Abstract
Genetic diversity in fungi and mammals is generated through mitotic double-strand break-repair (DSBR), typically involving homologous recombination (HR) or non-homologous end joining (NHEJ). Microhomology-mediated joining appears to serve a subsidiary function. The African trypanosome, a divergent protozoan parasite, relies upon rearrangement of subtelomeric variant surface glycoprotein (VSG) genes to achieve antigenic variation. Evidence suggests an absence of NHEJ but chromosomal repair remains largely unexplored. We used a system based on I-SceI meganuclease and monitored temporally constrained DSBR at a specific chromosomal site in bloodstream form Trypanosoma brucei. In response to the lesion, adjacent single-stranded DNA was generated; the homologous strand-exchange factor, Rad51, accumulated into foci; a G2M checkpoint was activated and >50% of cells displayed successful repair. Quantitative analysis of DSBR pathways employed indicated that inter-chromosomal HR dominated. HR displayed a strong preference for the allelic template but also the capacity to interact with homologous sequence on heterologous chromosomes. Intra-chromosomal joining was predominantly, and possibly exclusively, microhomology mediated, a situation unique among organisms examined to date. These DSBR pathways available to T. brucei likely underlie patterns of antigenic variation and the evolution of the vast VSG gene family.
INTRODUCTION
Trypanosomatids are protozoa that branched early from the eukaryotic lineage (1) and several are parasites of substantial medical and veterinary importance (http://www.who.int/tdr/diseases/). The African trypanosome, Trypanosoma brucei, is spread among mammalian hosts by the tsetse fly and causes Human African Trypanosomiasis, which is fatal if untreated, and represents the leading cause of mortality in some areas. Trypanosoma brucei also causes Nagana disease in cattle, rendering 10 million square kilometres of land unsuitable for livestock.
Trypanosoma brucei circulates free in the host bloodstream, escaping immune responses by a process of antigenic variation, which involves monoallelic expression of variant surface glycoprotein (VSG) genes (2). The expressed VSG is always adjacent to a telomere and homologous recombination (HR) is thought to be the major mechanism contributing to antigenic variation, occurring in up to 1% of cells per population doubling (3,4). Analysis of genome sequence data revealed a vast reservoir (>1500) of VSG genes clustered at subtelomeres (5). Most are pseudogenes that may be used to assemble intact genes using short, possibly imperfect stretches of sequence homology (6).
DNA double-strand-breaks (DSBs) typically occur during DNA replication and can also be brought about by other chemical and physical forces (7,8). Non-homologous end-joining (NHEJ) and HR are the major DSB repair (DSBR) pathways in mammals and unicellular eukaryotes, respectively and NHEJ also operates in many prokaryotes that encode a two-component, Ku/DNA ligase apparatus (9). HR-repair requires an undamaged homologous sequence in the same cell. When multiple potential templates are available, the choice may be governed by chromosome disposition prior to damage or, alternatively, damage may induce a homology search (10). Chromosome disposition likely leads to post-replicative preference for template sequences on sister chromatids (11,12), a process that requires cohesion (13). Other repair templates may be sequences nearby on the same chromosome (14), allelic sequence on a homologous chromosome (15) or homologous sequences on heterologous chromosomes (16). DSBs not repaired by HR or NHEJ may be repaired by microhomology-mediated joining (MMJ) which appears to serve as a back-up or salvage pathway (17–20,21). The DSBR pathways described above have been co-opted in several instances for ‘programmed’ DNA rearrangements. Prominent examples are immunoglobulin and T-cell receptor gene rearrangement (22) and mating-type switching (23) in vertebrates and fungi, respectively. The response to DNA damage is also the basis for experimental genetic manipulation.
Much of our current thinking regarding DSBR in T. brucei comes from the analysis of rare recombinants that integrate transfected linear DNA. This has revealed efficient HR (24) and MMJ (25). MMJ has also been reported using in vitro extracts while NHEJ has not been reported (26). In addition, several proteins have been shown to play a role in DSBR in T. brucei, including Mre11 (27,28), Rad51 (25) and related proteins (29) and a sirtuin (30). Antigenic variation can operate via Rad51-dependent or independent HR pathways (25,31) and a Rad51-related protein has also been shown to play a role (29).
DSBR takes place in the context of chromatin. Little is known about the DNA-damage response or chromosomal DSBR in trypanosomes, or how different pathways contribute to repair. Specific telomere removal was reported recently in T. brucei but this did not trigger a classical DNA-damage response (32). Rather, the terminally deleted chromosome was replicated and segregated without being repaired. We have now used conditional expression of the meganuclease, I-SceI, to generate a lesion in the core of a T. brucei chromosome. This has allowed investigation of the kinetics and pathways of chromosomal DSBR and represents the first report of a DNA damage checkpoint response in a trypanosomatid. HR occurs between homologous and heterologous chromosomes while, in contrast to the situation in other cells analysed to date, the dominant end-joining pathway uses microhomology with no evidence for NHEJ-mediated repair.
MATERIALS AND METHODS
Trypanosoma brucei growth and manipulation
Lister 427, MITat1.2 (clone 221a), bloodstream form cells were grown in HMI-11. Transformation was performed as described previously (33), cell density was determined using a haemocytometer and tetracycline (Tet) was from Sigma and was used at 1 μg/ml.
Plasmid construction
Plasmid constructs for expression of the Tet repressor from the TUB locus (TetR-BLE) and for tet-on expression of I-SceI with an N-terminal SV40 nuclear-localization-signal from a RRNA spacer locus (I-SceI-HYG) were described previously (32).
