Abstract
The A site of the small ribosomal subunit participates in the fidelity of decoding by switching between two states, a resting ‘off’ state and an active decoding ‘on’ state. Eight crystal structures of RNA duplexes containing two minimal decoding A sites of the Homo sapiens mitochondrial wild-type, the A1555G mutant or bacteria have been solved. The resting ‘off’ state of the mitochondrial wild-type A site is surprisingly different from that of the bacterial A site. The mitochondrial A1555G mutant has two types of the ‘off’ states; one is similar to the mitochondrial wild-type ‘off’ state and the other is similar to the bacterial ‘off’ state. Our present results indicate that the dynamics of the A site in bacteria and mitochondria are different, a property probably related to the small number of tRNAs used for decoding in mitochondria. Based on these structures, we propose a hypothesis for the molecular mechanism of non-syndromic hearing loss due to the mitochondrial A1555G mutation.
INTRODUCTION
Fidelity of decoding during protein biosynthesis can be divided into two steps; (i) initial selection and (ii) proofreading (1,2). In the first step, the non-cognate tRNA is discriminated by the difference in free energy of base pairings between the codon and cognate tRNA anticodon. In the second step, the aminoacyl-tRNA decoding site (or the A site) acts as an RNA molecular switch upon recognition of the codon–anticodon pair and provokes an irreversible step of GTP hydrolysis. These two steps of decoding guarantee the high fidelity of translation. Crystal structures of various bacterial 30S and 70S ribosomes have revealed the molecular mechanisms of the proofreading step at atomic level (3–10). When the cognate tRNA is delivered to the A site, the A-site molecular switch changes its conformation from an ‘off’ state to an ‘on’ state. These two local states are linked to the ‘open’ and ‘closed’ global states of the ribosome during decoding, as observed by Ramakrishnan and coworkers (1,2,6), but the connecting mechanisms are not fully described yet. In the ‘off’ state, two adenine residues, A1492 and A1493, do not make any interaction with codon–anticodon helix. On the other hand, in the ‘on’ state, these two adenines fully bulge out and come together with G530 in the shoulder domain of the 30S subunit to recognize the first two Watson-Crick base pair of codon–anticodon mini-helix and induce ribosomal transitions from the ‘open’ to the ‘closed’ forms (1,2,6). Recently, functional characteristics and tertiary structures of the A-site molecular switch in Homo sapiens cytoplasms have been revealed by X-ray analyses of RNA fragments containing the minimal A site (11), but those in H. sapiens mitochondria are still unknown.
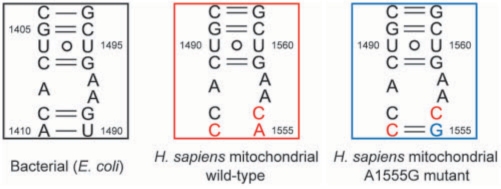
The secondary structure of the H. sapiens mitochondrial A site is very similar to that of the bacterial A site (Figure 1) (12). Differences are found at residue 1556 (1491 in bacterial numbering), which is a C in mitochondria and a G in bacteria, and at the base pair between 1494 and 1555 (1410 and 1490 in bacterial numbering), which is a CoA opposition in mitochondria and an A-U (66.5%) or a G=C (26.4%) base pair in bacteria (13). It is important to note that residue 1492 (1408 in bacterial numbering) is an A in the H. sapiens mitochondrial as in bacterial A sites, while it is a G in the H. sapiens cytoplasmic A site (11,12). Because of the similarity in the secondary structure of the A site, it has long been believed that the H. sapiens mitochondrial A site has functional characteristics and tertiary structure similar to those of the bacterial A site.
Figure 1.
Secondary structures of the bacterial, H. sapiens mitochondrial wild-type and its A1555G mutant A sites. The nucleotides different from those in the bacterial A site are colored in red. The G1555 residue in the A1555G mutant is colored in blue.
However, the structural components of the H. sapiens mitochondrial ribosome are noticeably different from those of the bacterial ribosome. The mitochondrial ribosome is formed by a small 28S subunit and a large 39S subunit. The small subunit consists of a 12S rRNA and 29 proteins (14–16), while the large subunit consists of a 16S rRNA and 48 proteins (14,17,18). The 5S rRNA, which is present in the bacterial ribosome, is absent in the mitochondrial ribosome. The ratio of protein to rRNA mass in the mitochondrial ribosome (2:1) is inverted from the ratio found in the bacterial ribosome (1:2). A surprising feature of the H. sapiens mitochondrial system is the use of an extended decoding mechanism, which allows translation of all 60 codons with only 22 tRNA species (19–23). Mammalian mitochondrial translation does not use nucleus-encoded tRNAs, although mitochondrial import of some tRNAs occurs in eukaryotic microorganisms (protozoa, fungi, algae), in some plants and in a few animals (24,25). In addition, tRNA modifications at the wobble position of the anticodon, where post-transcriptionally modified nucleotides often play an essential role in the precise decoding of the genetic code in bacteria and eukaryotic cytoplasms (26), are less frequent in mitochondria (23,27). In other words, in contrast to Escherichia coli with 45 tRNA species (or 41 anticodons) and H. sapiens cytoplasms with 46 tRNA species (28,29), mitochondria must gain flexibility (each tRNA decodes two or four codons) in the decoding process (20–22) without loosing fidelity. Therefore, it is possible that mitochondria have unique proofreading mechanism in the decoding process to accommodate flexibility and fidelity.
The A1555G mutant of the H. sapiens mitochondrial A site has been found in more than 120 families throughout the world and is one of the most common genetic causes of non-syndromic hearing loss (30–32). In the A1555G mutant A site, the G1555 residue is expected to form a Watson-Crick base pair with the C1494 residue on the opposite strand rendering it closer to the bacterial A site (Figure 1).
In the present study, we have solved eight crystal structures of RNA fragments containing two minimal A sites of the H. sapiens mitochondrial wild-type, the A1555G mutant or bacteria. Comparative structural analyses provided us with insights into the decoding mechanisms in the mitochondrial and bacterial ribosomes. These observations lead to a hypothesis for the molecular mechanism of non-syndromic hearing loss due to the A1555G mutation in the mitochondrial A site.
