Abstract
MicroRNAs (miRNAs) have recently been proposed as a versatile class of molecules involved in regulation of a variety of biological processes. However, the role of miRNAs in TGF-β-regulated biological processes is poorly addressed. In this study, we found that miR-24 was upregulated during myoblast differentiation and could be inhibited by TGF-β1. Using both a reporter assay and Northern blot analysis, we showed that TGF-β1 repressed miR-24 transcription which was dependent on the presence of Smad3 and a Smads binding site in the promoter region of miR-24. TGF-β1 was unable to inhibit miR-24 expression in Smad3-deficient myoblasts, which exhibited accelerated myogenesis. Knockdown of miR-24 led to reduced expression of myogenic differentiation markers in C2C12 cells, while ectopic expression of miR-24 enhanced differentiation, and partially rescued inhibited myogenesis by TGF-β1. This is the first study demonstrating a critical role for miRNAs in modulating TGF-β-dependent inhibition of myogenesis, and provides a novel mechanism of the genetic regulation of TGF-β signaling during skeletal muscle differentiation.
INTRODUCTION
The development of skeletal muscle (myogenesis) is a highly regulated, multi-step process in which pluripotent mesodermal cells give rise to myoblasts that subsequently withdraw from the cell cycle and differentiate into myotubes (1,2). Myogenesis is orchestrated by the basic helix-loop-helix myogenic regulatory factors (MRFs), Myf5, MyoD, Myogenin and MRF4. These factors control myoblast commitment and differentiation via regulating the gene transcription by binding to sequence-specific DNA elements (E box, CANNTG) in the promoters of muscle genes (3,4). Activation of muscle gene expression by MRFs is also dependent on their association with other molecules such as members of the MEF2 family of transcription factors, MEF2A-D (5,6). Several studies have shown that myogenesis is also regulated by a number of growth factors, including fibroblast growth factor, insulin-like growth factor and transforming growth factor-β (TGF-β) (7–9).
TGF-β signaling, an important and complex signaling pathway, controls a diverse set of cellular processes (10,11). TGF-β signaling is transmitted from cell surface to nucleus by Smad proteins (12), of which there are three functional classes. The receptor-regulated Smads, Smad2 and 3, transduce signals from TGF-βs and Activins, while Smads 1, 5 and 8 mediate BMP signaling pathways. As the common mediator, Smad4 complexes with receptor-regulated Smads to transcribe target genes in response to either BMPs or TGF-β/Activin signals. Finally, the inhibitory Smads, Smad6 and Smad7, are negative regulators of TGF-β signaling (12).
Members of TGF-β have been shown to potently inhibit terminal differentiation of cultured myoblasts by downregulating MRFs (13–16). Myostatin, a member of the TGF-β family, has been shown to be a negative regulator of muscle differentiation and growth (17,18). Recent studies revealed that Smad3 mediates the suppression of myogenesis by TGF-β through suppressing the expression of E-box muscle genes and MEF2 proteins (19,20), while Smad7 has been implicated in promoting and enhancing myogenesis by interacting with MyoD and abrogating of Myostatin signaling (21).
miRNAs comprise a large family of ∼22-nucleotide, single-stranded RNAs that silence gene expression by translational repression and degradation of cognate mRNAs by binding to the 3′ untranslated region of target mRNAs (22–24). Accumulating evidence supports a role for miRNAs in the regulation of myogenesis (25). miR-1/-206 and miR-133 play opposing roles in modulating skeletal muscle proliferation and differentiation. While miR-1 and miR-206 promote myogenesis, miR-133 inhibits myoblast differentiation and promotes proliferation by repressing serum response factor and a key splicing factor (26–29). miR-181 has been shown to promote myoblast differentiation by downregulating a myogenic inhibitor, the homeobox protein Hox-A11 (30).
Recently, several reports have implicated miRNAs in TGF-β signaling. miR-192 was found to be increased by TGF-β in mouse mesangial cells, and play an important role in the kidney and diabetic nephropathy in controlling TGF-β-induced Col1a2 expression by downregulating Smad-interacting protein 1 (31). Zebrafish miR-430 was found to be able to dampen and balance the expression of the Nodal agonist squint and the TGF-β Nodal antagonist lefty (32). Xenopus laevis miR-15 and miR-16 could restrict the size of the organizer by targeting the Nodal type II receptor Acvr2a (33). However, the miRNA which functions in TGF-β-inhibited myogenesis has not been reported.
In this work, we identified miR-24 as a miRNA that involved in the inhibition of skeletal muscle differentiation by TGF-β, thus revealing a new level of regulation in TGF-β signaling-regulated myogenesis.
MATERIALS AND METHODS
Mouse strains
To obtain mice with targeted deletion of Smad4 gene in heart, homozygous mice for the floxed Smad4 allele (Smad4Co/Co) (34) were bred with αMHC-Cre transgenic mice in which the expression of Cre recombinase is driven by α-MHC promoter (35). The genotyping was performed as described (35). The generation and genotyping of Smad3ex8/ex8 null mice, in which the 8th exon of Smad3 gene is completely deleted by homologous recombination, were described previously (36).
Vector construction
For the expression of miR-24, genomic fragment of mouse miR-24 precursor was amplified by PCR using the primer pairs: 5′-GACCCCATCTCCTCAGGCC-3′ and 5′-CAGCTGTTTGGACACCCAGATG-3′. The PCR product was first cloned into pIRES2-EGFP (Invitrogen, Carlsbad, CA, USA). The fragment containing CMV promoter and miR-24-2 precursor was then cloned into pINCO retroviral vector (37) to generate pINCO-miR24.