To generate pRSP2110, primers for an I-SceI site (lower case), SceFb (GGCCCCGCGGAtagggataacagggtaatA) and SceRb (GGCCTattaccctgttatccctaTCCGCGG), were annealed and ligated at the NotI site in pESPiRFP:PAC (32). The entire RSP cassette, including TUB processing signals, was then amplified (Phusion DNA Pol, Finnzymes Diagnostics) using the TUBIR5Xcm (AGCTccaGTCCTTGTGtggGTCCCATTGTTTGCCT) and TUBIR3Xcm (GATCccaCACAAGGACtggCCCCTCGACTATTTTCTTTG) primers, digested with XcmI (lower case) and ligated to similarly digested pARD (33). pRSP2110 was digested with BamHI/Bsp120I prior to introduction into T. brucei.
DNA analysis
For Southern blot analysis of DSBR, purified genomic DNA was digested with HindIII and processed according to standard protocols. Gels were washed in 0.25 M HCl for 15 min followed by two washes in H2O. The RFP probe was a 687-bp HindIII/NotI fragment encompassing the full ORF; the 2110.1 probe was a 699-bp SacI fragment from pARD (33); the αTUB probe was a 516-bp XcmI/StuI fragment and the 7240 probe was a 731-bp HindIII/XhoI coding region fragment.
For slot-blot analysis, 3 μg of each DNA sample was added to 200 μl of 10× SSC. Twenty micro litres were removed, added to 200 μl of 0.4 M NaOH and denatured at RT for 5 min. Hybond N (Amersham), supported by one layer of Whatman 3MM paper, was soaked in 20× SSC followed by H2O and placed in a slot-blot manifold. DNA samples were then loaded into the slots and drawn onto the membrane using a vacuum pump followed by washing each well with ∼250 μl of 10× SSC. Membranes were processed as for Southern blotting. The RFP and 7240 probes were the same as used for Southern blotting; the 2110.2 probe was a 680-bp NarI/XcmI fragment from pARD (33) and the 2100 probe was a 681-bp PstI/NcoI fragment from pARD.
A Typhoon TRIO phosphorimager (Amersham) was used to quantify signals. For slot-blots, values for native DNA samples were corrected for background (signal from cells lacking DSBs), ssDNA versus dsDNA (the probe was double stranded), loading (90% of the DNA was native) and the presence of additional alleles for the 2110.2 and 2100 probes. Thus, percentage values were derived as follows: adjusted native value divided by the denatured value multiplied by 22.2 (×2/9 × 100) for RFP and by 44.4 (×2/9 × 100 × 2) for 2110.2 and 2100.
A series of RFP and PAC-specific primers were used to amplify and sequence repair junctions.
Microscopy and protein analysis
Western blotting and fluorescence microscopy were carried out using standard protocols as previously described (30). Briefly, for immunofluorescence analysis, cells were labelled using rabbit-anti-Rad51 (29) primary antibody at 1:500 and fluorescein-conjugated goat- anti-rabbit secondary antibody (Pierce) at 1:2000. Samples were mounted in Vectashield (Vector Laboratories) containing 4, 6-diamidino-2-phenylindole (DAPI). Cells were scored for Rad51 foci and cell cycle phase by two of us to generate mean values ± standard deviation. Images were captured using an Eclipse E600 microscope with digital camera (Nikon). We employed Metamorph software for image processing and deconvolution; all settings were identical for each data set. For western blotting, rabbit- anti-Rad51 was used at 1/200 and blots were developed using an ECL kit (Amersham).
RESULTS
An experimental system to explore pathways of DSBR in T. brucei
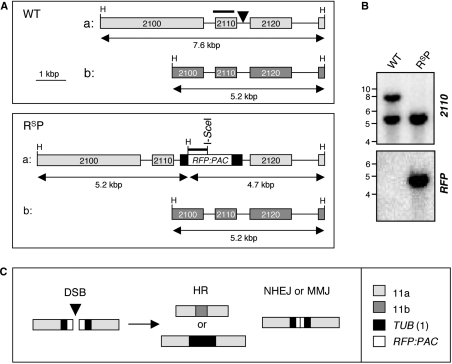
We have used the I-SceI meganuclease that cleaves a specific 18-bp sequence to produce single DSBs in T. brucei chromosomes. Theoretically, a site of this length would occur only once in 7 × 1010 bp, equivalent to a genome >2000 times the size of that in T. brucei. Formation of single chromosomal DSBs was temporally and spatially controlled by placing the I-SceI gene under the control of an inducible promoter and by introducing a single chromosomal I-SceI cleavage site (32). For this study, we introduced DSBs at a locus >1 Mbp from the nearest telomere on chromosome 11, the largest T. brucei chromosome at ∼ 5.7 Mbp in length. Most genes are tightly clustered and co-transcribed on T. brucei chromosomes and the locus chosen is within the 452-bp segment between the Tb11.02.2110 and 2120 genes (Figure 1A and see www.genedb.org). We previously sequenced the region flanking the 2110, Nα-acetyltransferase gene in the Lister 427 strain, showed that the gene is essential for growth and identified a polymorphic HindIII site nearby (33). The sequenced allele was designated ‘a’ and we mapped the additional HindIII site at allele ‘b’ (Figure 1A and data not shown). This HindIII site was important to allow us to distinguish between alleles on Southern blots (Figure 1B).
Figure 1.