MATERIALS AND METHODS
Crystallization
Two internal loops of the bacterial A site, the H. sapiens mitochondrial A site or its A1555G mutant were inserted between Watson-Crick pairs in sequences designed to fold as a double helix (BACT, WT and MUT, respectively) (Supplementary Figures 1, 2 and 4). The RNA oligomers were chemically synthesized by Dharmacon (Boulder, CO) and purified by HPLC and reverse phase chromatography. To resolve phase, bromine derivatives of WT and MUT with 5-bromouracil residues (BrU) at the central stem were also synthesized (WT-Br, MUT-Br1 and MUT-Br2, respectively) (Supplementary Figures 3, 5–8). Before crystallization, 4 mM paromomycin or tobramycin solution containing 50 mM sodium cacodylate buffer (pH 6.5) was prepared, and 2 mM RNA solutions containing 100 mM sodium cacodylate (pH 6.5) and 25 mM sodium chloride were annealed by heating at 85°C for 2 min followed by slow cooling to 37°C. Same volumes of RNA solution and paromomycin or tobramycin solution were mixed at 37°C and then cooled slowly to room temperature (21–25°C). Crystallizations were performed by the hanging-drop vapor diffusion method at 20°C and 37°C by mixing 1 μl of RNA/aminoglycoside solution and 1 μl of crystallization solution containing 50 mM sodium cacodylate (pH 7.0), 0–2 mM spermine tetrahydrochloride, 50–200 mM potassium chloride, 20–200 mM strontium chloride, 0–5% (v/v) glycerol and 1–5% (v/v) 2-methyl-2,4-pentanediol. Crystals suitable for X-ray experiments were obtained after optimization of crystallization conditions. For the bacterial A-site RNA, the BACT-Co crystal was obtained in the condition containing hexammine cobalt chloride (Table 1 and Supplementary Figure 1). For the mitochondrial A-site RNA, two types of crystals (WT-K and WT-Br-K) were obtained in conditions containing potassium chloride (Table 1 and Supplementary Figures 2 and 3). And for the A1555G mutant RNA, five types of crystals (MUT-K, MUT-Br1-Sr, MUT-Br1-K1, MUT-Br1-K2 and MUT-Br2-Co) were obtained in conditions containing potassium chloride, strontium chloride or hexammine cobalt chloride (Table 1 and Supplementary Figures 4–8).
Table 1.
Crystal data, statistics of data collections and statistics of structure refinements
| Bacterial | H. sapiens mitochondrial wild type | H. sapiens mitochondrial A1555G mutant | ||||||
|---|---|---|---|---|---|---|---|---|
| Crystal code | BACT-Co | WT-K | WT-Br-K | MUT-K | MUT-Br1-Sr | MUT-Br1-K1 | MUT-Br1-K2 | MUT-Br2-Co |
| Aminoglycoside | Paromomycin | Paromomycin | Paromomycin | Paromomycin | Paromomycin | Paromomycin | Tobramycin | Paromomycin |
| PDB ID | 3BNL | 3BNN | 3BNO | 3BNP | 3BNQ | 3BNR | 3BNS | 3BNT |
| Crystal data | ||||||||
| Space group | I4 | P1 | P1 | P3121 | C2 | C2 | C2 | P64 |
| Unit cell (Å) (°) | a=b=53.6, c=63.5 | a=34.6, b=44.5, c=52.4 | a=35.2, b=45.5, c=52.2 | a=b=66.6, c=57.6 | a=71.3, b=76.1, c=55.9 | a=71.2, b=77.4, c=56.3 | a=72.7, b=77.1, c=56.5 | a=b=74.9, c=22.5 |
| α=68.8, β=71.3, γ=67.7 | α=71.4, β=73.4, γ=74.0 | β=117.1 | β=115.1 | β=115.7 | ||||
| Za | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 0.5 |
| Data collection | ||||||||
| Beamline | BM30 of ESRF | PX of SLS | ID23-1 of ESRF | PX of SLS | ID23-1 of ESRF* | ID23-1 of ESRF | ID14-2 of ESRF | BM30 of ESRF* |
| Wavelength (Å) | 0.91636 | 0.9999 | 0.8856 | 0.9801 | 0.92070/0.92110/0.91625 | 0.91625 | 0.933 | 0.91950/0.91984/0.91637 |
| Resolution (Å) | 36.8–2.6 | 39.6–2.0 | 29.0–2.35 | 57.7–2.7 | 49.8–2.0/49.8–2.0/49.8–2.0 | 51.0–2.1 | 35.4–1.9 | 37.4–2.3/37.4–2.4/37.4–2.3 |
| Of the outer shell (Å) | 2.7–2.6 | 2.1–2.0 | 2.4-2.35 | 2.9–2.7 | 2.1–2.0/2.1–2.0/2.1–2.0 | 2.2–2.1 | 2.0–1.9 | 2.4–2.3/2.5–2.4/2.4–2.3 |
| Unique reflections | 5295 | 17270 | 11751 | 4263 | 17465/17512/17519 | 16388 | 21598 | 3368/2981/3374 |
| Completeness (%) | 99.9 | 97.0 | 98.0 | 99.4 | 97.4/97.4/97.4 | 99.8 | 97.4 | 100.0/99.9/99.9 |
| In the outer shell (%) | 100.0 | 92.6 | 97.7 | 100.0 | 85.6/85.4/85.5 | 100.0 | 97.4 | 100.0/100.0/100.0 |
| Rmergeb (%) | 5.1 | 9.6 | 9.9 | 7.8 | 4.9/4.9/5.0 | 11.6 | 3.1 | 8.4/9.5/7.2 |
| In the outer shell (%) | 28.6 | 36.9 | 39.0 | 36.4 | 28.0/33.2/37.3 | 37.8 | 37.8 | 30.4/26.9/25.7 |
| Ranomc (%) | – | – | – | – | 5.1/4.0/4.5 | – | – | 6.6/6.5/5.2 |
| In the outer shell (%) | – | – | – | – | 24.6/29.0/33.1 | – | – | 19.0/16.0/15.5 |
| Redundancy | 7.1 | 2.7 | 7.7 | 9.9 | 3.5/3.5/3.5 | 6.9 | 3.8 | 9.9/9.9/9.9 |
| In the outer shell | 7.3 | 2.6 | 8.0 | 10.2 | 2.6/2.6/2.6 | 7.1 | 3.8 | 10.5/10.5/10.5 |
| Structure refinement | ||||||||
| Resolution range (Å) | 36.0–2.6 | 39.6–2.0 | 36.9–2.35 | 40.0–2.7 | 49.8–2.0 | 51.0–2.1 | 35.4–1.9 | 37.4–2.3 |
| Used reflections | 5278 | 17269 | 11718 | 4204 | 17514 | 16147 | 21594 | 3371 |
| R-factord (%) | 22.0 | 23.8 | 22.6 | 23.1 | 22.9 | 21.5 | 24.7 | 21.7 |
| Rfreee | 25.1 | 26.4 | 25.5 | 27.0 | 26.9 | 24.5 | 26.0 | 25.5 |
| Number of DNA atoms | 940 | 1788 | 1788 | 908 | 1942 | 1884 | 1884 | 464 |
| Number of cations | 8 [Co(NH3)6]3+ | – | – | – | 8 Sr2+, 2 K+ | 2 K+ | 2 K+ | 1 [Co(NH3)6]3+, 1 Na+ |
| Number of water | 38 | 114 | 31 | 13 | 191 | 183 | 102 | 55 |
| Number of aminoglycoside | None | None | None | None | 1 (non-specific) | 1 (non-specific) | None | None |
| RMSD | ||||||||
| Bond length (Å) | 0.004 | 0.005 | 0.005 | 0.005 | 0.004 | 0.004 | 0.004 | 0.004 |
| Bond angles (°) | 0.9 | 0.9 | 0.9 | 1.0 | 0.8 | 0.9 | 0.8 | 0.8 |
| Improper angles (°) | 1.3 | 0.7 | 0.7 | 0.9 | 0.7 | 0.7 | 0.7 | 1.3 |
aNumber of dsRNA in the asymmetric unit.
