability averaged over a window size of 21 codon shown as blue line (right-hand y-axis). A higher-resolution vector diagram of the figure is available from Supplementary Data (Figure S2).
Figure 4.
Predicted RNA secondary structures using Alifold and PFOLD (see legend to Figure 2) in (A) 2C/CRE region human enterovirus species A and (B–F) regions of maximum SSSV in different calicivirus genera. For human enterovirus sequences, separate plots of synonymous variability were shown for species A–C. The positions of the CRE in 2C and the arrangement of NS (green) and S (red) gene reading frames in caliciviruses shown to scale on the x-axis, with the sub-genomic transcript shown in pink. A higher-resolution vector diagram of the figure is available from Supplementary Data (Figure S3).
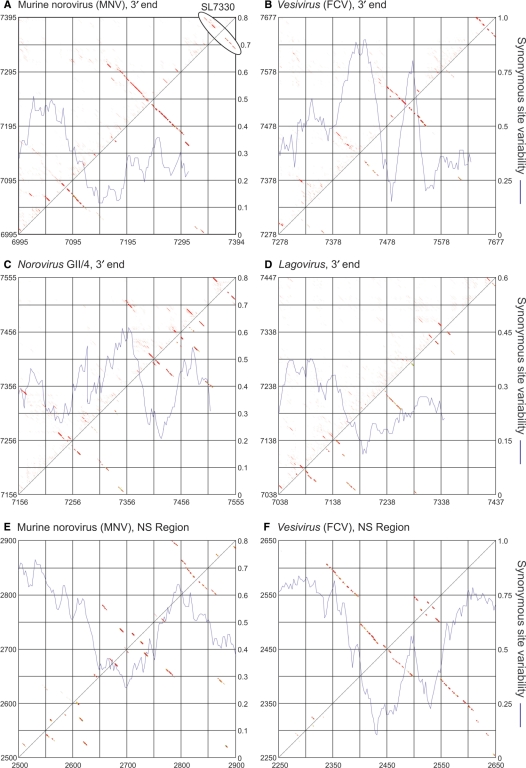
Figure 5.
(A–D) Predicted RNA secondary structures for 3′ ends of MNV, norovirus, vesivirus and lagovirus sequences, and (E, F) regions in the NS genes of MNV and Vesiviruses showing SSSV (Figure. 1B and D). A higher-resolution vector diagram of the figure is available from Supplementary Data (Figure S4).
The ability of the two programs to predict RNA secondary structures were assessed by analysing human enterovirus 5′UTR and 2C/CRE sequences (Figures 3A and 4A). In the 5′UTR, predictions from Alifold and PFOLD were highly concordant, and both identified the six main stem–loops [labelled I–VI; (37)], including the 5′ proximal cloverleaf (stem–loop I) and the IRES structure, stem–loop IV (Figure 3A). Predictions for species B and C 5′UTR sequences were comparable to those of species A (data not shown). Program output from both the methods was in all cases characterized by a virtual absence of predicted alternative or long-range predicted pairings in the 5′UTR. In the 2C/CRE region, a single well-defined stem–loop was predicted by both methods corresponding to the previously described CRE between positions 4442 and 4493 (Figure 4A). For all three enterovirus species, the position of the CRE co-localized precisely to a region of suppression of synonymous variability.
Alifold and PFOLD were used to detect RNA secondary structures in different caliciviruses groups in regions showing SSSV and large folding energy differences between native and control sequences [5′ end (Figure 3B–F), NS/S junction (Figures 4B–5F) and the 3′ end (Figure 5A–D)]. Regions of SSSV in MNV and vesiviruses within the NS gene were additionally examined (Figure 5E and F). In the 5′ 400 bases of each virus group, two or more discrete, highly conserved RNA stem–loops were detected, with highly concordant results between the two prediction methods (Figure 3B–F). Analysis was repeated using anti-genomic sequences, and produced comparable results but with generally less support for the identified structures [fewer base pairings and lower probability values for identified pairings (data not shown)].