Experimental system to study DSBR. (A) The schematic illustrates Tb11.02.2110/2120 loci on chromosome 11a and b (WT: top) and the same loci after insertion of the RsP cassette (RsP: bottom). The arrowhead in the top panel indicates the RsP insertion site, 209-bp downstream of the 2110 stop codon. Bars above the maps indicate the locations of the 2110 and RFP probes. The sizes of the HindIII fragments expected on Southern blots are indicated below the maps. Black boxes indicate tubulin (TUB) sequences flanking the RsP cassette. H, HindIII sites. (B) Southern blot analysis indicated RsP insertion on chromosome 11a. DNA was digested with HindIII. See fragment sizes in (A). (C) Schematic illustration of possible DSBR mechanisms. HR requires extensive homologous sequence; MMJ requires only short stretches and NHEJ requires little or none. HR can use templates from chromosome 11b (top) or from TUB genes on chromosome 1 (bottom). Portions of RFP and PAC would be expected to be retained following NHEJ or MMJ.
We embedded an I-SceI site within a red fluorescent protein (RFP)–puromycin N-acetyltransferase (PAC) fusion gene and added the appropriate 2110/2120 targeting sequences to generate the pRSP2110 construct. The RFP/I-SceI/PAC (RSP) cassette was flanked by (TUB) sequences that served a dual-purpose, to promote pre-mRNA trans-splicing and polyadenylation and as templates for HR with chromosome 1 (see below). When the pRSP2110 construct was introduced into T. brucei, PAC served as a selectable marker and recombinant ‘Sce2110’ cells were resistant to puromycin, but RFP-fluorescence was not detected. This is likely due to insufficient expression at the pol II-transcribed locus since we saw only weak red fluorescence in cells where the RSP cassette was inserted at a highly transcribed pol I locus (data not shown). Genomic DNA was extracted for Southern analysis and this indicated that the cassette had integrated at allele ‘a’ (Figure 1B).
An I-SceI-induced lesion in the hemizygous RSP cassette may be repaired via a number of pathways (Figure 1C). HR is known to be efficient in T. brucei and can repair a break by gene conversion using genetic information from an undamaged sister chromatid, the homologous chromosome 11b or up to 40 ectopic copies of the TUB sequence from the tandem arrays on chromosomes Ia or b. Since we use continuous expression of I-SceI, a site regenerated by identical sister chromatid repair can be cleaved again until repair mutates the recognition site and hence, this repair route would not be observed using our assay. In contrast, use of the 11b template would remove RSP, while TUB recombination would replace RSP with a αTUB gene, changes that can be detected. NHEJ or MMJ are error-prone and are characterized by deletions at the break-site. In such cases, analysis of the junction sequences allowed us to distinguish between these alternative end-joining pathways (see below). Thus, the approach was designed to allow the analysis of competition between repair pathways that use homologous templates on different chromosomes and NHEJ or MMJ pathways.
Physical monitoring of DNA resection and repair
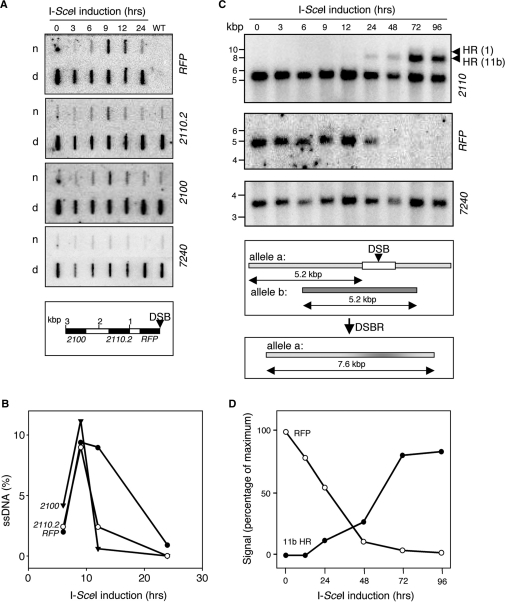
Introduction of a DSB on T. brucei chromosome 11 was controlled by placing the I-SceI gene downstream of a Tet-inducible promoter. Upon addition of Tet to the medium, the enzyme generated a DSB at the RSP locus. To monitor the kinetics of repair, we extracted DNA from cells at different time points following I-SceI-induction. Prior to initiation of mitotic HR, a DSB must be processed by degradation of the 5' strand in a process known as resection to generate single-stranded DNA (ssDNA) with a 3′ end. The ssDNA is thought to generate the signal for the DNA damage checkpoint, and is the substrate for Rad51 binding to initiate a search for a suitable homologous repair template (7). To physically monitor ssDNA adjacent to the lesion on chromosome 11, hybridization analysis was applied to native chromosomal DNA samples (Figure 2A and B); hybridization to native DNA will occur only if ssDNA is present. Denatured samples were analysed in parallel to control for loading. To increase sensitivity, we loaded nine times more native DNA relative to denatured DNA for each time point.
Figure 2.
Physical monitoring of DNA resection and repair. (A) Monitoring ssDNA adjacent to the lesion by slot-blot analysis. Genomic DNA samples were extracted at various times following I-SceI-induction. Ninety percent of each sample was ‘native’ (n) and the remainder denatured (d). The probes used on each blot are indicated on the right. 7240 is a distal, chromosome 11 control. The schematic map indicates the location of the probes (black) in relation to the lesion (DSB). (B) Kinetics of ssDNA formation. Phoshorimager analysis was used to quantify the signals in (A). (C) Monitoring repair by Southern blot analysis. Genomic DNA extracted at various times following I-SceI-induction was digested with HindIII and subjected to Southern blot analysis using the probes indicated. Arrowheads indicate the fragments expected following HR between chromosome 11a and 11b or the TUB locus on chromosome 1 (see Figure 1). For chromosome 11, 7240 served as a loading control. The schematic illustrates dominant allelic HR with chromosome 11b. (D) Kinetics of repair by HR with chromosome 11b. Phoshorimager analysis was used to quantify the signals in (C).