For promoter assay, 1040-bp genomic fragment upstream of the transcriptional start site of miR-24-2 precursor was amplified by PCR using the primer pairs: 5′-CCCTGGTACGGGTGCTAAATAC-3′ and 5′-AGGCACAGTGAGGGGGGCA-3′. To mutate potential Smads-binding site in the promoter region, an overlapping PCR was performed with two additional primers (5′-AAGGATCCACACCCTAGCCCTC-3′, 5′-AAGGATCCTTTTGCTGAGTTTAGATTCTGTCCCCA-3′) in addition to above primers. The PCR product was cloned into pGL3-basic (Promega, Madison, WI, USA) to generate pGL3-P24 and pGL3-P24-mut.
For the evaluation of ectopic expressed miR-24, a synthesized linker (5′-GATCTCTGTTCCTGCTGAACTGAGCCAA-3′ and 5′-CGCGTTGGCTCAGTTCAGCAGGAACAGA-3′) perfectly complementary to mature miR-24 was inserted into pGL3-CM, in which the multiple cloning site of the pGL3-control vector (Promega, Madison, WI, USA) was removed and placed downstream of the luciferase gene as described (29,38), to generate miR-24 ‘sensor’.
Cell culture and luciferase reporter assay
Primary myoblast cells were obtained from the gastrocnemius muscles of Smad3–/– mice (36) and their wild-type littermates as described (39). The cells were propagated on culture plates coated with collagen (Sigma-Aldrich, St Louis, MO, USA) in growth medium (GM). C2C12 myoblast cells were cultured as described (40). To induce myogenesis in vitro, the proliferating myoblasts were shifted from GM into differentiation medium (DM) and harvested at the indicated times. rhTGF-β1 (R&D Systems, Inc., Minneapolis, MN, USA) was added into DM at the final concentration of 5 ng/ml to inhibit myogenesis if needed. GM for isolated primary myoblasts consists of DMEM supplemented with 15% fetal bovine serum (GIBCO BRL, USA), 1% glutamine and penicillin-streptomycin (HyClone, USA), and 2.5 ng/ml of bFGF (R&D Systems, Inc., Minneapolis, MN, USA). GM for C2C12 consists of DMEM supplemented with 10% newborn calf serum (GIBCO BRL, USA), 1% glutamine and penicillin-streptomycin. DM for both C2C12 and primary myoblasts consists of DMEM supplemented with 2% horse serum (HyClone, USA), 1% glutamine and penicillin-streptomycin.
For luciferase assay, reporter plasmids were co-transfected with phRG-TK vectors (Promega, Madison, WI, USA), which express synthetic renilla luciferase to normalize transfection efficiency, using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). Luciferase activities were detected with Dural-luciferase® reporter assay reagents (Promega, Madison, WI, USA) using LB 960 Centro XS3 luminometer (Berthold Technologies, GmbH & Co. KG, Germany).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP was performed according to the recommendation of Chromatin Immunoprecipitation Assay Kit of Upstate Biotechnology, Charlottesiville, VA. Antibodies used for ChiP were all purchased from Santa Cruz Biotechnology. The genomic region of miR-24-2 flanking the potential Smads-binding site was amplified with the following primer paires: 5′-AGTAGAGGAGGGCTAGGGTGTGGA-3′, 5′-TCTTGCTTGCCTGCCTATCTTGAC-3′.
Retroviral infection and electroporation
To express miR-24 in C2C12 cells, retroviral vector pINCO-miR24 was transfected into PLAT-E cells with FuGene6 (Roche Ltd, Switzerland) to be packaged. Two days later, the cell culture medium was collected and filtered with 0.45 μm filter (Pall Corporation, East Hills, NY, USA) to get virus-containing medium. The medium was mixed with fresh cultural medium of equal volume containing 8 μg/ml polybrene (Sigma-Aldrich, St Louis, MO, USA), and then used to infect C2C12 cells. 24 h later, the cells were transferred to DM to induce myogenesis for indicated times, or used for electroporation.
To inhibit the function of miR-24, 2′-O-methyl antisense inhibitory oligoribonucleotides targeted towards miR-24 (5′-CUGUUCCUGCUGAACUGAGCCA-3′) and scrambled sequence (5′-CCUUCGACUUCGCGUGAUCAGA-3′) were purchased from GeneChem (GeneChem Co., Ltd, Shanghai, China) and introduced into C2C12 cells by electroporation with Gene Pulser (Bio-rad Lab., Hercules, CA, USA) (50 nM/each reaction)
Northern blot
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) based on the suggested protocol. RNA quality was assessed by 1% agarose gel electrophoresis in the presence of ethidium bromide. Northern blot analysis was performed as described (41) using 20 μg total RNA from each sample. Probes were synthesized in Invitrogen Biotechnology Co., Ltd (Beijing) as following: miR-24 (MI0000572), 5′-CTGTTCCTGCTGAACTGAGCCA-3′; miR-23a (MI0000571), 5′-GGAAATCCCTGGCAATGTGAT-3′; miR-27a (MI0000578), 5′-GCGGAACTTAGCCACTGTGAA-3′. Probes were labeled with 32P γ-ATP using T4 polynucleotide kinase (New England Biolabs, Inc., Ipswich, MA, USA).