Other than a tendency for the predicted structures to be situated in the first 150 nt of the genomes, there was little similarity in shape or position of the individual structures between groups, or even within a genus. For example, the predicted structure for the genogroup II noroviruses showed no resemblance to the structure prediction for MNV (Figure 3B and C) and were similarly distinct from those previously predicted using Foldrna for the genogroup I sequence M87661 (38). However, there was a correlation between the location of predicted stem–loops and marked regions of SSSV (blue line), within the first 100–170 nt of the genome. Beyond position 150, the occurrence and conservation of predicted RNA secondary structures was highly variable. However, MNV showed an uninterrupted series of stem–loops throughout the analysed region, although this was not associated with SSSV. These additional structures likely comprise elements of genome-scale ordered RNA structure (GORS) found specifically in viruses causing persistent infections (30). While the ability of the bioinformatic prediction methods to identify phylogenetically conserved structures is influenced by the degree of naturally occurring sequence diversity within the analysed groups, this was not the explanation for the differing extents of structure observed in the 5′ region. For example, the sequence diversity of lagoviruses and human noroviruses used in the analysis is comparable to that of MNV, while vesiviruses were more divergent than all three (Table 1).
In the NS/S junction region, suppression of synonymous variability localized to a region just upstream of the NS/S junction in human norovirus, vesivirus, lagovirus and sapovirus sequences (Figure 4C–F). MNV differed in showing a more generalized suppression in variability throughout and 3′ to the junction region, potentially reflecting further sequences constraints (see Discussion section). In the region of every localized suppression, one or more stem–loops were predicted (almost invariably concordantly by Alifold and PFOLD) in each virus group, with a stem–loop localized just before the transcription start site (pink box). The absence of consistently predicted structures in sapoviruses may have been the result of the high divergence of sequences used for analysis (see below).
The structures predicted 5′ to the transcription site in each genus comprised unbranched stem–loops of varying duplex length and sizes of terminal loops (Figure 6). Because of the frequently observed greater folding energy differences (MFEDs) for anti-genomic sequences in this region (Figure 1), the structures have been depicted in their anti-sense orientation. The occurrence of several G:U pairs in the stems (and absence of opposed C–A mismatches) in sapoviruses, MNV and vesiviruses provides further evidence that this represents the functional orientation of the RNA structures, and is supported by prediction of the depicted structures using AliFold and PFOLD (data not shown). Although no structures were predicted using the total dataset of sapoviruses, structurally related stem–loops were found on analysis of subsets of sapoviruses restricted to individual genogroups. For example, the five available genotype II (GII) sequences were predicted to form a stem–loop with a terminal unpaired region of 6 nt. The same structure was predicted for GIV (n = 5), but with several G:C/G:U semi-co-variant substitutions in the stem (data not shown). Predicted stem–loops for GI and GV were similar, even though there was little or no sequence conservation of nucleotides forming the stem. In each case, however, the terminal loop sequence, UUC(G/A)U was highly conserved between genogroups, an observation also consistent with the identity of this region between MNV and human viruses in the Norovirus genus (UGCG).
Figure 6.
Consensus RNA secondary structures upstream of the sub-genomic transcript (grey filled boxes; initiating anti-codons blocked) for the five calicivirus genera predicted by Alifold, shown in anti-sense orientation in a 3′ to 5′ left to right direction. Separate structure predictions were made for different human norovirus and sapovirus genogroups and for Newbury1- and NB-like beco/naboviruses because of their high sequence variability in this region (see text). The unpaired 6-base sequences found in all genera/groups between the predicted RNA structures and the start of the transcript are underlined.
Analysis of the 400-base fragment at the NS/S junction of genogroup II human noroviruses in its anti-sense orientation did not predict RNA secondary structures in the same region as found in other calicivirus genera. However, a stem–loop comparable in size and containing the same UGCG tetraloop as MNV could potentially form, and was the most energetically favoured structure predicted in separate analysis using individual sequences with MFOLD (data not shown). The potential additional U:A pairing at the base of the stem (Figure 6) was not predicted because the A base-paired downstream as part of another stem–loop. To investigate whether other human norovirus variants formed structures comparable to those of genogroup I and MNV, MFOLD was used to predict secondary structures in sequences of the available genogroup I sequences. For these sequences, a more stable stem–loop was predicted with an uninterrupted duplex stem of 11 bp, and a terminal loop containing the UGCG unpaired terminal loop (Figure 6).
Despite the lack of comparative sequence data for the beco/naboviruses, similarly positioned although structurally different stem–loops were found in the Newbury1 and NB genogroups, both with a terminal CAUGU pentanucleotide terminal sequence.