We used an RFP probe and a series of probes at different locations on chromosome 11. The RFP probe allowed resection to be monitored immediately adjacent to the lesion, while the 2110.2 and 2100 probes monitored resection within 1 kb and 2.4 kb. The 7240 probe served as a control to monitor DNA ∼2.8 Mbp from the lesion. The analysis indicated DNA resection across all three regions tested within 3 kb of the lesion (Figure 2A) while the distal, control probe showed no evidence of resection. Thus, resection was sufficiently extensive to facilitate ectopic HR or HR with chromosome 11b (see Figure 1C). After correction for loading, background, DNA conformation and gene copy number (see Materials and methods section), we saw that ∼10% of the DNA at each of the three sites adjacent to the lesion was in the single-stranded conformation 9 h after I-SceI induction (Figure 2B). This effect was transient with little ssDNA detected after 24 h. Single-stranded RFP was detected at 12 h however, after adjacent single-stranded sequences were apparently sequestered, presumably reflecting the formation of Holliday junction intermediates (34).
To physically monitor DNA repair, we used Southern blot analysis (Figure 2C). First, loss of the RFP signal indicated that few cells escaped the action of I-SceI (Figure 2C and D). With regard to repair, the predominant pathway involved use of chromosome 11b as HR template to regenerate the ‘7.6 kb’ allele. We also saw evidence for ectopic recombination with the TUB array on chromosome 1. These events generated a fragment of 9.5 kb (7.6 kb + 1.9 kb of αTUB sequence) and, though the signal was weak, we were able to independently confirm these ectopic recombination events (see below and Figure 5B, lane 9). Thus, although the 2110/2120 sequence was more distal to the break relative to the TUB sequence, it was favoured in a competition between the two sequences. NHEJ or MMJ were expected to generate RFP fragments of <4.7 kb. Such events may not have been detected using this assay due to size heterogeneity and/or loss of RFP sequence (but see below). Since HR with chromosome 11b predominated, we plotted RFP-loss and 11b-mediated repair against time (Figure 2D). The 2110 signal, representing 11b repair, was normalized to the ratio of 2110 signals representing each allele in wild-type cells. The analysis indicated that chromosome 11b-mediated HR represented ∼85% of the repair events.
Figure 5.
Survivors display repair via homologous recombination and microhomology-mediated joining. (A) Survivorship. To generate mostly clonal populations, we distributed ∼96 cells over 288 wells (three 96-well plates). This yielded 93 (no Tet) and 53 (+Tet) wells, respectively with live cells detected after 1 week of growth. (B) Genomic DNA was extracted from survivors of I-SceI-induction, digested with HindIII and subjected to Southern blot analysis using the probes indicated (see Figure 2C for other details). In the αTUB panel, the intense band in every sample represents the tandem array on chromosome 1; the bands in the un-induced (U) and all survivor lanes represent the TetR insertion at the TUB locus and the additional 9.5-kb band in track 9 (arrowhead) confirmed ectopic recombination with chromosome 1. (C) Joined junctions. The PCR, end-joining survey employed primers at the extreme ends of the RFP/PAC ORF and DNA from a large population of cells after 7 days of Tet exposure (Tet+). DNA from cells prior to Tet exposure (Tet−) served as a control. Junction sequences from two major PCR products (arrowheads 1 and 2) and from four independent cloned survivors are shown on the right. The RFP (top row), PAC (bottom row) and junction sequences (middle row) are aligned with microhomology highlighted. The number of nucleotides deleted in each case and the number of clones representing each sequence are indicated.
Rad51 assembles into foci in response to a single DSB
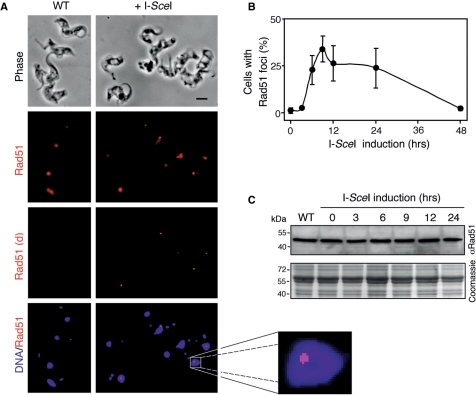
Rad51 (RecA in bacteria and RadA in archaebacteria) forms helical filaments on ssDNA and catalyses homologous strand exchange (35). Several recombination proteins, including Rad51, show diffuse localization in undamaged cells, but localize to sites of DNA damage forming sub-nuclear foci detectable by microscopy. Since we demonstrated the processing of single DSBs to generate ssDNA, we also wanted to determine whether these DSBs could trigger the assembly of Rad51 foci. We carried out immunofluorescence analysis using anti-Rad51 to compare wild-type cells to cells with I-SceI-induced lesions. Figure 3A shows that Rad51 was enriched in the nuclei of wild-type cells but the signal was typically diffuse (left-hand panels) in contrast to the situation in cells with a lesion, where a substantial proportion displayed Rad51 foci (right-hand panels). We used deconvolution to enhance foci and these images more clearly show nuclear foci in many induced cells and the absence of foci in the majority of wild-type cells. Consistent with previous work (29), foci were detected in only ∼1% of wild-type nuclei, likely reflecting the recruitment of repair proteins to sites of spontaneous DNA damage. We then counted Rad51 foci in cells induced for different periods of time (Figure 3B). In cells with a lesion on chromosome 11, there was a rapid increase in the proportion with nuclear foci, peaking at ∼30% 9 h after induction. A high proportion of cells with foci was still detected after 24 h but had diminished to background after 48 h (Figure 3B).
Figure 3.