Semi-quantitative and real-time RT-PCR
Reverse transcription PCR was performed essentially as described (42) using the mRNA selective PCR kit (DRR025A, TaKaRa, Dalian, China). Briefly, 2 μg total RNA was reversely transcribed by AMV reverse transcriptase XL with Oligo dT primer. Primer pairs used were listed as following: MEF2d (NM_133655), 5′-CATCAGCATCAAGTCAGAACC-3′ and 5′-GAGGGAGTGGGAACCATC-3′; Myf5 (NM_008656), 5′-AAG AACAGCAGCTTTGAC and A-3′, 5′-CGTGATAGATAAGTCTGGAGC-3′; MyoD (NM_010866), 5′-GCTACGACACCGCCTACTACA-3′ and 5′-GGGTCTGGGTTCCCTGTTC-3′; Myogenin (NM_031189), 5′-TGAATGCAACTCCCACAGC-3′ and 5′-CACCCAGCCTGACAGACAA-3′; MHC (NM_010855), 5′-CTTGGTGGACAAACTACAGACT-3′ and 5′-TGCAGAATTTATTTCCGTGAT-3′; β-actin (NM_007393), 5′-CTGGCTGGCCGGGACCTGAC-3′ and 5′-CCGCTCGTTGCCAATAGTGATGAC-3′.
Real-time PCR was performed using the LightCycler system (Roche Ltd, Switzerland) with the FastStart DNA Master SYBR Green. The standard curve method of quantification was used to calculate the expression of target genes relative to the housekeeping gene β-actin. The control expression level was set to 1 as described (43). Experiments were repeated at least three times. The following primers were used: MEF2d (NM_133655), 5′-CGGGGACCGGGATGATGGAC-3′ and 5′-GAGCGGGAGGAGCCCAGGATAA-3′; Myogenin (NM_031189), 5′-AGCGGCTGCCTAAAGTGGAGAT-3′ and 5′-AGGAGGCGCTGTGGGAGTTG-3′; MHC (NM_010855), 5′-CGCCCACCTGGAGCGGATGA-3′ and 5′-CTTGCGGTCCTCCTCGGTCTGGT-3′; β-actin (NM_007393), 5′-CTGGCTGGCCGGGACCTGAC-3′ and 5′-ACCGCTCGTTGCCAATAGTGATGAC-3′.
Immunoblotting and immunostaining
Immunoblotting were carried out mainly as described (44) using antibodies against Smad4 (provided by Dr Ye-Guang Chen), Smad2/3 (#3102, Cell Signal Technology, Inc.), eIF5 (sc-282, Santa Cruz Biotechnology, Inc.), MEF2 (sc-313, Santa Cruz Biotechnology), Myogenin (QC1653, Yuanpinghao Biotechnology, Beijing, China), caveolin3 (ab2912, Abcam plc, Cambridge, UK) and α-actin (BM0001, Boster Biological Technology, Wuhan, China). Immunostaining was carried out mainly as described (45). Briefly, C2C12 cells were fixed with 4% formaldehyde for 30 min at 4°C and then treated with 0.5% Triton-X 100 in PBS for 5 min at room temperature. After that, cells were incubated with primary antibody against skeletal Myosin (fast) (BM0096, Boster Biological Technology, Wuhan, China) at 4°C overnight at the concentration of 1:100 dilutions, followed by incubation with TRITC-conjugated secondary antibody (1:100, Zhongshan, Beijing, China). The slides were stained with 4′, 6′-diamidino-2-phenylindole (DAPI) to visualize the nuclei. Photo capture was performed using a Nikon laser microscope (Eclipse E600, Nikon Instruments Inc., Japan). The fluorescence densities were measured with Image-Pro plus 5.0 (Media Cybernetics, Inc., USA) referring to (29). For each sample, more than eight fields covering the whole slide were picked and red fluorescence-positive cells and total cells with DAPI staining were counted.
Statistical analysis of data
All values are reported as means ± SD. Differences were assessed by two-tailed Student t test using Excel software. P < 0.05 was considered to be statistically significant.
RESULTS AND DISCUSSIONS
miR-24 is upregulated in differentiated myoblasts and downregulated by TGF-β
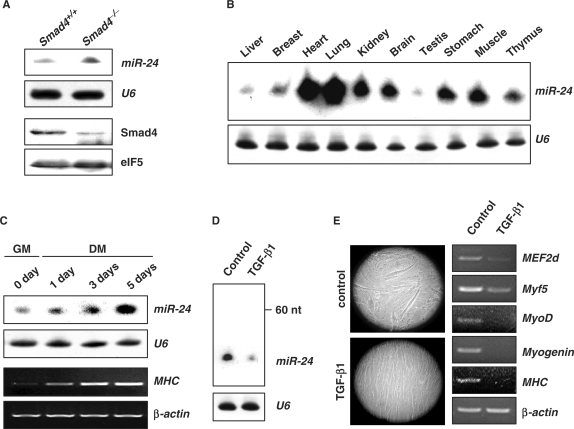
To identify miRNAs regulated by TGF-β, we performed microarray analyses of heart tissues in which Smad4 had been specifically knocked out (35) (See ‘Materials and methods’ section; Smad4–/– hereafter) and heart tissues of wild-type littermates, using a microarray that represented 347 different miRNAs and 814 predicted miRNAs (data not show). miR-24 was found to be upregulated in Smad4–/– mouse heart tissues, compared with littermates, and was confirmed by Northern blot (Figure 1A). miR-24 was therefore considered a candidate miRNA regulated by TGF-β. miR-24 was detected ubiquitously with relatively high levels of expression in the heart and muscle of 2-month old mice (Figure 1B), suggesting a role for miR-24 in the development and functional maintenance of these tissues.