Despite the diversity of structures between genera, there was a remarkably conserved positional relationship between the 5′ limit of the predicted stem–loops and the start of the anti-genomic template for the sg transcript (boxed). Although the start of the sg transcript of beco/naboviruses has not yet been mapped experimentally, the boxed region depicted starts with the canonical CACU sequence typical of the 5′ ends of transcripts in other calicivirus genera. Observations that first 12 bases of the proposed transcript were an exact repeat of the first 12 bases at the 5′ end of the beco/nabovirus genome, and the identity of 17 from 22 bases before the start codon of the putative sg RNA (sgRNA), further supports this proposed assignment. For all five calicivirus genera, there was a gap (invariably of 6 nt) that was predicted to be unpaired in each virus group. This positional relationship was shared between genera despite the lack of any obvious sequence homology of the six unpaired bases, nor between sequences or structural features of the upstream stem–loops. It likely reflects some commonality in the mechanism of transcription initiation of the sgRNA, perhaps also reflected in the high degree of sequence conservation of the initial 4–6 bases of the sgRNA, and the occurrence of repeated sequences at the start of the genomic RNA in all five genera.
Conserved secondary structures were detected in the 3′ ends of MNV, human noroviruses, vesiviruses and sapoviruses (Figure 5). Sequences from sapoviruses were highly divergent and aligned poorly in this region and were therefore excluded from analysis, as were beco/naboviruses because of lack of comparative sequence data. Similar problems with high diversity and problematic alignment were encountered for the human norovirus GII sequences which were highly divergent (∼29%) in the ORF2/3 region (Table 1); analysis was therefore restricted to the 11 available sequences in the GII/4 subset. Alifold and PFOLD prediction algorithms detected a 3′ terminal hairpin in human norovirus sequences (GII/4) similar in position and size but with an entirely different primary nucleotide sequences to the previously reported RNA secondary structure in human norovirus GI [Figure 5C; (39)]. Terminal stem–loop structures were additionally detected in vesivirus and lagovirus sequences (Figure 5B and D), consistent with previous predictions (40). There were also a number of other upstream structures; particularly striking was the large predicted stem–loop between positions 7478 and 7554 in vesiviruses, associated with suppression of variability in the terminal coding region. This structure was distinct from the small stem–loop previously predicted immediately downstream of the frameshift site (at position 7314) between the capsid gene and ORF2 (41). It did however, co-localize precisely with the previously described cis-acting replication element in the ORF3 coding sequence of FCV (42) (see Discussion section)
MNV sequences showed evidence for two similarly positioned large stem–loops, including a terminal hairpin from positions 7330 to the end of the genome (Figure 7), and a large stem–loop of 164 bases centring around position 7239. The unpaired region in the MNV 3′ terminal stem–loop accommodates a variable number of inserted pyrimidines in many naturally occurring sequence variants of MNV without disrupting the overall structure (Figure 7).
Figure 7.
Upper panels: Alifold predictions of RNA secondary structures in 5′, NS/S junction and 3′ terminal region of MNV for sequences in sense (lower right quadrant), and anti-sense (upper left quadrant). Lower figure: predicted RNA secondary structures for the prototype MNV-1 sequence (NC_008311), showing positions of introduced substitutions in mutants m50, m51, m53 and m54 to disrupt secondary structure (filled and unfilled circles) and compensatory changes in m53r to restore pairings (unfilled and filled squares; see Legend). SL8, SL29 and SL7330 were drawn in their sense orientation; SLa5045 in anti-sense orientation (see text). The position of naturally occurring insertions of pyrimidines in the 3′ terminal loop (SL7330) is indicated.
Finally, a limited analysis of RNA secondary structure was carried out in the NS regions of MNV and vesiviruses in which further regions of marked SSSV were detected around positions 2700 and 2400, respectively (Figure. 5E and F). In both cases, there was evidence for conserved secondary structures coinciding with the region of SSSV. Most notably, the vesivirus sequences formed a long duplex stem bounding a large internal stem–loop and second double loop, a structural complex occupying nearly 350 nt.
Functional investigation of predicted RNA secondary structures in MNV
The recent development of a reverse genetic system for MNV (17) allowed RNA secondary structures described earlier to be functionally analysed. Substitutions were introduced into secondary structures predicted for the 5′ end of the genome (stem–loops starting at positions 8 and 29, SL8 and SL29), in the predicted anti-genomic structure upstream of the sgRNA transcript (SLa5045), and in the 3′ terminal hairpin (SL7330) to generate mutants m50, m51, m53 and m54, respectively (Figure 7). SL8, altered in the mutant m50 and SL29 (m51) and SL7330 (m54) have been drawn in their sense (genomic) orientation while the predicted RNA structure at the NS/S junction in MNV should be considered as an anti-sense structure (SLa5045, as altered in m53).