Rad51 accumulates at sub-nuclear foci in response to DSBs. (A) Immunofluorescence analysis of Rad51 in wild-type (WT) cells and in Sce2110 cells 9 h after I-SceI-induction. Rad51 signals are shown before and after deconvolution (d). DNA was counter-stained with DAPI. Scale bar, 5 μm. An expanded view of a nucleus with a prominent Rad51 focus is shown to the right. (B) Rad51 foci kinetics. The proportion of nuclei with Rad51 foci were counted at different times after I-SceI-induction. n = 200 at each time point. Error bars, SD. (C) Rad51 levels remain constant during DSBR. Western blotting with anti-Rad51 and a series of protein samples extracted at different times after I-SceI-induction. An equivalent Coomassie-stained gel served as a loading-control. The predicted Mwt of TbRad51 is ∼41 kDa.
In the related trypanosomatids, Trypanosoma cruzi and Leishmania major, Rad51 expression is increased following chemical or radiation-induced DNA damage (36,37). To determine whether I-SceI-induced lesions lead to increased expression of Rad51, we carried out western blotting using anti-Rad51 and a series of whole-cell protein extracts representing different times following I-SceI-induction (Figure 3C). We saw no evidence for an increase in Rad51 expression indicating that the pre-existing cellular pool of Rad51 was redistributed into foci in response to DNA damage.
Rad51 foci and the G2M cell cycle checkpoint
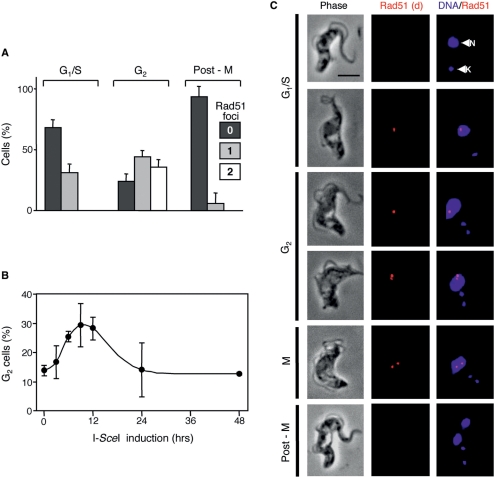
A single DNA-DSB can trigger a DNA damage checkpoint that arrests the cell cycle and allows time for repair prior to further progression thereby suppressing deleterious genome rearrangements (7). Efficient and temporally constrained introduction of DSBs in T. brucei provided an opportunity to explore DNA damage checkpoint control. No method is available to synchronize bloodstream form T. brucei, but nuclear and mitochondrial (kinetoplast) DNA, stained with DAPI, provide excellent cytological markers that define position in the cell cycle (38). In bloodstream form cells, ∼80% of cells display a single nucleus and a single kinetoplast (1N1K) indicating earlier phases of the cell cycle (G1/S). A single nucleus and two kinetoplasts (1N2K) indicate late nuclear G2 and two nuclei and two kinetoplasts (2N2K) indicate completion of mitosis. First, we examined the proportion of cells with Rad51 foci in these three categories after 12 h of I-SceI induction. We had also noted that some cells had two Rad51 foci, so we subdivided each category into cells with zero, one or two foci (Figure 4A).
Figure 4.
Rad51 foci and the G2M cell cycle checkpoint. Cells were processed for Rad51 immunofluorescence microscopy and DNA was counter-stained with DAPI. (A) The bar-chart shows the proportion of Sce2110 cells at different phases of the cell cycle with zero, one or two Rad51 foci 12 h after I-SceI-induction. Cell cycle phase was defined by the number of nuclei (N) and kinetoplasts (K) as determined by DAPI staining. n = 50 at each cell cycle phase. Error bars, SD. (B) G2M phase (1N2K) kinetics. The proportion of 1N2K cells was counted at different times after I-SceI-induction. n = 200 at each time point. Error bars, SD. (C) Immunofluorescence analysis of Rad51 in Sce2110 cells after I-SceI-induction. Rad51 signals are shown after deconvolution (d). N, nucleus; K, kinetoplast. Scale bar, 5 μm.
Approximately 30% of cells in G1/S phases (1N1K) had a single focus and ∼75% of cells in G2 phase (1N2K) had foci, with about half of these displaying two. Very few post-mitotic cells (2N2K) had foci and this category of cells gave similar results 24 h following I-SceI-induction indicating that the paucity of foci is not simply because cells with a lesion had insufficient time to progress to mitosis. Rad51 foci were of varying degrees of brightness with the brightest foci predominantly observed in G2 phase nuclei. This could reflect sister chromatids with lesions in close juxtaposition due to cohesion or differences in resection or Rad51-loading. Cells with two foci were exclusively in the G2/M phase of the cell cycle consistent with the idea that many cells do indeed bear lesions on both sister chromatids and also that cohesion can be lost in the absence of repair.
To determine whether a G2M cell cycle checkpoint was triggered by the lesions on chromosome 11, we counted the proportion of cells in the late G2 phase of the cell cycle. 1N2K-cells constituted ∼12% of wild-type cultures and a similar proportion was detected among Sce2110 cells prior to I-SceI-induction (Figure 4B). Late G2 phase cells rapidly accumulated following I-SceI induction indicating that a G2M cell cycle checkpoint was triggered in response to the lesion (Figure 4B).
The cells shown in Figure 4C are arranged in order of progression through the cell cycle as defined by DAPI staining and reflect Rad51 assembly and, possibly, disassembly. The top two 1N1K cells were at earlier points in the cell cycle when Rad51 foci were more commonly not detected (cell at top), but ∼30% had a single focus (second cell from top). The third, fourth and fifth 1N2K cells indicate progression through G2/M. These cells more commonly had one (third cell from top) or two Rad51 foci (fourth and fifth cells from top). The lower images indicate a 2N2K cell that had completed mitosis. Rad51 foci were rarely detected in these cells.