bRmerge=100 × Σhklj|Ihklj – <Ihklj>| / Σhklj < Ihklj>.
cRanom=100 × Σhklj|Ihklj(+) – Ihklj(−)| / Σhklj[Ihklj(+) + Ihklj(−)].
dR-factor=100 × Σ||Fo| – |Fc|| / Σ|Fo|, where |Fo| and |Fc| are optimally scaled observed and calculated structure factor amplitudes, respectively.
eCalculated using a random set containing 10% of observations that were not included throughout refinement (58).
*For phase determination with the multiple anomalous diffraction (MAD) method, three datasets were collected with three wavelengths. Statistics from left to right are of peak, edge and remote data, respectively.
Data collection, structure determination and refinement
X-ray data of eight crystals were collected at 100 K with synchrotron radiation at the ID14-2, ID23-1 or BM30 beamline in the European Synchrotron Radiation Facility (ESRF; Grenoble, France), or at the PX beamline in the Swiss Light Source (SLS; Villingen, Switzerland). Each dataset was processed by using the program Crystalclear (Rigaku/MSC) or MOSFLM (33,34), and was scaled and merged using Crystalclear or SCALA from the CCP4 suite of crystallographic programs (35).
The BACT-Co crystal has I4 spacegroup (Table 1), which is different from any bacterial A-site crystals obtained previously in conditions without hexammine cobalt chloride (P212121, P21 or P41) (36–41). Two crystals of the mitochondrial A-site RNA, WT-K and WT-Br-K, have the same spacegroup P1 with similar unit cell dimensions, suggesting their isomorphism (Table 1). Three different crystal forms were obtained for the A1555G mutant-RNA, P3121, C2 and P64 (Table 1).
For MAD phasing using the anomalous scattering of bromine atoms, X-ray data of MUT-Br1-Sr and MUT-Br2-Co were taken with three different wavelengths based on XAFS measurements (Table 1). Initial phases were estimated by the MAD method using the program SOLVE (42) with figure-of-merit of 0.40 and 0.46, respectively. Electron densities, modified by solvent flattening with the program CNS (43), showed clearly the phosphate-ribose backbone with the individual bases. The molecular structures of MUT-Br1-Sr and MUT-Br2-Co were constructed on a graphic workstation with the program O (44). Initial phases of other crystals were determined by the Molecular Replacement method with the program AMoRe (45) using the bulk-solvent technique (46,47). The details of phase determinations will be discussed elsewhere.
The atomic parameters of each structure were refined with the program CNS (43) through a combination of simulated-annealing, crystallographic conjugate gradient minimization refinements and B-factor refinements, followed by interpretations of the omit map at every nucleotide residue. The statistics of structure refinements are summarized in Table 1. Electron density indicating specific binding of aminoglycosides to the A site could not be observed in any of the crystal structures, indicating that complex formation did not occur under the present conditions. In MUT-Br1-Sr and MUT-Br1-K1, a non-specific binding of paromomycin between two symmetrically related RNA duplexes was observed (Supplementary Figures 5 and 6).
The adiabatic morphing calculations between the ‘off’ and ‘on’ states of the A-site molecular switches were performed with the program CNS (43) using a programmed input file morph_dist.inp (48,49). All figures were drawn using the PyMOL Molecular Graphics system (2002) DeLano Scientific, San Carlos, CA (http://www.pymol.org).
Coordinates
The atomic coordinates have been deposited in the Protein Data Bank (PDB) with the ID codes 3BNL (BACT-Co), 3BNN (WT-K), 3BNO (WT-Br-K), 3BNP (MUT-K), 3BNQ (MUT-Br1-Sr), 3BNR (MUT-Br1-K1), 3BNS (MUT-Br1-K2) and 3BNT (MUT-Br2-Co).
RESULTS
Overview of the A-site structures
In all cases, the two RNA strands form a duplex (Supplementary Figures 1–8). At the center of the duplex, four contiguous Watson-Crick G=C base pairs are formed. In the case of bromine derivatives, two of four G=C base pairs are replaced with the Watson-Crick A-BrU base pairs. Three Watson-Crick G=C base pairs close the stem at both ends of the duplex. Some of the terminal overhanging residues at the 5′-end (UU or C) are involved in crystal packing interactions and others are disordered in the solvent channel.
The BACT-Co crystal contains an asymmetrical RNA duplex (Supplementary Figure 1). Two A sites in the duplex have almost identical conformations except for the A1492 residue (Supplementary Figure 9a), and are similar to the bacterial ‘off’ state with tucked-in A1492 and A1493 found in the Thermus thermophilus 30S ribosomal particle [PDB ID: 1J5E in (3)].