In these preliminary investigations, mutations introduced in all four regions disrupted structure in either orientation of the sequence (data not shown). To determine whether these alterations in the RNA stem–loops influenced infectivity or phenotype, virus recovery was attempted from the WT and mutant cDNA constructs (17) by transfection of cDNA constructs under the control of fowl-pox virus expressed T7 RNA polymerase [FPV-T7; (43)] into BHK cells which support genomic replication, assembly and release of infectious virions but not MNV infection. The yield of virus obtained after 24 h therefore reflects a single round of virus replication.
Virus yield from the m50 and m51 mutants showed relatively modest 15–21-fold reductions in virus titre after 24 h respectively, and in the case of m51, no effect on plaque size. In contrast, mutants m53 and m54 produce no detectable virus as determined by either TCID50 or plaque assay (log10 reductions of >2.9; Figure 8B). To determine whether it was disruption of the secondary structure or changes to the primary sequence that was responsible for the reduction in replication of the mutants, a further mutant, m53r was constructed that contained compensatory mutations at synonymous sites that restored RNA base pairings (Figure 7). Mutant m53r showed replication titres restored to close to wild-type levels, with log10 reductions in recovery of –0.62 (compared to an absence of replication of the original mutant m53). In all cases, western blot analysis of transfected cells demonstrated that the levels of the RdRp NS7 expressed during virus recovery was unaffected by the introduction of the mutations in m50, m51, m53 or m54 (Figure 8A).
Figure 8.
Replication ability of MNV with mutations that disrupted predicted RNA secondary structures in the 5′ end (m50, m51), NS/S junction (m53) and 3′ end (m54) of the genome (Figure 6) and MNV with compensatory mutations that restores base pairing in SLa5045. (A, C) Western blot analysis of NS7 levels during MNV recovery. Cells were infected with FPV-T7 and subsequently transfected with cDNA constructs containing WT or mutant MNV genomes under control of a T7 RNA polymerase promoter. Twenty-four hours post-transfection cells were harvested and the levels of NS7 determined by immunoblot using rabbit polyclonal anti-sera to MNV NS7. (B) Virus yield and plaque phenotype of viruses generated from either WT or mutant MNV cDNA constructs. Virus yields of mutants were expressed as log10 reductions in TCID50 per 35 mm dish from the WT control (mean value 50 100 ± 18 200). All virus recoveries were performed five times and mean reductions and standard deviations shown; the limit of detection was 60 TCID50/35 mm dish. (D) Virus yield and plaque phenotype of WT or mutant (m53 and m53r) cDNA constructs. Virus recoveries were performed five times with a mean recovery from the WT control of 14 160 ± 2500, and a limit of detection of 60 TCID50/35 mm dish.
DISCUSSION
Bioinformatic RNA secondary structure prediction
This study used a variety of established and newly developed RNA structure prediction methods to identify and characterize regions of secondary structure formation in each genus of the Caliciviridae. These approaches offer profound advantages over simple energy-minimizing algorithms such as MFOLD on single sequences used for previous RNA structure predictions in caliciviruses (40,38). The limitations of MFOLD and other energy-minimizing algorithms to predict RNA structure arise from difficulties in determining correct structures from a frequently wide range of alternative, similarly energetically favoured configurations. This problem is compounded by the observation that RNA may indeed not even adopt the most energetically favoured structure after transcription (44,45), particularly if chaperoned by proteins (such as the poly-pyrimidine tract-binding protein) into functionally active secondary and tertiary RNA structures that would otherwise not spontaneously form (46–49). Furthermore, the algorithms used by almost all RNA structure prediction methods cannot predict tertiary structure elements (pseudo-knots, kissing loops), yet these may contribute to the stability of secondary structures (50,51).
For all these reasons, we have been necessarily cautious and conservative in our structure predictions for caliciviruses. Predictions represent, in almost all cases, a consensus of several algorithmically independent methods that can exploit the increasing amount of comparative sequence data for each genus. Whole-genome sequences were initially analysed to localize sites and favoured orientation of RNA structures using high-resolution thermodynamic scanning. Although RNA structure is clearly not the only cause of suppression of sequence variability at synonymous sites, occurring for example also in overlapping gene sequences (52), there was an impressive co-localization of the major sites of suppression of synonymous site variability and thermodynamically predicted structure, that occurred in homologous positions in different calicivirus genera. These methods also identified the CRE of enteroviruses, the existence of which is defined both genetically and biochemically, and which therefore forms an effective ‘positive control’ for the methods used.