HR dominates DSB repair
We employed ‘clonogenic’ assays to determine the proportion of cells that survive I-SceI-induced lesions and also to further explore the quantitative contribution of different chromosomal DSBR pathways in T. brucei. Clonogenic analysis under standard or I-SceI-inducing conditions indicated 57 ± 11% survival following I-SceI induction (Figure 5A). We expected most of these survivors to revert to puromycin sensitivity due to loss, or error-prone repair, of the RsP cassette (see Figures 1C and 2C) and screening with puromycin confirmed this prediction. Twenty four of the 93 uninduced sub-clones were tested and were all puromycin resistant, while all but three of 24 I-SceI-survivors were sensitive.
To distinguish between different repair mechanisms, 22 survivors were analysed by Southern blotting; half of the samples are shown in Figure 5B. The analysis shown in Figure 2C and D indicates that HR with chromosome 11b was the predominant repair mechanism and, consistent with this, 19 survivors reflected this mechanism (nine shown in Figure 5B). Figure 2C also indicates ectopic HR with the TUB locus on chromosome 1 and use of this pathway was confirmed by the survivor in lane nine (Figure 5B, band at 9.5 kb in the 2110 blot) and two survivors analysed on a second blot (data not shown). Re-hybridization with a αTUB probe (Figure 5B) and PCR analysis (data not shown) confirmed that a αTUB ORF was copied from the tandem array on chromosome 1 as predicted for ectopic HR.
Gene conversion during allelic recombination can lead to loss of heterozygosity. Using the current assay system, we were unable to accurately assess the length of the gene conversion tract but, regeneration of the 7.6 kb fragment showed that the tract was restricted to <2.5 kb on one side of the lesion in 19 cases of allelic recombination (Figure 5B and see polymorphic HindIII site in Figure 1A). One survivor (clone 15, not shown) may have arisen due to a longer gene conversion tract, but another possible explanation is NHEJ or MMJ with loss of RFP. Indeed, we saw three examples of apparent end joining with retention of RFP sequence (Figure 5B). Finally, we quantified the chromosome 11 signals, relative to chromosome 1, and found no evidence for chromosome loss. Thus, HR dominated DSBR while, in a competition between allelic and ectopic inter-chromosomal recombination, allelic recombination was strongly favoured.
Microhomology-mediated joining contributes to chromosomal DSBR
Of 22 I-SceI-survivors analysed by Southern blotting, three appeared to reflect NHEJ or MMJ based on the presence of residual RFP signal on truncated HindIII fragments (Figure 5B, lanes 4, 6 and 9). Two of these were ‘mixed’ and also reflected HR with chromosome 11 (lane 6) or with chromosome 1 (lane 9, band at 9.5 kb). We saw two additional ‘mixed’ survivors on the second blot that reflected HR with both chromosomes 11 and 1 (data not shown). Detection of four survivors that exhibit evidence of multiple repair pathways was explained by I-SceI-induction in cells that had already replicated their nuclear genome prior to DSBR. To examine repair junctions in examples of end joining, we amplified junction fragments from genomic DNA using polymerase chain reaction (PCR) and then sequenced the products. Among samples shown in Figure 5B (lanes 6 and 9) and additional samples from earlier preliminary clonogenic analysis, we identified two distinct junctions from four independent clones. Sequence alignment clearly revealed that end joining was microhomology mediated (Figure 5C). We then used a PCR assay designed to survey large numbers (ca. ≫ 10 000) of end-joining events. This yielded only two major, reproducible products (Figure 5C) that were directly sequenced. As expected, one of the junctions was identical to the one identified in three independent clones while the other was unique (Figure 5C). The three junctions identified (six independent sequences in total) reflected microhomologies of 11–13 bp with one or two mismatches and loss of 72–278 bp (Figure 5C). Interestingly, junction 1 retains an intact RFP/PAC ORF explaining why three clonogenic survivors, including clones 6 and 9, displayed puromycin resistance (see above).
DISCUSSION
The introduction of chromosomal DSBs in a spatially and temporally co-ordinated manner has been an extremely powerful approach for investigating DSBR (39). We have taken this approach in T. brucei and demonstrate a number of major advantages over previous approaches used to examine DSBR pathways, which have relied upon the introduction of linear constructs and analysis of rare recombinants. First, the method allows introduction of single DSBs at specific loci. Second, temporal control and the efficiency of cleavage and repair allows for physical monitoring of DSBR. Third, we are able to compare allelic and ectopic recombination and chromosomal MMJ.
The specific induction of a single DNA DSB in T. brucei reveals DNA resection and accumulation of Rad51 foci, the timing of which is consistent with the idea that the former triggers the latter. Rad51 foci are sites of active DNA recombination as demonstrated in yeast (40) and are seen in the nuclei of T. brucei treated with the DNA-damaging agent, phleomycin (29). The current results now suggest that a single DNA lesion can also be visualized using Rad51 as a molecular marker in T. brucei. Rad51 may not accumulate into foci in the G1 phase of the cell cycle, however, reminiscent of the situation in yeast (41). We also show that a single lesion activates a G2M DNA damage checkpoint in T. brucei. This checkpoint arrests cell cycle progression prior to mitosis and is particularly important to allow time to repair stalled or broken replication forks, the major source of lesions requiring recombinational repair.