Figure 1.
Suppressed expression of miR-24 by TGF-β1. (A) The expression of miR-24 in Smad4+/+ and Smad4–/– mouse heart tissues was detected by Northern blot. The expression of Smad4 was detected by Western blot. U6 RNAs detected by Northern blot and eIF5 were used as loading controls respectively. (B) Expression profile of miR-24 in 2-month old mouse tissues detected by Northern blot. (C) Northern blot analysis of miR-24 expression using total RNA isolated from C2C12 myoblasts cultured in growth medium (GM) or differentiation medium (DM) for 0, 1, 3 and 5 days. RT-PCR analysis of MHC expression to monitor the differentiation status at indicated times. (D) Northern blot analysis of miR-24 expression in C2C12 cells transferred to DM with or without TGF-β1 (5 ng/ml) for 3 days. (E) Morphological demonstration and differentiation marker expression (MEF2d, Myf5, MyoD, Myogenin, MHC) of C2C12 cells transferred to DM with or without TGF-β1 (5 ng/ml) for 3 days.
To investigate the potential involvement of miR-24 in myogenesis, we examined miR-24 expression during the induced differentiation of C2C12 myoblast cells. We found that miR-24 was upregulated in differentiated C2C12 myoblast cells as compared with undifferentiated cells (Figure 1C). The expression level of miR-24 was closely associated with differentiation status as demonstrated by the expression of differentiation-specific myogenic genes such as myosin heavy chain (MHC) (Figure 1C). Since TGF-β1 has been reported to inhibit the induced differentiation of C2C12 myoblast cells, we next examined the consequences of TGF-β1 on miR-24 expression in differentiating C2C12 cells. The expression of miR-24 was found to be dramatically reduced upon TGF-β1 treatment (Figure 1D), while myotube formation and expression of myogenic genes were likewise inhibited (Figure 1E). Taken together, we propose that miR-24 is a TGF-β-regulated miRNA involved in regulation of skeletal muscle differentiation.
TGF-β regulates expression of miR-24 at the transcriptional level
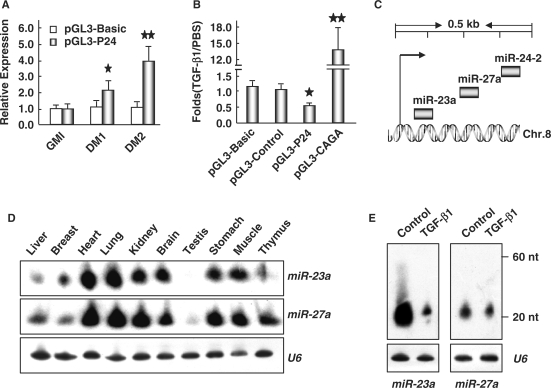
The expression of muscle miRNAs has been shown to be regulated at transcriptional and/or post-transcriptional levels (46). However, we did not detect an increase in levels of the miR-24 pre-transcript upon treatment with TGF-β1 (Figure 1D). We propose that miR-24 regulation therefore occurs at the transcriptional level. To test this hypothesis, we used the promoter region 1040-bp upstream of the transcriptional start site of miR-24-2 (47) to drive the expression of a luciferase reporter gene in C2C12 cells. The activity of this promoter increased with induced differentiation of C2C12 (Figure 2A), suggesting that this promoter region recapitulated expression of the miR-24 gene. Treatment with TGF-β1 inhibited promoter activity (Figure 2B), supporting the proposal that TGF-β1 regulates transcription of miR-24. Since miR-23a and miR-27a were clustered with miR-24-2 in a narrow region of mouse chromosome 8 (Figure 2C), they should share a similar regulation mechanism, which was supported by their similar expression patterns (Figure 2D). We further examined the effects of TGF-β1 on the expression of miR-23a and miR-27a, and found that TGF-β1 also inhibited expression of miR-23a and miR-27a without an elevation of their pre-transcripts (Figure 2E). These data suggest that TGF-β inhibits the expression of miR-24 at the transcriptional but not post-transcriptional level.
Figure 2.
TGF-β1 inhibits the transcription of miR-24 and clustered genes. (A) C2C12 cells transfected with reporter vector containing miR-24 promoter were first cultured in GM for 24 h, and then transferred into DM for 1 and 3 days, the luciferase activities were monitored at indicated time points with pGL-3 basic vector as a negative control. The results were shown as the relative expression of firefly luciferase against renila luciferase. One star means P < 0.05, two stars mean P < 0.01. (B) C2C12 cells transfected with reporter vector containing miR-24 promoter were transferred into DM with or without TGF-β1 (5 ng/ml) for 1.5 days, the luciferase activities were monitored with pGL-3 basic and control vector as negative controls as well as pGL3-CAGA as a positive control. The results were shown as the ratio of relative luciferase activities in TGF-β1-treated cells against those in DM cultured cells. One star means P < 0.05, two stars mean P < 0.01. (C) Schematic structures of the miR-24 cluster in mouse Chromosome 8. (D) Expression profile of miR-23a and miR-27a in 2-month old mouse tissues detected by Northern blot. (E) Northern blot analysis of the expression of miR-23a and miR-27a in C2C12 cells transferred to DM with or without TGF-β1 (5 ng/ml) for 3 days. U6 RNAs detected by Northern blot were used as loading controls.