Prediction of specific pairings within regions of suspected RNA structure formation used the comparative thermodynamic prediction algorithm, Alifold, in which secondary structure predictions from RNAFold were weighted by their phylogenetic conservation and occurrence of co-variant sites (53). These predictions were compared directly with a non-thermodynamic-based method (PFold), in which pairing rules and prediction of specific pairings were determined using a reconstructed evolutionary history and stochastic context-free grammar to give a probability distribution of structures (27). Both methods produced strikingly concordant predictions during the analysis of divergent sequence datasets (Table 1).
RNA structure predictions were consistent with published analyses based on thermodynamic folding of individual sequences (40,38,39). This includes predictions for 3′ terminal hairpins in RHDV and FCV (40) and genogroup I noroviruses (39), a finding which we can generalize to all calicivirus groups and genera where suitable comparative data were available (Figure 5). Although frequently variable in position and size, structures at the 5′ end of the genome were a consistent feature within the first 150 bases of each analysed sequence alignment, again consistent with the position of a paired stem–loop found in group I noroviruses (38).
Notably, we also demonstrate the marked suppression of variability at synonymous sites which precisely coincides with predicted structures in the 5′ end of the genome, the NS/S region and—to a lesser extent—the 3′ end of the genome. SSSV would be expected wherever functions additional to encoding a single polypeptide occur, for example the presence of conserved binding motifs for cellular or viral proteins or overlapping (out-of-frame) alternative reading frames (ARFs). ARFs have been reported in sapovirus genogroups I and IV or V (54) of 161 aa or 155 aa (nt 5183–5665 or 5183–5647) respectively (but are absent from genogroup II) and in the Lagoviruses and Vesiviruses. Among noroviruses, MNV exhibits a 213 aa conserved ARF (35) in an analogous position (5069–5707 nt) and characterized by marked suppression of synonymous variation (ORF4, Figure 2B), while there is no comparable coding sequence in genogroups I and II. The beco/naboviruses also possess a similarly positioned +1 ARF of 87 amino acids conserved in all 31 sequences available for analysis (data not shown).
In the future, the availability of more sequences within the main genogroups of sapoviruses, genogroup I of noroviruses, sequences from the ocean virus group of vesiviruses and of beco/naboviruses should help complete the structure predictions for this virus family. Nevertheless, the data obtained here provides a valuable catalogue of bioinformatically well-supported RNA secondary structure predictions for future functional investigations in calicivirus cell culture models (FCV and MNV).
Role(s) of RNA secondary structure in calicivirus replication
It is likely that all positive-strand RNA viruses exhibit an underlying conservation of replication mechanisms, reflected in the caliciviruses for example in the common NS gene core and features of the RdRp (55). As a general phenomenon, positive-strand mammalian viruses use a wealth of RNA secondary structures to control or promote different steps in their replication cycles (20,56–59). In picornaviruses, CRE elements comprising stem–loops of varying lengths in different genome regions template the uridylylation of VPg protein (60), while 5′ UTRs contain extensive RNA secondary structures with demonstrated roles in translation initiation and replication (61–67). Replication and control structures are increasingly recognized in flaviviruses, including the well-characterized IRES of HCV and pestiviruses (68–70), and secondary structures required for genome replication within NS5B and perhaps the core coding sequences of HCV (23–25), as well as in the 3′UTRs of vector-borne flaviviruses (71,72) and pestiviruses (73). It is therefore unsurprising to identify conserved concordantly predicted RNA structures in caliciviruses, and encouraging that functional analysis demonstrates an essential role in replication.
To date, our understanding of the RNA structures or sequences required for calicivirus translation and replication has been very limited. Previous reports have identified a number of host cell factors, including the poly-pyrimidine tract-binding protein and La autoantigen, which interact with the 5′ and 3′ extremities of the Norwalk virus genome (39,42). A role for these interactions which were observed in vitro has yet to be described, although our recent work demonstrated that the interaction of the poly-pyrimidine tract-binding protein with the FCV genome is required for efficient virus replication in a temperature-dependent manner (75). This fits with the hypothesis that it functions as an RNA chaperone aiding the folding of the viral RNA into a conformation required for efficient virus replication.