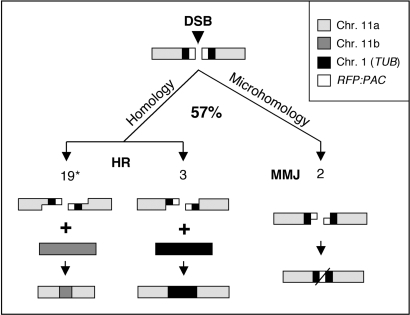
Following the induction of a DSB, we monitored chromosomal DSBR and assessed the contribution of different mechanisms (summarized in Figure 6). Lesions are generated in most, if not all, cells and successful repair generated viable cells in ∼60% of cases. HR clearly dominated DSBR with the allelic sequence on the homologous chromosome favoured over a homologous sequence at an ectopic location. Monitoring of the entire population or clonogenic survivors suggested that ∼85% or 75% of cells used allelic recombination, respectively. Under-representation of allelic recombination in the survivor assay is explained by repair after nuclear genome replication in some cases. Thus, allelic recombination was responsible for ∼85% of repair. If we calculate the number of repair events as a function of total cells instead of survivors, we see ∼50% allelic HR and 5% ectopic HR and MMJ (Figure 6).
Figure 6.
Summary of double-strand break-repair mechanisms in T. brucei. Approximately 57% of cells recover from a double-strand break (DSB) on chromosome 11a. Three distinct modes of repair were detected among at least 26 independent repair events. Resection is illustrated and the numbers of events detected are shown in each case. Asterisk denotes ∼85% of repair occurs via allelic recombination with chromosome 11b (see the text).
HR repair requires that the lesion associates with undamaged homologous sequence. Our results indicated that allelic recombination is preferred over ectopic recombination, even when the allelic-HR-substrate is >1 kb from the break and beyond the ectopic-HR substrate. A similar search for chromosomal break-distal homology has been reported in Saccharomyces cerevisiae (42) which, interestingly, contrasts with inefficient recombination with non-terminal homologous sequences on DNA constructs introduced into T. brucei (24). This remarkable preference for allelic recombination raised the question of how homologous sequences are ‘found’ and ‘selected’ in a chromosomal context. First, the length of homologous segments may impact on donor choice. In our analysis, the ectopic HR substrates are 240 bp and 323 bp while, in a T. brucei transformation assay, maximal transformation was observed when using substrates of 200 bp or longer (24). This argues against the idea that the ectopic homologous sequence is insufficient to compete with the allelic sequence. Second, donor sequence copy number may have an impact, as demonstrated in S. cerevisiae (43), but this would have favoured ectopic recombination since there are up to 40 potential donor templates on chromosome 1. Homologous chromosome were reported to be co-aligned along their lengths via multiple interstitial interactions during G1 and G2 in S. cerevisiae and Drosophila (44) but this view has been disputed (45). Thus, favoured allelic recombination in T. brucei may reflect either chromosome disposition prior to damage or a damage-induced homology search (10).
Homologues of factors required for NHEJ are absent or diverged in trypanosomatids (26) and Ku and Rad51-independent MMJ has been reported in T. brucei following the introduction of linear constructs (25) and using cell extracts (26). Disruption of factors required for NHEJ reveal MMJ as a subsidiary repair pathway in mammalian cells (46) and the genetic requirements for MMJ in yeast suggest that it is a form of ‘micro single-strand annealing’ (18). Following the induction of a DSB, we looked for evidence of intra-chromosomal end joining, either NHEJ or MMJ, and found several examples or MMJ but no evidence for NHEJ. Thus, HR and MMJ dominate chromosomal DSBR in T. brucei, but, since NHEJ can occur in as few as 0.33% of S. cerevisiae with a lesion (47), we are unable to exclude the possibility that NHEJ also operates at a low level in T. brucei.
Recombination plays a specialized role in VSG gene rearrangement and host immune evasion in T. brucei and can bring about antigenic variation in 1% of cells per population doubling. The telomeric VSG expression sites may select from among a vast variety of subtelomeric donors for translocation of a new VSG or formation of VSG mosaics (6). Repetitive flanking sequences are important for this switching and diversification and these rearrangements can operate via Rad51-dependent or independent pathways (25,31). Below, we briefly consider our results in relation to antigenic variation, the potential role of microhomology and the choice of recombination partners.
NHEJ is important for immunoglobulin class switch recombination in mammalian B cells. When the NHEJ pathway is eliminated, however, by Ku disruption for example, the programmed DSBs are channelled into an end-joining pathway that uses stretches of microhomology and often involves chromosomal translocation (17,19–21). Antigenic variation appears to be unaffected in Ku-deficient T. brucei, consistent with the idea that NHEJ is not required to generate VSG diversity (48). We now show that NHEJ plays, at most, a minor role in chromosomal DSBR in T. brucei. In the case of VSG recombination, flanking, imperfect, so-called 70-bp repeats, present an abundance of microhomologous recombination substrates and assembly of mosaic VSG genes (6) may also be driven by microhomology. We suggest that, with a deficiency in NHEJ, MMJ could play an important role in VSG rearrangement and expression in T. brucei.
We previously reported use of I-SceI to remove the telomere at a silent VSG expression site, but this failed to trigger a DNA damage response or, in most cases, to trigger recombination (32). This dramatic difference, relative to a lesion in the core of the chromosome reported here, may reflect the suppression of subtelomeric recombination by telomere-binding proteins, as demonstrated in mammalian cells (49) or, more likely, by the phylogenetically restricted DNA base modification, β-d-glucosyl-hydroxymethyluracil, found throughout silent VSG expression sites (50,51). It will now be important to analyse active expression sites that may display enhanced recombination due to transcription (52) and the absence of DNA base modification.