For the H. sapiens mitochondrial A site, two types of crystals with P1 spacegroup, WT-K and WT-Br-K, were obtained (Table 1). In each crystal, two RNA duplexes are in the asymmetric unit (Supplementary Figures 2 and 3). Therefore, a total of eight internal loops are observed. Although four of these eight A-site internal loops are free from any intermolecular interactions in crystalline state and four others are involved in crystal packing, all of them have identical conformations with a bulged-out residue, A1558 (A1493 in bacterial numbering), and a tucked-in residue, A1557 (A1492 in bacterial numbering) (Supplementary Figure 9b), suggesting that there is no crystal-packing effect on this conformation of the A site. Since this conformation is different from the ‘on’ states of the bacterial and H. sapiens cytoplasmic A sites with bulged-out A1492 and A1493 [PDB ID: 1IBM in (5); 2FQN in (11)], it should correspond to the ‘off’ state of the mitochondrial wild-type A site.
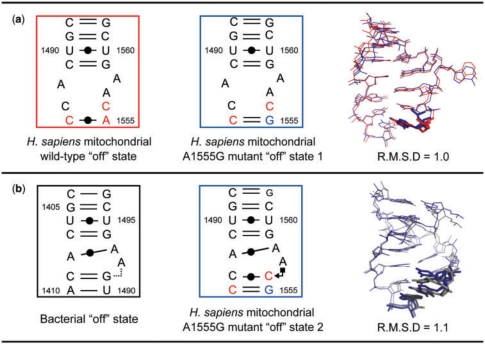
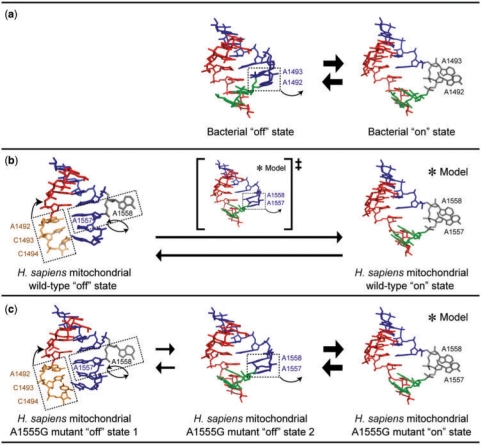
For the A1555G mutant of the H. sapiens mitochondrial A site, five types of crystal structures were solved (Table 1). Eight unique duplexes (i.e. 16 A sites) were identified in these structures (Supplementary Figures 4–8). According to their structural properties, they are divided into two types of conformations: (i) a conformation with bulged-out A1558 and tucked-in A1557 (Supplementary Figure 9c) and (ii) a conformation with both A1558 and A1557 tucked-in (Supplementary Figure 9d). These two conformations are observed in crystals with different spacegroups, again suggesting that there is no crystal-packing effect on these conformations of the A site. The former one is almost identical with the ‘off’ state of the mitochondrial wild-type A site mentioned above (Figure 2a). On the other hand, the latter one is almost identical with the bacterial ‘off’ state found in the BACT-Co crystal mentioned above (Figure 2b). Therefore, both conformations should correspond to the ‘off’ state of the mitochondrial A1555G mutant. For convenience, we name them the first and second ‘off’ states of the mitochondrial A1555G mutant, respectively.
Figure 2.
Superimpositions between the ‘off’ state of the H. sapiens mitochondrial wild-type A site (red) and the first ‘off’ state of the A1555G mutant A site (blue) (a) and between the second ‘off’ state of the A1555G mutant A site (blue) and the ‘off’ state of the bacterial A site (gray) (b). The nucleotides different from those in each other are drawn in stick. In figures, geometric nomenclature and classification of nucleic acid base pairs are according to the Classification in (57).
Hereafter, we will detail each conformation of the A site using the best defined structure with lower temperature factor and better identified solvent peaks (Figures 3–6).
Figure 3.
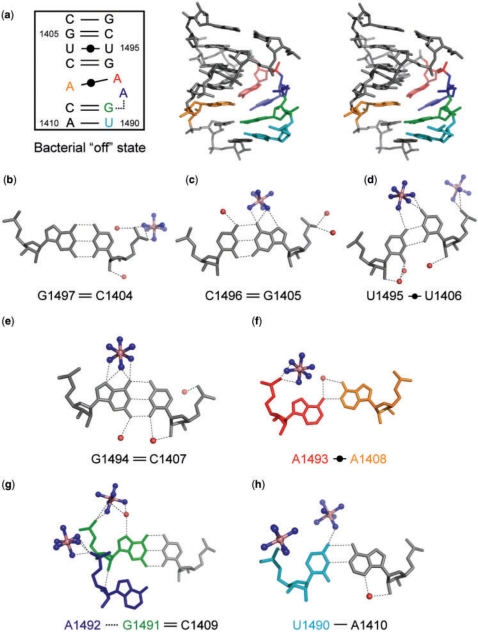
The bacterial A site in the ‘off’ state. (a) Secondary structure and stereoview. (b–h) Atomic details of each base pair of the A site. Three universally conserved adenine residues, A1408, A1492 and A1493, are colored in orange, blue and red, respectively. Two nucleotides different from those in the mitochondrial A site, U1490 and G1491, are colored in cyan and green, respectively. The hydrogen bonds are represented by black dashed lines.
Figure 4.
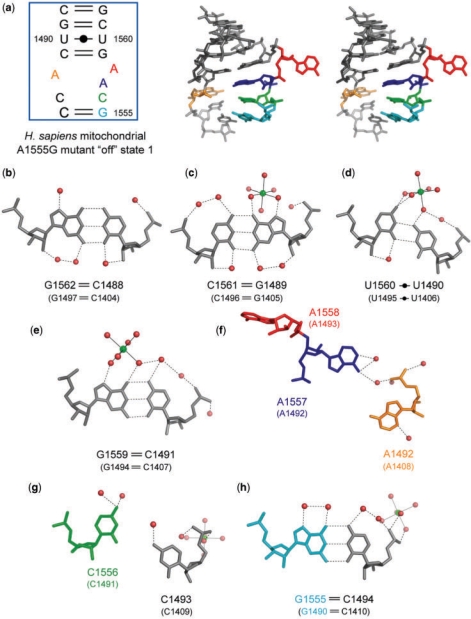
The H. sapiens mitochondrial A site in the ‘off’ state. (a) Secondary structure and stereoview. (b–h) Atomic details of each base pair of the A site. Three universally conserved adenine residues, A1492, A1557 and A1558 (A1408, A1492 and A1493 in bacterial numbering), are colored in orange, blue and red, respectively. Two nucleotides different from those in the bacterial A site, A1555 and C1556 (A1490 and C1491 in bacterial numbering), are colored in cyan and green, respectively. The hydrogen bonds are represented by black dashed lines.
Figure 5.