Calicivirus genomes have short UTRs preceding coding regions, only 5 and 4 nt in the case of the MNV genomic and sgRNAs, respectively. UTRs in picornaviruses and other plus strand RNA viruses are much larger and are known to interact with the cellular translation machinery and/or the viral replicase components to control translation and/or replication. Given the lack of such UTRs in calicivirus genomes, it is likely the stem–loops present within the coding region play similar roles. We have previously determined that caliciviruses use a novel protein-directed translation initiation mechanism which relies on the interaction of eIF4E with the VPg protein covalently linked to the 5′ end of the viral genomic and sgRNA (76,77). It is possible that the stem–loop sequences within the 5′ end of the coding regions contribute to viral translation through interactions with either canonical or non-canonical translation initiation factors to direct ribosome assembly and translation initiation. However, although associated with SSSV, the actual predicted structures were highly variable both between genera, and in the case of noroviruses, also between genogroups. Furthermore, disruption of the two predicted stem–loops in the 5′ region of MNV showed only a limited effect on replication, with ∼20 fold reduction in titre, and a reduced plaque size for m50, phenotypes, observations that do not indicate a critical role in translation initiation. Although the m50 and m51 mutants showed similar expression of NS7 (Figure 8), the positions of SL8 and SL29 immediately downstream of the ribosomal binding site may indicate a role in controlling the rate of translation processivity; potential roles in translation and RNA replication that underlie the observed reductions in virus titre of the mutants can be readily dissected in the future using the MNV system.
RNA structures at the 3′ end of the genome were similarly variable in size between genogroups and genera, although each sequence set analysed showed evidence for phylogenetically conserved terminal stem–loops, consistent with previous predictions for individual sequences (40,39). Despite the variability between different genogroups within noroviruses [Figure 5; (39)], disruption of SL7330 (mutant m54) entirely destroyed its replication ability, implying an essential role of RNA structure and/or primary sequence, for example during the initiation of minus-strand replication.
Upstream from the 3′ terminal hairpin, MNV, vesiviruses, norovirus GII/4 show evidence for further stem–loops. Although we have not attempted to assess the functional role of the (extremely large) penultimate MNV RNA structure, previously published data reveal a potential role for stem–loops in the analogous position in FCV (42). While the FCV ORF3 protein is required for the generation of infectious FCV virions, the underlying RNA sequence also contains an essential cis-acting role for FCV replication, mapped by deletion analysis to a 5′ boundary between 7482 and 7512, and a 3′ boundary between 7542 and 7572 (using numbering in the current study). These experimentally determined limits align almost exactly to the stem–loop predicted for FCV sequences, and the associated region of SSSV (Figure 5B), strongly suggesting that the function of the replication element is mediated though interactions with a structured RNA sequence.
Overall, the most consistently detected RNA secondary structures were found around the NS/S junction within each genus of noroviruses (Figures 5 and 6). Unlike SL7330, mutagenesis studies in MNV were able to demonstrate a functional role for RNA structure rather than primary sequence; restoration of replication ability was observed for the m53r mutant containing compensatory changes that restored base pairing in the duplex region of the stem–loop (Figures 7 and 8).
Three features of the MNV structure and homologues in other calicivirus genera provide compelling evidence that they are components of a sg transcriptional promoter; (i) their predicted favoured location on the anti-genomic strand, (ii) their highly conserved positional relationship with the sg transcript and (iii) the unpaired nature of the 6-nt spacer between the structure and the sg transcript (actual or inferred). The stem–loops may for example interact directly with the viral RdRp complex to initiate transcription from a site made accessible by the unpaired spacer. The structures may mediate transcriptional control associated with the switch from genome replication to virus assembly and release through the synthesis of encoded structural proteins. The common features of the structure and location suggest that these viruses all utilize a conserved mechanism for initiation of the sg transcript.
The RNA structure prediction for lagoviruses (Figure 6) is moreover entirely consistent with the only published functional analysis of the sg promoter in caliciviruses (78). In vitro transcription using a recombinant RHDV RdRp from anti-genomic template sequences mapped the minimal promoter region to between positions –40 and –60 (between positions 5266 and 5246) and position +1 (i.e. 5296, the first base of the transcript) for correct initiation of sgRNA synthesis. This upstream boundary coincides precisely with the 5′ base of the predicted stem–loop on the anti-genomic strand of lagoviruses (Figure 6). These data suggest that the stem–loop and downstream 6 base unpaired region may be necessary and sufficient for sgRNA transcription in vivo in lagoviruses and perhaps in other genera too. Our mutational analysis of the stem–loop in MNV indeed demonstrates that it is critical for replication (Figure 7). Further studies are underway to characterize the nature of the defect in the m53 virus.