Smad3 is required for the inhibition of miR-24 expression by TGF-β
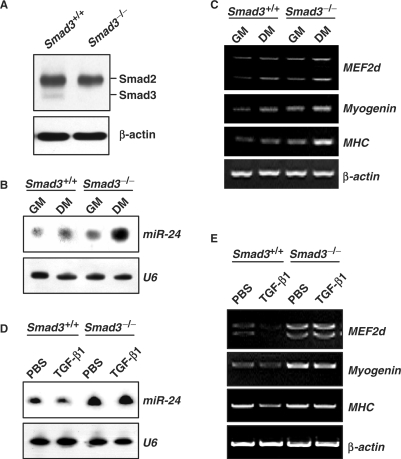
Smad3 is central in transducing TGF-β signaling during myogenesis (19,20). Therefore, we hypothesized that inhibition of miR-24 by TGF-β is also mediated by Smad3. To test this hypothesis, we compared expression of miR-24 in primary myoblast cells isolated from Smad3 complete knockout (Smad3–/–) mice (36) and non-mutant littermates. Smad3 was only detected in wild-type but not in Smad3–/– myoblasts (Figure 3A). Expression of miR-24 in Smad3–/– myoblasts cultured in both growth and differentiation media increased significantly (Figure 3B). This was concomitant with increased expression of differentiation-specific myogenic genes such as MEF2d, Myogenin, and MHC (Figure 3C) and enhanced myotube formation (data not shown). Notably, TGF-β1 could inhibit the expression of miR-24 (Figure 3D) along with myogenic genes, MEF2d, Myogenin and MHC, in wild-type primary myoblasts (Figure 3E), but failed to do so in Smad3–/– primary myoblasts. These data suggest a requirement for Smad3 in the inhibition of miR-24 expression by TGF-β.
Figure 3.
Smad3 is required for the transcriptional inhibition of miR-24 by TGF-β1. (A) Expression of Smad3 detected by Western blot in Smad3–/– primary myoblast cells. β-actin was used as a loading control. (B) Enhanced expression of miR-24 in Smad3–/– primary myoblast cells in both GM and DM for 2 days. U6 RNAs detected by Northern blot were used as loading controls. (C) Enhanced expression of differentiation markers (MEF2d, Myogenin and MHC) in Smad3–/– primary myoblast cells in both GM and DM. β-actin was used as a loading control. (D) The expression of miR-24 expression in Smad3+/+ and Smad3–/– primary myoblast cells transferred into DM with or without TGF-β1 (5 ng/ml) for 2 days. (E) Expression of differentiation markers (MEF2d, Myogenin and MHC) in Smad3–/– primary myoblast cells transferred into DM with or without TGF-β1 (5 ng/ml) for 2 days.
A Smads-binding site in miR-24 promoter is essential for the inhibition of miR-24 expression by TGF-β1
In silico analysis (rVista, http://rvista.dcode.org/) indicated that a potential Smads-binding site (GTGGATCAGCAAGCT) was found between (−241)∼(−255) bp in the miR-24-2 promoter region, and the site is evolutionally conserved. This suggested us that the site might be the TGF-β responding element through Smads in the miR-24-2 promoter region. To prove this, a ChIP assay was performed to examine the interaction of Smads proteins with the miR-24-2 promoter region flanking this site. Both antibodies against Smad3 and Smad4 could successfully immunoprecipitate the DNA fragment containing the potential Smads-binding site (Figure 4B), supporting that both Smad3 and Smad4 could physically interact with this miR-24-2 promoter region. Furthermore, we examined the effects of TGF-β1 on the activity of miR-24-2 promoter with a mutated Smads-binding site (Figure 4A). As shown in Figure 4C and D, TGF-β1 failed to inhibit the transcription of miR-24-2 promoter carrying a mutated Smads-binding site in both NIH-3T3 and C2C12 cells. These results suggest that this Smads-binding site is essential for TGF-β1-induced inhibition of miR-24 expression.
Figure 4.
Inhibition of miR-24 expression by TGF-β1 depends on a Smads-binding site. (A) A schematic illustration for mutated (P24-mut) and native sequences (P24) of the potential Smads-binding site. Arrow heads demonstrate the position of primers for ChIP assay. (B) ChIP assay to evaluate the interaction of Smad3 and Smad4 with the promoter region of miR-24-2. C2C12 cells were cultured in DM with PBS or TGF-β1 (5 ng/ml) for 24 h and harvested for ChIP assay. Target ChIPed DNA amplification was found after IP with antibodies against Smad3 (Ab-Smad3) and Smad4 (Ab-Smad4). Immunoprecipitation with no antibody (control) or pre-immunized serum (serum) served as negative controls. DNAs without IP were used as input controls. (C) NIH-3T3 cells transfected with reporter vectors containing native or mutated miR-24 promoter were treated with or without TGF-β1 (5 ng/ml) for 24 h, and then harvested for luciferase assay. pGL-3 Basic and pGL3-CAGA were used as negative and positive controls respectively. (D) C2C12 cells transfected with reporter vector containing native or mutated miR-24 promoter were transferred into DM with or without TGF-β1 (5 ng/ml) for 2 days, the luciferase activities were monitored with pGL-3 Basic and pGL3-CAGA as negative and positive controls respectively. The results were shown as the ratio of firefly to renila luciferase activities. Two stars mean P < 0.01.