The A1555G mutant of the H. sapiens mitochondrial A site in the first ‘off’ state. (a) Secondary structure and stereoview. (b–h) Atomic details of each base pair of the A site. Three universally conserved adenine residues, A1492, A1557 and A1558 (A1408, A1492 and A1493 in bacterial numbering), are colored in orange, blue and red, respectively. Two nucleotides different from those in the bacterial A site, G1555 and C1556 (G1490 and C1491 in bacterial numbering), are colored in cyan and green, respectively. The hydrogen bonds are represented by black dashed lines.
Figure 6.
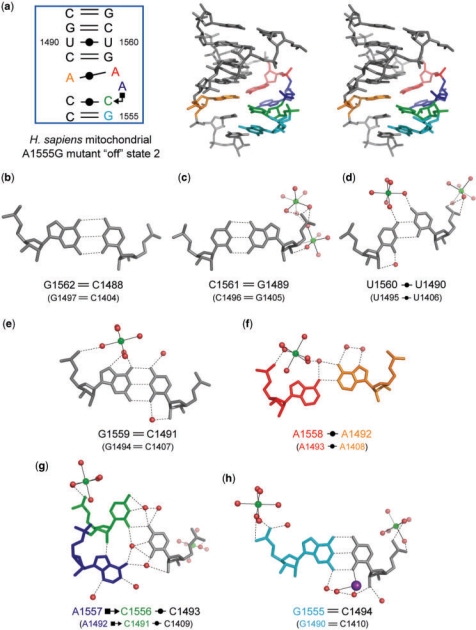
The A1555G mutant of the H. sapiens mitochondrial A site in the second ‘off’ state. (a) Secondary structure and stereoview. (b–h) Atomic details of each base pair of the A site. Three universally conserved adenine residues, A1492, A1557 and A1558 (A1408, A1492 and A1493 in bacterial numbering), are colored in orange, blue and red, respectively. Two nucleotides different from those in bacterial A site, G1555 and C1556 (G1490 and C1491 in bacterial numbering), are colored in cyan and green, respectively. The hydrogen bonds are represented by black dashed lines.
Structure of the bacterial A site in the ‘off’ state
A stereoview with the secondary structure of the bacterial A site in the ‘off’ state found in the BACT-Co crystal is shown in Figure 3a. Its conformation is similar to the bacterial ‘off’ state found in the T. thermophilus 30S ribosomal particle with a root mean square deviation (RMSD) of 1.1 Å [PDB ID: 1J5E in (3)]. In addition, all base pairs and base-stacking columns except residues A1492 and A1493 are present in these two bacterial ‘off’ states and in the bacterial ‘on’ state found in the T. thermophilus 30S ribosomal particle in complex with the cognate tRNA [PDB ID: 1IBM in (5)].
At the top side of the A-site internal loop, four Watson-Crick base pairs, C1404=G1497, G1405=C1496, U1406oU1495 and C1407=G1494, form (Figure 3b–e). At the bottom of the A-site internal loop, the Watson-Crick C1409=G1491 and A1410-U1490 base pairs form (Figure 3g and h). The A1493 residue stays inside the A-site helix and forms a Watson-Crick AoA base pair with the universally conserved A1408 residue through a single hydrogen bond N6-H…N1 (Figure 3f). On the other hand, the A1492 residue adopts different conformations in the two A sites of the asymmetrical duplex; one occupies the shallow/minor groove of the C1409=G1491 base pair and interacts with G1491 through a single hydrogen bond O4′…H-O2′ (Figure 3g), and the other one is fully bulged out to the solvent region (see red-line strand in Supplementary Figure 9a). There are three different base-stacking columns; one contains all bases on the short strand, the others are U1490-G1491 and A1492-A1493-G1494-U1495-C1496-G1497 on the long strand (Supplementary Figure 10a).
Structure of the H. sapiens mitochondrial A site in the ‘off’ state
A stereoview with the secondary structure of the H. sapiens mitochondrial A site in the ‘off’ state is shown in Figure 4a. Its conformation is surprisingly different from that of the bacterial ‘off’ state found in the BACT-Co crystal mentioned above (RMSD = 5.3 Å).
At the top side of the A-site internal loop, four Watson-Crick base pairs, C1488=G1562, G1489=C1561, U1490oU1560 and C1491=G1559, form as observed in the bacterial ‘off’ state mentioned above (Figure 4b–e). However, the geometry of the U1490oU1560 base pair (U1560 points into the deep/major groove site) is different from that in the bacterial ‘off’ state (compare Figure 4d with Figure 3d). At the bottom of the A-site internal loop, the Watson-Crick A1555oC1494 base pair with only one hydrogen bond N1…H-N4 closes the internal loop (Figure 4h). The universally conserved A1492 residue (A1408 in bacterial numbering) bulges out from the Watson-Crick stem, and does not form any base pair (Figure 4f). The subsequent C1493 residue is sandwiched between A1492 and C1494, and also does not have any partner to form a base pair in the internal loop of the A site (Figure 4g). Consequently, the short RNA strand of the A-site internal loop has two different stacking columns; C1488-G1489-U1490-C1491 and A1492-C1493-C1494 (Supplementary Figure 10b). On the long RNA strand of the A-site internal loop, the two adenine residues (which recognize the first two Watson-Crick base pairs of the codon–anticodon helix), A1558 and A1557 (A1493 and A1492 in bacterial numbering), have different conformations (Figure 4f). The A1558 residue is fully bulged out to the solvent region. On the other hand, the A1557 residue is pushed in the A-site helix and is stabilized by the C1556-A1557-G1559 stacking interaction. Although A1557 does not form any base pair, water mediated hydrogen bonds are observed between the Watson-Crick edge of A1557 and the O2P atom of A1492. As a result, all residues on the long strand of the A-site loop except A1558 are involved in the same stacking column (Supplementary Figure 10b). To form these bulged conformations, A1557 and A1558 adopt C2′-endo sugar puckers and have characteristic torsion angles around the P-O5′ (α), C5′-C4′ (γ) and O3′-Pn+1 (ζ). Small α (92°) and ζ (121°) angles and large γ (126°) angle allow the A1558 residue to protrude from the A-site helix into the solvent region, and small α (62°) and γ (50°) angles and large ζ (308°) angle allow the A1557 residue to be bulged into the A-site helix.