In summary, this study represents the first attempt to comprehensively map RNA secondary structures in caliciviruses. The development of more effective bioinformatic prediction methods that exploit the increasing amount of comparative sequence data for the different genera and genogroups of viruses in this family, combined with the development of experimental systems to study calicivirus replication in vitro provides the starting materials for future studies of the mechanisms of and molecular interactions in calicivirus replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The work was funded by a Wellcome Senior Fellowship awarded to Ian Goodfellow. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
REFERENCES
- 1.Thiel HJ, Konig M. Caliciviruses: an overview. Vet. Microbiol. 1999;69:55–62. doi: 10.1016/s0378-1135(99)00088-7. [DOI] [PubMed] [Google Scholar]
- 2.Green KY, Ando T, Balayan MS, Berke T, Clarke IN, Estes MK, Matson DO, Nakata S, Neill JD, Studdert MJ, et al. Taxonomy of the caliciviruses. J. Infect. Dis. 2000;181(Suppl 2):S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- 3.Oliver SL, Asobayire E, Dastjerdi AM, Bridger JC. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology. 2006;350:240–250. doi: 10.1016/j.virol.2006.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neill JD, Meyer RF, Seal BS. Genetic relatedness of the caliciviruses: San Miguel sea lion and vesicular exanthema of swine viruses constitute a single genotype within the Caliciviridae. J. Virol. 1995;69:4484–4488. doi: 10.1128/jvi.69.7.4484-4488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smiley JR, Chang KO, Hayes J, Vinje J, Saif LJ. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 2002;76:10089–10098. doi: 10.1128/JVI.76.20.10089-10098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smiley JR, Hoet AE, Traven M, Tsunemitsu H, Saif LJ. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J. Clin. Microbiol. 2003;41:3089–3099. doi: 10.1128/JCM.41.7.3089-3099.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 8.Hsu CC, Riley LK, Wills HM, Livingston RS. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 2006;56:247–251. [PubMed] [Google Scholar]
- 9.Hsu CC, Riley LK, Livingston RS. Molecular characterization of three novel murine noroviruses. Virus Genes. 2007;34:147–155. doi: 10.1007/s11262-006-0060-1. [DOI] [PubMed] [Google Scholar]
- 10.Clarke IN, Lambden PR. The molecular biology of caliciviruses. J. Gen. Virol. 1997;78:291–301. doi: 10.1099/0022-1317-78-2-291. [DOI] [PubMed] [Google Scholar]
- 11.Asanaka M, Atmar RL, Ruvolo V, Crawford SE, Neill FH, Estes MK. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl Acad. Sci. USA. 2005;102:10327–10332. doi: 10.1073/pnas.0408529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama K, Hansman GS, Oka T, Ogawa S, Takeda N. Investigation of norovirus replication in a human cell line. Arch. Virol. 2006;151:1291–1308. doi: 10.1007/s00705-005-0720-9. [DOI] [PubMed] [Google Scholar]
- 13.Clarke IN, Lambden PR. Organization and expression of calicivirus genes. J. Infect. Dis. 2000;181(Suppl 2):S309–S316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- 14.Straub TM, Honer zu BK, Orosz-Coghlan P, Dohnalkova A, Mayer BK, Bartholomew RA, Valdez CO, Bruckner-Lea CJ, Gerba CP, Abbaszadegan M, et al. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect Dis. 2007;13:396–403. doi: 10.3201/eid1303.060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sosnovtsev S, Green KY. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 16.Sosnovtsev SV, Garfield M, Green KY. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 2002;76:7060–7072. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhry Y, Skinner MA, Goodfellow I. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007;88:2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohll JB, Percy N, Ley R, Evans DJ, Almond JW, Barclay WS. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rieder E, Paul AV, Kim DW, van Boom JH, Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond JW, Barclay W, Evans DJ. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKnight KL, Lemon SM. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witwer C, Rauscher S, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 2001;29:5079–5089. doi: 10.1093/nar/29.24.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Shin H, Wimmer E, Paul AV. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 2004;78:10865–10877. doi: 10.1128/JVI.78.20.10865-10877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl Acad. Sci. USA. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B coding region sequences of hepatitis C virus by RNAse cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen B, Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- 28.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 30.Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Workman C, Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clote P, Ferre F, Kranakis E, Krizanc D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, MacKenzie JM, Green KY, Virgin HW. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thackray LB, Wobus CE, Chachu KA, Liu B, Alegre ER, Henderson KS, Kelley ST, Virgin HW. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen JS, Meyer IM, Forsberg R, Simmonds P, Hein J. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 2004;32:4925–4936. doi: 10.1093/nar/gkh839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart SR, Semler BL. RNA Determinants of Picornavirus Cap-Independent Translation Initiation. Semin. Virol. 1997;8:242–255. [Google Scholar]