TGF-β-regulated miR-24 promotes myoblast differentiation
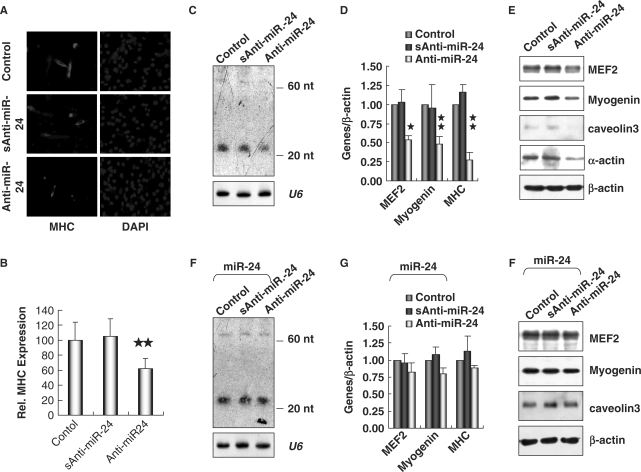
To address the role of miR-24 during myogenesis, we transfected C2C12 myoblasts with 2′-O-methyl antisense inhibitory oligoribonucleotides, which have been shown to inhibit the function of miRNAs (48,49). Treatment with miR-24 antisense oligoribonucleotides dramatically inhibited C2C12 myoblast differentiation, as indicated by reduced myotube formation (Figure 5A and B). The reduced expression of miR-24 was confirmed by Northern blot (Figure 5C). A decrease in both early and late myogenic markers, Myogenin and MHC, as well as MEF2, α-actin and caveolin3 upon the treatment of miR-24 antisense oligoribonucleotides was also detected by real-time PCR (Figure 5D) and Western blot (Figure 5E). Synthetic control oligoribonucleotides did not significantly affect differentiation (Figure 5A–E). Furthermore, we overexpressed miR-24 using the retroviral vector pINCO (Figure 6A) (37) in C2C12 cells treated with miR-24 antisense oligoribonucleotides, and found that overexpression of miR-24 almost restored the expression of myogenic markers (Figure 5F–H). All these results suggested that miR-24 is a promoting factor of myogenesis.
Figure 5.
Inhibition of miR-24 suppressed myogenesis in C2C12 cells. (A) Knockdown of miR-24 inhibited the expression of MHC in myoblasts. C2C12 myoblasts were electroporated with 2′-O-methyl antisense inhibitor of miR-24 (Anti-miR-24) with scrambled sequence (sAnti-miR-24) as a negative control. The myoblasts were then cultured in GM for 24 h and transferred into DM for 36 h before immunofluorescence staining of MHC. DAPI staining was performed to visualize the nuclei. (B) The expression of MHC shown in (A) was quantified after standardization with the expression level of MHC in controls. Two stars mean P < 0.01. (C) The expression of miR-24 detected by Northern blot in C2C12 myoblasts electroporated with Anti-miR-24. U6 RNAs detected by Northern blot were used as loading controls. (D) and (E) Knockdown of miR-24 suppressed the expression of myogenic factors, which was assessed by real-time RT-PCR (D) and Western blot analyses (E). (F) C2C12 myoblasts were first infected with retroviral vector pINCO-miR24 and then electroporated with Anti-miR-24. The expression of miR-24 was detected by Northern blot. U6 RNAs detected by Northern blot were used as loading controls. (G) and (H) The expression of myogenic factors assessed by real-time RT-PCR (G) and Western blot analyses (H) in cells treated as (F). One star means P < 0.05, two stars mean P < 0.01.
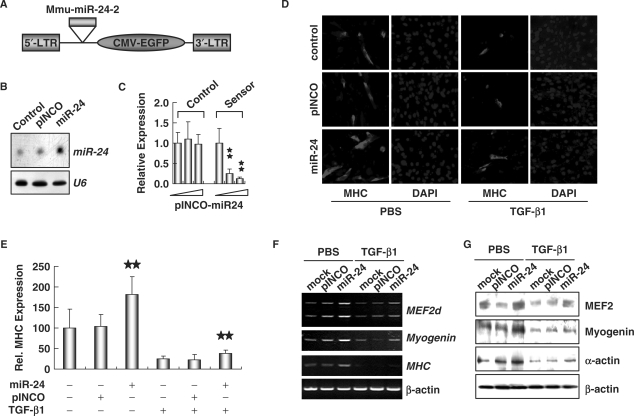
Figure 6.
Enhanced myogenesis in C2C12 cells ectopically expressing miR-24. (A) Schematic structures of the retroviral vector pINCO-miR24. (B) Northern blot analysis of miR-24 expression in C2C12 cells infected with pINCO-miR24. U6 RNAs detected by Northern blot were used as loading controls. (C) Activity detection of ectopically expressed miR-24. Reporter vector of miR-24 ‘sensor’, in which the complementary sequence of miR-24 was cloned downstream of luciferase coding sequences, was co-transfected with pINCO-miR24. pGL3-control vector was used as a negative control. Luciferase activity was measured 2 days after transfection. Two stars mean P < 0.01. (D) C2C12 cells infected with pINCO-miR24 or pINCO were transferred into DM with or without TGF-β1 (5 ng/ml) for 36 h. Immunofluorescence staining was performed to detect the expression of MHC. DAPI staining was done to visualize the nuclei. (E) The expression of MHC shown in (D) was quantified after standardization with the expression level of MHC in controls. Two stars mean P < 0.01. (F) and (G) Overexpression of miR-24 elevated the expression of myogenic factors, which was assessed by RT-PCR (F) and Western blot analyses (G).