Structure of the A1555G mutant of the H. sapiens mitochondrial A site in the first ‘off’ state
A stereoview with the secondary structure of the A1555G mutant of the H. sapiens mitochondrial A site in the first ‘off’ state is shown in Figure 5a. Its overall conformation is very similar to that of the ‘off’ state of the mitochondrial wild-type A site mentioned above (RMSD = 1.0Å) (Figure 2a). Only one difference between them is found at the bottom of the A-site internal loop. As expected, a Watson-Crick G1555=C1494 base pair is observed in the A1555G mutant (Figure 5h) instead of the A1555oC1494 base pair in the wild-type (Figure 4h). As observed in the wild-type ‘off’ state, there are three stacking columns; one contains all residues on the long strand except A1558, and others are C1488-G1489-U1490-C1491 and A1492-C1493-C1494 on the short strand (Supplementary Figure 10c).
Structure of the A1555G mutant of the H. sapiens mitochondrial A site in the second ‘off’ state
A stereoview with the secondary structure of the A1555G mutant of the H. sapiens mitochondrial A site in the second ‘off’ state is shown in Figure 6a. Its conformation is completely different from that of the first ‘off’ state of the A1555G mutant just discussed (RMSD = 4.5 Å). On the other hand, it is almost identical with the bacterial ‘off’ state found in the BACT-Co crystal (RMSD = 1.1 Å) (Figure 2b). Differences between them are found only at the bottom of the A-site internal loop. The C1493 residue (C1409 in bacterial numbering) forms a Watson-Crick C1493oC1556 base pair with only one hydrogen bond N4-H…O2 (Figure 6g) instead of the Watson-Crick C=G base pair in bacteria (Figure 3g). Since C1556 points into the deep/major groove side, there is a large space at the shallow/minor groove edge of C1556 occupied by A1557 (A1492 in bacterial numbering). The A1557 residue forms a cis Hoogsteen/Sugar-edge base pair with C1556 through a direct hydrogen bond N7…H-O2′ and a water-mediated hydrogen bond N6-H…W…O2 (Figure 6g). Two water-mediated hydrogen bonds, N6-H…W…O2 and N6-H…W…N3, are observed between A1557 and C1493 (Figure 6g). At the bottom of the A-site helix, the Watson-Crick C1494=G1555 base pair forms (Figure 6h) instead of the Watson-Crick A-U base pair in bacteria (Figure 3h). Therefore, as observed in the bacterial ‘off’ state found in the BACT-Co crystal, there are three different base-stacking columns. All bases on the short strand make one stacking column. On the other hand, the long strand has two different base-stacking columns; G1555-C1556 and A1557-A1558-G1559-U1560-C1561-G1562 (Supplementary Figure 10d).
The ‘on’ states of the H. sapiens mitochondrial wild-type and A1555G mutant A sites
According to observations by Ramakrishnan and coworkers on the bacterial A-site molecular switch on the 30S and 70S ribosomes (3–10), the two adenine residues, A1492 and A1493, bulge out of the A-site helix and decode the first two base pairs of the codon–anticodon minihelix by recognizing their shallow/minor grooves and forming A-minor contacts. This conformation of the A site, called the ‘on’ state, has been observed also in several crystal structures of RNA duplexes containing the bacterial A site even in the absence of aminoglycosides (50,51). In these cases, two bulged-out adenines make the A-minor contacts with a Watson-Crick stem of a neighboring duplex, which mimics the cognate codon–anticodon stem. By analogy with the secondary structure of the bacterial A site, the H. sapiens mitochondrial wild-type and A1555G mutant may have the ‘on’ states with two bulged-out adenines, A1557 and A1558, as shown in Figure 7b and c and Supplementary Figure 11. However, such an ‘on’ state could not be observed in the present structures, suggesting that the free energy minima of the ‘off’ states are either lower than those of the ‘on’ states or that the free energy barriers between the ‘off’ and ‘on’ states are high compared with those of the bacterial A site.
Figure 7.
Overview of the A-site molecular switches of the bacterial, mitochondrial wild-type and its A1555G mutant. Base-stacking columns are colored in red, blue, orange and green, respectively. Bases, which are not involved in any stacking column, are colored in gray. Except when indicated, all structures are crystallographically determined.
DISCUSSION
Bacteria have a ‘soft’ A-site molecular switch
For the bacterial A site, four conformations of the ‘off’ states have been reported so far (Supplementary Figure 11a) (11,52). Differences between them are found only at the two adenine residues, A1492 and A1493; (i) both are tucked-in, as observed in the present BACT-Co crystal; (ii) only A1492 is bulged-out; (iii) only A1493 is bulged-out; (iv) both adenines are bulged-out but do not recognize the codon–anticodon stem. These multiple conformations of the bacterial ‘off’ state suggest high flexibility of the bacterial A-site molecular switch. On the other hand, the bacterial ‘on’ state is only observed in a single conformation, the one with the two bulged-out adenines (Figures 7a and 8a and Supplementary Figure 11a). Since all base pairs and base-stacking columns except the A1492 and A1493 residues are conserved in the bacterial ‘off’ and ‘on’ states, conformational adaptations between these two states can easily occur from any of the ‘off’ states (Figure 7a, Supplementary Figure 11a and Supplementary movie ‘bacteria’). Recently, replica molecular dynamics simulations have confirmed that the free energy barrier between the ‘off’ and ‘on’ states of the bacterial A site is sufficiently low for aa-tRNA binding to shift the equilibrium (53). For these reasons, we suggest that bacteria present an energetically ‘soft’ A-site molecular switch, which achieves high speeds of translation, probably at a slight cost of proofreading accuracy.
Figure 8.
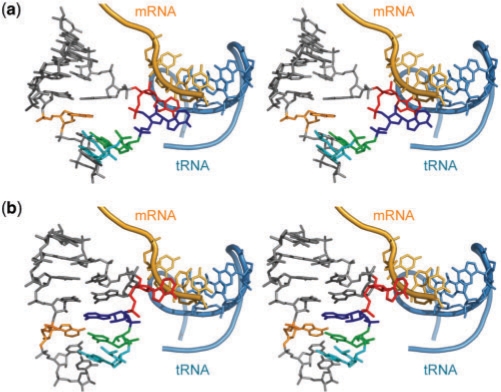
Stereoviews of interactions among the A site, mRNA (orange) and tRNA (sky blue). (a) The T. thermophilus ribosome [PDBID: 2j00 in (8)]. (b) The H. sapiens mitochondrial A site superposed on the T. thermophilus ribosomal A site. The A sites have the ‘on’ and ‘off’ states in (a) and (b), respectively. Three universally conserved adenine residues, A1492, A1557 and A1558 (A1408, A1492 and A1493 in bacterial numbering), are colored in orange, blue and red, respectively. Two nucleotides different in the bacterial and mitochondrial A sites, residues 1555 and 1556 (1490 and 1491 in bacterial numbering), are colored in cyan and green, respectively.