- 38.Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 39.Gutierrez-Escolano AL, Vazquez-Ochoa M, Escobar-Herrera J, Hernandez-Acosta J. La, PTB, and PAB proteins bind to the 3(′) untranslated region of Norwalk virus genomic RNA. Biochem. Biophys. Res. Commun. 2003;311:759–766. doi: 10.1016/j.bbrc.2003.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seal BS, Neill JD, Ridpath JF. Predicted stem-loop structures and variation in nucleotide sequence of 3′ noncoding regions among animal calicivirus genomes. Virus Genes. 1994;8:243–247. doi: 10.1007/BF01704518. [DOI] [PubMed] [Google Scholar]
- 41.Neill JD, Reardon IM, Heinrikson RL. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 1991;65:5440–5447. doi: 10.1128/jvi.65.10.5440-5447.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sosnovtsev SV, Belliot G, Chang KO, Onwudiwe O, Green KY. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Britton P, Green P, Kottier S, Mawditt KL, Penzes Z, Cavanagh D, Skinner MA. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J. Gen. Virol. 1996;77:963–967. doi: 10.1099/0022-1317-77-5-963. [DOI] [PubMed] [Google Scholar]
- 44.Batey RT, Doudna JA. The parallel universe of RNA folding. Nat. Struct. Biol. 1998;5:337–340. doi: 10.1038/nsb0598-337. [DOI] [PubMed] [Google Scholar]
- 45.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 46.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 47.Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 49.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 50.Leontis NB, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 51.Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30:395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizokami M, Orito E, Ohba K, Ikeo K, Lau JY, Gojobori T. Constrained evolution with respect to gene overlap of hepatitis B virus. J. Mol. Evol. 1997;44(Suppl 1):S83–S90. doi: 10.1007/pl00000061. [DOI] [PubMed] [Google Scholar]
- 53.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu BL, Clarke IN, Caul EO, Lambden PR. Human enteric caliciviruses have a unique genome structure and are distinct from the Norwalk-like viruses. Arch. Virol. 1995;140:1345–1356. doi: 10.1007/BF01322662. [DOI] [PubMed] [Google Scholar]
- 55.Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 56.Joost Haasnoot PC, Olsthoorn RC, Bol JF. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA. 2002;8:110–122. doi: 10.1017/s1355838202012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiang W, Paul AV, Wimmer E. RNA signals in Entero- and Rhinovirus genome replication. Semin. Virol. 1997;8:256–273. [Google Scholar]
- 58.Huthoff H, Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41:10439–10445. doi: 10.1021/bi025993n. [DOI] [PubMed] [Google Scholar]
- 59.Schlesinger S, Makino S, Linial ML. Cis-Acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin. Virol. 1994;5:39–49. doi: 10.1006/smvy.1994.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodfellow IG, Kerrigan D, Evans DJ. Structure and function of the poliovirus cis-acting replication element (CRE) RNA. 2003;I9:124–137. doi: 10.1261/rna.2950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 62.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 63.Xiang W, Harris KS, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 65.Mirmomeni MH, Hughes PJ, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mellits KH, Meredith JM, Rohll JB, Evans DJ, Almond JW. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J. Gen. Virol. 1998;79(Pt 7):1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- 67.Rohll JB, Moon DH, Evans DJ, Almond JW. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poole TL, Wang CY, Popp RA, Potgieter LND, Siddiqui A, Marc S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 69.Tsukiyama Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen CJ, Kuo MD, Chien LJ, Hsu SL, Wang YM, Lin JH. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khromykh AA, Kondratieva N, Sgro JY, Palmenberg A, Westaway EG. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 2003;77:10623–10629. doi: 10.1128/JVI.77.19.10623-10629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isken O, Grassmann CW, Yu H, Behrens SE. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu HY, Grassmann CW, Behrens SE. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: Functional role during RNA replication. J. Virol. 1999;73:3638–3648. doi: 10.1128/jvi.73.5.3638-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karakasiliotis I, Chaudhry Y, Roberts LO, Goodfellow IG. Feline calicivirus replication: requirement for polypyrimidine tract-binding protein is temperature-dependent. J. Gen. Virol. 2006;87:3339–3347. doi: 10.1099/vir.0.82153-0. [DOI] [PubMed] [Google Scholar]
- 76.Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006;281:25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- 77.Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morales M, Barcena J, Ramirez MA, Boga JA, Parra F, Torres JM. Synthesis in vitro of rabbit hemorrhagic disease virus subgenomic RNA by internal initiation on (-)sense genomic RNA: mapping of a subgenomic promoter. J. Biol. Chem. 2004;279:17013–17018. doi: 10.1074/jbc.M313674200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.