In order to further prove the involvement of miR-24 in TGF-β-regulated myogenesis, we overexpressed miR-24 in C2C12 cells (Figure 6A) (37) and treated them with TGF-β1. Expression of miR-24 was examined by Northern blot analysis (Figure 6B). The activity of miR-24 was validated using a miR-24 ‘sensor’ (38), in which the complementary sequence was cloned downstream of the luciferase gene (Figure 6C). C2C12 cells overexpressing miR-24 showed accelerated myotube formation and increased expression of myogenic marker MHC, and this was inhibited by addition of TGF-β1 (Figure 6D and E). RT-PCR and Western blot analyses confirmed that overexpression of miR-24 in C2C12 resulted in upregulation of muscle transcriptional factors MEF2d and Myogenin, and myoblast differentiation markers MHC and α-actin, which was suppressed by treatment with TGF-β1 (Figure 6F and G). Transfection of the pINCO vector did not affect expression of these genes (Figure 6F and G). In addition, we found that overexpressed miR-24 was also able to partially restore expression of muscle transcriptional factors and myogenic markers normally inhibited by TGF-β (Figure 6F and G). These data demonstrate that miR-24 is a downstream target of TGF-β, and suppressed expression of miR-24 may constitute a mechanism by which TGF-β inhibits myogenesis.
Taken together, we have discovered a new mechanism for the inhibition of myogenesis by TGF-β that involves the suppression of a non-muscle-specific miRNA. We found that miR-24 was upregulated in differentiated myoblasts and could be inhibited by TGF-β at the transcriptional level (Figures 1A, D and 2B). Moreover, Smad3 ablation completely blocked inhibition of miR-24 expression by TGF-β (Figure 3D). Furthermore, ectopic expression of miR-24 enhanced differentiation of C2C12 myoblasts, and partially rescued inhibited myogenesis by TGF-β (Figure 6). Thus, we have shown that miR-24 is an essential miRNA for the modulation of TGF-β-inhibited myogenesis. Since changed expression of miR-24 affects both early and late myogenic markers, the underlying mechanism might be that miR-24 regulates an unknown upstream signal which then affects the expression of both early and late myogenic markers. It's also possible that miR-24 first regulates the expression of early genes (myogenin and MEF2), which then affects the expression of late genes (MHC, α-actin and caveolin3 et al.). More experiments are needed to address the detailed mechanisms.
We also noticed that miR-24 clustered closely with miR-23, miR-27 and miR-189 at two genomic loci. miR-24-2, miR-23a and miR-27a cluster in a narrow intergenic region of ∼400 bp on Chromosome 8 (Figure 2C), while miR-24-1, miR189, miR-23b and miR27b are located in an intronic region of <800 bp on chromosome 13. Moreover, miR-24-1 and miR-189 are generated from a common pre-transcript. We showed that miR-23a and miR-27a shared expression patterns with miR-24 and were also inhibited by TGF-β (Figures 1B, 2D and E), suggesting that the miRNAs in the miR-24 cluster might cooperatively regulate TGF-β-mediated inhibition of myogenesis. This proposal is consistent with the opinion that polycistronic miRNAs regulate common molecular events cooperatively (28,50,51). Recent studies revealed that clustered miR-1 and miR-133 have distinct functions in regulating skeletal muscle proliferation and differentiation (29). Whether the miRNAs in the miR-24 cluster play different roles in modulating myoblast proliferation and differentiation in response to TGF-β signaling needs to be further clarified.
Current data on the roles of miRNAs in myogenesis has been obtained largely from studies on muscle-specific miR-1, miR133 and miR-206 (29,52,53). However, the roles of non-muscle-specific miRNAs in myogenesis have not been thoroughly examined. A recent study showed that the non-muscle-specific miRNA, miR-181 is involved in the establishment of myoblast differentiation (33). Here, we identified miR-24 as another non-muscle-specific miRNA involved in myogenesis. In addition to being strongly induced during myogenesis (Figure 1C), miR-24 expression is maintained at high levels in terminally differentiated muscle tissues including heart and skeletal muscle (Figure 1B). These observations suggest that miR-24 might function during both differentiation and homeostatic maintenance of cardiac and skeletal muscle tissues. Indeed, previous studies have shown that miR-24 is upregulated during cardiac hypertrophy and is able to induce hypertrophic growth when overexpressed in primary cardiomyocytes (54). Considering that miR-24 is broadly expressed in multiple tissues (Figure 1C), we speculate that miR-24 might play different roles in the homeostasis of these tissues. Analysis of a tissue-specific knockout of miR-24 will help to address this issue.
In conclusion, we demonstrated that miR-24, and possibly its clustered miRNAs, is required for the modulation of TGF-β-inhibited myoblast differentiation, providing new clues for better understanding the molecular mechanisms underlying the physiological roles of TGF-β during myogenesis.
ACKNOWLEDGEMENTS
This work was supported by Chinese National Key Program on Basic Research (2005CB522506; 2005CB724600; 2006CB943501; 2006BAI23B01-3), National Natural Science Foundation of China (30430350; 30671078; 30470833), National High-Tech Research and Development Program (2006AA02Z168), The Chinese Space Medico-Engineering Pre-Research Project (2005SY5206006) and the grants (H030230280410, Z0006303041231 and 06Q085). We thank Dr Dazhi Wang at University of North California for pGL3-CM vector, Prof. Jincai Luo at Peking University for retroviral vector and cell lines, Prof. Yeguang Chen at Tsinghua University for TGF-β responsible reporter vector (pGL3-CAGA). We also thank Dr Jianguang Zhou at Institute of Biotechnology for pGL3-contol vector, Dr Xiaowei Zhou at Institute of Biotechnology for luciferase assay. Funding to pay the Open Access publication charges for this article was provided by Chinese National Key Program on Basic Research (2006BAI123B01-3).