H. sapiens mitochondria have a ‘hard’ A-site molecular switch
In contrast to the flexible bacterial ‘off’ state, the H. sapiens mitochondrial ‘off’ state is only observed in a single conformation, which is completely different from any of the bacterial ‘off’ state (Supplementary Figure 11). By analogy with the secondary structure of the bacterial A site, the H. sapiens mitochondrial wild-type may adopt the ‘on’ state with two bulged-out adenines, A1557 and A1558, as shown in Figure 7b and Supplementary Figure 11b. Superimposition between the mitochondrial ‘off’ state and the crystal structure of the T. thermophilus 70S ribosome in complex with tRNA and mRNA (8) suggests that the switching from the ‘off’ state to the ‘on’ state needs a global conformational change (Figure 7b, 8b and Supplementary movie ‘mitochondria-WT’). On the short RNA strand of the A-site internal loop, the bulged-out A1492 and the subsequent C1493 and C1494 residues (1408, 1409 and 1410 in bacterial numbering) have to move within the loop in order to form the long base-stacking column between all bases of the strand (Figure 7b). On the long RNA strand, the tucked-in A1557 residue (A1492 in bacterial numbering) has to bulge out into the shallow/minor groove side, and the bulged-out A1558 residue (A1493 in bacterial numbering) has to move a long distance towards the shallow/minor groove of the codon–anticodon stem (Figure 7b and 8b). Such conformational transitions are accompanied by drastic changes of torsion angles and sugar puckers. For example, the sugar puckers of A1557 and A1558 (A1492 and A1493 in bacterial numbering) must be altered from the C2′-endo to C3′-endo conformations. Therefore, the free energy barrier between the ‘off’ and ‘on’ states may be high. In other words, the mitochondrial A-site molecular switch may be energetically harder to achieve than the bacterial one. It is important to remember that the H. sapiens mitochondria with 22 tRNA species must gain some flexibility (each tRNA decodes two or four codons) in the decoding process (20–22) without loosing fidelity in contrast to E. coli with 45 tRNA species (or 41 anticodons) and H. sapiens cytoplasms with 46 tRNA species (28,29). However, mitochondria need to synthesize only 13 encoded protein species during their long life cycle (23). It is thus possible that mitochondria require a harder A-site molecular switch in order to gain the necessary high accuracy of proofreading probably at the cost of translation speed.
Hypothesis for the molecular mechanism of non-syndromic hearing loss due to the A1555G mutation in the mitochondrial A-site molecular switch
The A1555G mutant of the H. sapiens mitochondrial A site has two different ‘off’ states; one named the first ‘off’ state is almost identical with the mitochondrial wild-type ‘off’ state (Figure 2a), and the other one named the second ‘off’ state is very similar to the bacterial ‘off’ state found in the present BACT-Co crystal (Figure 2b). The latter conformation may be the intermediate state between the first ‘off’ state and the ‘on’ state of the A1555G mutant, which could not be observed in the mitochondrial wild-type. In other words, the A1555G mutation renders the mitochondrial A-site molecular switch close to the bacterial one. By analogy with the secondary structure of the bacterial A site, the A1555G mutant may have the ‘on’ state with bulged-out A1557 and A1558, as shown in Figure 7c and Supplementary Figure 11c. Conformational changes between the first and second ‘off’ states as well as between the second ‘off’ and ‘on’ states may reversibly occur (see Supplementary movie ‘mitochondria-A1555G’). Since the latter case would be easier to occur than the former one, the A1555G mutant molecular switch is closer to the bacterial A-site molecular switch (compare Figure 7c with Figure 7a and Supplementary Figure 11c with Supplementary Figure 11a). As discussed above, although the decoding process in mitochondria is more flexible than that in bacteria, the proofreading step in mitochondria with the ‘hard’ A-site molecular switch may be more accurate than that in bacteria. However, the decoding process in mitochondria with the A1555G mutation (containing the ‘soft’ bacterial-type A-site molecular switch) should become more inaccurate. Since all 13 mitochondrial proteins are subunits of the respiratory chain complexes and are involved in synthesis of ATP by oxidative phosphorylation (23), non-syndromic hearing loss related to the A1555G mutation may be caused by interfering with energy production through protein mistranslation inducing excess superoxide production resulting in oxidative damage to mitochondria as proposed for aminoglycoside action on mitochondria (54) and bacteria (55).