In addition to the avenues outlined above, the system reported here will allow for a genetic dissection of the chromosomal DSBR pathways available to T. brucei. This will lead to a better understanding of chromosomal recombination and repair and of the mechanisms underlying antigenic variation in this divergent eukaryote and important pathogen.
ACKNOWLEDGEMENTS
This work was funded by The Wellcome Trust (069909). We thank Sam Alsford, Martin Taylor and John Kelly (LSHTM) for comments on the draft manuscript and Mark Mitchell (Bristol, UK) for help with graphics. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr. Mol. Med. 2004;4:563–576. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- 3.Morrison LJ, Majiwa P, Read AF, Barry JD. Probabilistic order in antigenic variation of Trypanosoma brucei. Int. J. Parasitol. 2005;35:961–972. doi: 10.1016/j.ijpara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callejas S, Leech V, Reitter C, Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 2006;16:1109–1118. doi: 10.1101/gr.5147406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 8.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 10.Barzel A, Kupiec M. Finding a match: how do homologous sequences get together for recombination? Nat. Rev. Genet. 2008;9:27–37. doi: 10.1038/nrg2224. [DOI] [PubMed] [Google Scholar]
- 11.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 14.Smith JA, Bannister LA, Bhattacharjee V, Wang Y, Waldman BC, Waldman AS. Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2. Mol. Cell Biol. 2007;27:7816–7827. doi: 10.1128/MCB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moynahan ME, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc. Natl Acad. Sci. USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 18.Decottignies A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics. 2007;176:1403–1415. doi: 10.1534/genetics.107.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 21.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 23.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 24.Barnes RL, McCulloch R. Trypanosoma brucei homologous recombination is dependent on substrate length and homology, though displays a differential dependence on mismatch repair as substrate length decreases. Nucleic Acids Res. 2007;35:3478–3493. doi: 10.1093/nar/gkm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway C, Proudfoot C, Burton P, Barry JD, McCulloch R. Two pathways of homologous recombination in Trypanosoma brucei. Mol. Microbiol. 2002;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- 26.Burton P, McBride DJ, Wilkes JM, Barry JD, McCulloch R. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot. Cell. 2007;6:1773–1781. doi: 10.1128/EC.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson NP, McCulloch R, Conway C, Browitt A, Barry JD. Inactivation of Mre11 does not affect VSG gene duplication mediated by homologous recombination in Trypanosoma brucei. J. Biol. Chem. 2002;277:26185–26193. doi: 10.1074/jbc.M203205200. [DOI] [PubMed] [Google Scholar]
- 28.Tan KS, Leal ST, Cross GA. Trypanosoma brucei MRE11 is non-essential but influences growth, homologous recombination and DNA double-strand break repair. Mol. Biochem. Parasitol. 2002;125:11–21. doi: 10.1016/s0166-6851(02)00165-2. [DOI] [PubMed] [Google Scholar]
- 29.Proudfoot C, McCulloch R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res. 2005;33:6906–6919. doi: 10.1093/nar/gki996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alsford S, Kawahara T, Isamah C, Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol. Microbiol. 2007;63:724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 31.McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glover L, Alsford S, Beattie C, Horn D. Deletion of a trypanosome telomere leads to loss of silencing and progressive loss of terminal DNA in the absence of cell cycle arrest. Nucleic Acids Res. 2007;35:872–880. doi: 10.1093/nar/gkl1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingram AK, Cross GA, Horn D. Genetic manipulation indicates that ARD1 is an essential Nα-acetyltransferase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;111:309–317. doi: 10.1016/s0166-6851(00)00322-4. [DOI] [PubMed] [Google Scholar]
- 34.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 36.McKean PG, Keen JK, Smith DF, Benson FE. Identification and characterisation of a RAD51 gene from Leishmania major. Mol. Biochem. Parasitol. 2001;115:209–216. doi: 10.1016/s0166-6851(01)00288-2. [DOI] [PubMed] [Google Scholar]
- 37.Regis-da-Silva CG, Freitas JM, Passos-Silva DG, Furtado C, Augusto-Pinto L, Pereira MT, DaRocha WD, Franco GR, Macedo AM, Hoffmann JS, et al. Characterization of the Trypanosoma cruzi Rad51 gene and its role in recombination events associated with the parasite resistance to ionizing radiation. Mol. Biochem. Parasitol. 2006;149:191–200. doi: 10.1016/j.molbiopara.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- 39.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki T, Bressan DA, Shinohara M, Haber JE, Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell Biol. 1999;19:4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melamed C, Kupiec M. Effect of donor copy number on the rate of gene conversion in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1992;235:97–103. doi: 10.1007/BF00286186. [DOI] [PubMed] [Google Scholar]
- 44.Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenz A, Fuchs J, Burger R, Loidl J. Chromosome pairing does not contribute to nuclear architecture in vegetative yeast cells. Eukaryot. Cell. 2003;2:856–866. doi: 10.1128/EC.2.5.856-866.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verkaik NS, Esveldt-van Lange RE, van Heemst D, Bruggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 47.Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 2003;328:847–862. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- 48.Conway C, McCulloch R, Ginger ML, Robinson NP, Browitt A, Barry JD. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem. 2002;277:21269–21277. doi: 10.1074/jbc.M200550200. [DOI] [PubMed] [Google Scholar]
- 49.Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 50.Ekanayake DK, Cipriano MJ, Sabatini R. Telomeric co-localization of the modified base J and contingency genes in the protozoan parasite Trypanosoma cruzi. Nucleic Acids Res. 2007;35:6367–6377. doi: 10.1093/nar/gkm693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. β-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl Acad. Sci. USA. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alsford S, Horn D. RNA polymerase I transcription stimulates homologous recombination in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:77–79. doi: 10.1016/j.molbiopara.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]