Conflict of interest statement. None declared.
REFERENCES
- 1.Merlie JP, Buckingham ME, Whalen RG. Molecular aspects of myogenesis. Curr. Top. Dev. Biol. 1977;11:61–114. doi: 10.1016/s0070-2153(08)60743-7. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Gen. Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Sem. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Chanoine C, Della Gaspera B, Charbonnier F. Myogenic regulatory factors: redundant or specific functions? Lessons from Xenopus. Dev. Dyn. 2004;231:662–670. doi: 10.1002/dvdy.20174. [DOI] [PubMed] [Google Scholar]
- 5.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 6.Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 7.Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 8.Pirskanen A, Kiefer JC, Hauschka SD. IGFs, insulin, Shh, bFGF, and TGF-beta1 interact synergistically to promote somite myogenesis in vitro. Dev. Biol. 2000;224:189–203. doi: 10.1006/dbio.2000.9784. [DOI] [PubMed] [Google Scholar]
- 9.Joulia-Ekaza D, Cabello G. Myostatin regulation of muscle development: molecular basis, natural mutations, physiopathological aspects. Exp. Cell Res. 2006;312:2401–2414. doi: 10.1016/j.yexcr.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science (New York, N.Y) 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 11.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 12.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 13.Massague J, Cheifetz S, Endo T, Nadal-Ginard B. Type beta transforming growth factor is an inhibitor of myogenic differentiation. Proc. Natl Acad. Sci. USA. 1986;83:8206–8210. doi: 10.1073/pnas.83.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson EN, Sternberg E, Hu JS, Spizz G, Wilcox C. Regulation of myogenic differentiation by type beta transforming growth factor. J. Cell Biol. 1986;103:1799–1805. doi: 10.1083/jcb.103.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaidya TB, Rhodes SJ, Taparowsky EJ, Konieczny SF. Fibroblast growth factor and transforming growth factor beta repress transcription of the myogenic regulatory gene MyoD1. Mol. Cell. Biol. 1989;9:3576–3579. doi: 10.1128/mcb.9.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan TJ, Edmondson DG, Li L, Olson EN. Transforming growth factor beta represses the actions of myogenin through a mechanism independent of DNA binding. Proc. Natl Acad. Sci. USA. 1991;88:3822–3826. doi: 10.1073/pnas.88.9.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 18.Amthor H, Huang R, McKinnell I, Christ B, Kambadur R, Sharma M, Patel K. The regulation and action of myostatin as a negative regulator of muscle development during avian embryogenesis. Dev. Biol. 2002;251:241–257. doi: 10.1006/dbio.2002.0812. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Kang JS, Derynck R. TGF-beta-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J. 2004;23:1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kollias HD, Perry RL, Miyake T, Aziz A, McDermott JC. Smad7 promotes and enhances skeletal muscle differentiation. Mol. Cell. Biol. 2006;26:6248–6260. doi: 10.1128/MCB.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 31.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl Acad. Sci. USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science (New York, N.Y) 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 33.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Xu N, Feng X, Hou N, Zhang J, Cheng X, Chen Y, Zhang Y, Yang X. Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ. Res. 2005;97:821–828. doi: 10.1161/01.RES.0000185833.42544.06. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18:1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan GP, Pelicci PG. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 38.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 39.Qu-Petersen Z, Deasy B, Jankowski R, Ikezawa M, Cummins J, Pruchnic R, Mytinger J, Cao B, Gates C, et al. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. J. Cell Biol. 2002;157:851–864. doi: 10.1083/jcb.200108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 41.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science (New York, N.Y) 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 42.Sun Q, Zhang Y, Liu F, Zhao X, Yang X. Identification of candidate biomarkers for hepatocellular carcinoma through pre-cancerous expression analysis in an HBx transgenic mouse. Cancer Biol. Ther. 2007;6:e1–7. doi: 10.4161/cbt.6.10.4683. [DOI] [PubMed] [Google Scholar]
- 43.Tan X, Weng T, Zhang J, Wang J, Li W, Wan H, Lan Y, Cheng X, Hou N, et al. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J. Cell Sci. 2007;120:2162–2170. doi: 10.1242/jcs.03466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Q, Wang Y, Zhang Y, Liu F, Cheng X, Hou N, Zhao X, Yang X. Expression profiling reveals dysregulation of cellular cytoskeletal genes in HBx–induced hepatocarcinogenesis. Cancer Biol. Ther. 2007;6:668–674. doi: 10.4161/cbt.6.5.3955. [DOI] [PubMed] [Google Scholar]
- 45.Sun Q, Li J, Wang C, Huang X, Huang H, Du D, Liang Y, Han H. Overexpression of mouse GlcNAc-1-phosphotransferase-gamma subunit in cells induced an I-cell-like phenotype of mucolipidosis. Gene. 2005;347:55–64. doi: 10.1016/j.gene.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 46.Callis TE, Chen JF, Wang DZ. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26:219–225. doi: 10.1089/dna.2006.0556. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA (New York, N.Y) 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakajima N, Takahashi T, Kitamura R, Isodono K, Asada S, Ueyama T, Matsubara H, Oh H. MicroRNA-1 facilitates skeletal myogenic differentiation without affecting osteoblastic and adipogenic differentiation. Biochem. Biophys. Res. Commun. 2006;350:1006–1012. doi: 10.1016/j.bbrc.2006.09.153. [DOI] [PubMed] [Google Scholar]
- 53.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]