CONCLUSIONS
Due to the similarity of the secondary structure of the A site, it has been supposed that the functional characteristics and tertiary structure of the A-site molecular switch is basically conserved in bacteria, H. sapiens cytoplasms and mitochondria. However, these three cell types are noticeably different in their biological properties such as life cycle, genome size, structural component of ribosome and number of tRNA species. In our present and previous work (11), we have shown how a small difference of nucleotide sequences affects the dynamics of the A-site molecular switches underlying the decoding mechanisms adapted to their biological properties and environments. In addition, as observed in our previous work (56), it also affects binding of aminoglycosides to the A-site molecular switches. The actual mechanisms and the coupling between the observed substates of the A sites and the global ribosomal movements are certainly rather complex and will require further techniques and experiments for more extensive unraveling.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
During part of this work, J.K. was supported by the Japan Society for the Promotion of Science and the project ANR-O5-MIIM-026-01 from the Agence Nationale de la Recherche. We thank the European Synchrotron Radiation Facility and the Swiss Light Source for provision of synchrotron radiation facilities and acknowledge the people of beamline ID14-2, ID23-1 and BM30 in ESRF and PX in SLS. We are grateful to B. François for crystallization and data collection of the MUT-K crystal and to A. Urzhumtsev for helpful discussions and suggestions for solving structures. Funding to pay the Open Access publication charges for this article was provided by the CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structure. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 2.Ogle JM, Carter AP, Ramakrishnan V. Structural insights into translation fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 3.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 4.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 5.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 6.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 7.Murphy FV, 4th, Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 8.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 9.Dunham CM, Selmer M, Phelps SS, Kelley AC, Suzuki T, Joseph S, Ramakrishnan V. Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA. 2007;13:817–823. doi: 10.1261/rna.367307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weixlbaumer A, Murphy FV, 4th, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo J, Urzhumtsev A, Westhof E. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006;34:676–685. doi: 10.1093/nar/gkj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, MÜller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfister P, Hobbie S, Vicens Q, Böttger EC, Westhof E. The molecular basis for A-site mutations conferring aminoglycoside resistance: relationship between ribosomal susceptibility and X-ray crystal structures. Chembiochem. 2003;4:1078–1088. doi: 10.1002/cbic.200300657. [DOI] [PubMed] [Google Scholar]
- 14.O’Brien TW, Liu J, Sylvester JE, Mougey EB, Fischel-Ghodsian N, Thiede B, Wittmann-Liebold B, Graack H. Mammalian mitochondrial ribosomal proteins (4). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 2000;275:18153–18159. doi: 10.1074/jbc.M909762199. [DOI] [PubMed] [Google Scholar]
- 15.Koc EC, Burkhart W, Blackburn K, Moseley A, Koc H, Spremulli LL. A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J. Biol. Chem. 2000;275:32585–32591. doi: 10.1074/jbc.M003596200. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem. 2001;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 2001;276:21724–21736. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 18.Koc EC, Burkhart W, Blackburn K, Moseley A, Koc H, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 19.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 20.Barrell BG, Anderson S, Bankier AT, de Bruijn MH, Chen E, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, et al. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc. Natl Acad. Sci. USA. 1980;77:3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama S, Nishimura S. Modified nucleosides and codon recognition. In: Söll D, Raj-Bhandary U, editors. tRNA: Structure, Biosynthesis and Function. Washington, DC: ASM Press; 1995. pp. 207–223. [Google Scholar]
- 22.Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J. Mol. Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florentz C, Sohm B, Tryoen-Tóth P, Pütz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider A, Maréchal-Drouard L. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 25.Entelis NS, Kolesnilov OA, Martin RP, Tarassov IA. RNA delivery into mitochondria. Adv. Drug Deliv. Rev. 2001;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- 26.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc. Natl Acad. Sci. USA. 1980;77:3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J. Mol. Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 29.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.Prezant TR, Agapian JV, Bohlman MC, Bu X, Öztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 31.Hutchin TP, Cortopassi GA. Mitochondrial defects and hearing loss. Cell. Mol. Life Sci. 2000;57:1927–1937. doi: 10.1007/PL00000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischel-Ghodsian N. Mitochondrial deafness. Ear Hear. 2003;24:303–313. doi: 10.1097/01.AUD.0000079802.82344.B5. [DOI] [PubMed] [Google Scholar]
- 33.Leslie AGW. Molecular data processing. In: Moras D, Podjarny AD, Thierry JC, editors. Crystallography Computing 5, From chemistry to Biology. Oxford: Oxford University Press; 1992. pp. 50–61. [Google Scholar]
- 34.Rossmann MG, van Beek CG. Data processing. Acta Crystallogr. 1999;D55:1631–1640. doi: 10.1107/s0907444999008379. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Vicens Q, Westhof E. Crystal Structure of Paromomycin Docked into the Eubacterial Ribosomal Decoding A Site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 37.Vicens Q, Westhof E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- 38.Vicens Q, Westhof E. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J. Mol. Biol. 2003;326:1175–1188. doi: 10.1016/s0022-2836(02)01435-3. [DOI] [PubMed] [Google Scholar]
- 39.François B, Szychowski J, Adhikari SS, Pachamuthu K, Swayze EE, Griffey RH, Migawa MT, Westhof E, Hanessian S. Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site. Angew. Chem. Int. Ed. Engl. 2004;43:6735–6738. doi: 10.1002/anie.200462092. [DOI] [PubMed] [Google Scholar]
- 40.François B, Russell RJM, Murray JB, Aboul-ela F, Masquida B, Vicens Q, Westhof E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo J, François B, Russell RJ, Murray JB, Westhof E. Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the γ-amino-α-hydroxybutyryl (haba) group. Biochimie. 2006;88:1027–1031. doi: 10.1016/j.biochi.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 46.Fokine A, Urzhumtsev A. On the use of low-resolution data for translation search in molecular replacement. Acta Crystallogr. 2002;A58:72–74. doi: 10.1107/s0108767301016476. [DOI] [PubMed] [Google Scholar]
- 47.Fokine A, Capitani G, Grütter MG, Urzhumtsev A. Bulk-solvent correction for fast translation search in molecular replacement: service programs for AMoRe and CNS. J. Appl. Crystallogr. 2003;36:352–355. [Google Scholar]
- 48.Echols N, Milburn D, Gerstein M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 2003;31:478–482. doi: 10.1093/nar/gkg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shandrick S, Zhao Q, Han Q, Ayida BK, Takahashi M, Winters GC, Simonsen KB, Vourloumis D, Hermann T. Monitoring molecular recognition of the ribosomal decoding site. Angew Chem. Int. Ed. Engl. 2004;43:3177–3182. doi: 10.1002/anie.200454217. [DOI] [PubMed] [Google Scholar]
- 51.Zhao F, Zhao Q, Blount KF, Han Q, Tor Y, Hermann T. Molecular recognition of RNA by neomycin and a restricted neomycin derivative. Angew Chem. Int. Ed. Engl. 2005;44:5329–5334. doi: 10.1002/anie.200500903. [DOI] [PubMed] [Google Scholar]
- 52.Kondo J, Westhof E. Structural comparisons between prokaryotic and eukaryotic ribosomal decoding A sites free and complexed with aminoglycosides. In: Arya DP, editor. Aminoglycoside antibiotics, From Chemical Biology to Drug Discovery. Hoboken, NJ: Wiley-Interscience; 2007. pp. 209–223. [Google Scholar]
- 53.Sanbonmatsu KY. Energy landscape of the ribosomal decoding center. Biochimie. 2006;88:1053–1059. doi: 10.1016/j.biochi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 54.Hutchin T, Cortopassi G. Proposed molecular and cellular mechanism for aminoglycoside ototoxicity. Antimicrob. Agents Chemother. 1994;38:2517–2520. doi: 10.1128/aac.38.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 56.Kondo J, François B, Urzhumtsev A, Westhof E. Crystal structure of the Homo sapiens cytoplasmic ribosomal decoding site complexed with apramycin. Angew. Chem. Int. Ed. Engl. 2006;45:3310–3314. doi: 10.1002/anie.200600354. [DOI] [PubMed] [Google Scholar]
- 57.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structure. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.