Abstract
Recognition of tRNA by the cognate aminoacyl-tRNA synthetase during translation is crucial to ensure the correct expression of the genetic code. To understand tRNALeu recognition sets and their evolution, the recognition of tRNALeu by the leucyl-tRNA synthetase (LeuRS) from the primitive hyperthermophilic bacterium Aquifex aeolicus was studied by RNA probing and mutagenesis. The results show that the base A73; the core structure of tRNA formed by the tertiary interactions U8–A14, G18–U55 and G19–C56; and the orientation of the variable arm are critical elements for tRNALeu aminoacylation. Although dispensable for aminoacylation, the anticodon arm carries discrete editing determinants that are required for stabilizing the conformation of the post-transfer editing state and for promoting translocation of the tRNA acceptor arm from the synthetic to the editing site.
INTRODUCTION
An aminoacyl-tRNA synthetase (aaRS) catalyzes the activation of its cognate amino acid and transfers it to the corresponding tRNA. tRNA identity is governed by positive (determinants) and negative (anti-determinants) elements that respectively trigger specific aminoacylation and prevent false charging (1).
The tRNALeu identity is of particular interest because of the following reasons: (i) tRNALeu, along with tRNASer, tRNASec and prokaryotic tRNATyr, belongs to class II tRNAs in which the variable arm consists of more than 10 nucleotides, while class I tRNAs contain a variable arm with only 4–5 nucleotides (2); (ii) usually, there are five to six tRNALeu isoacceptors corresponding to the six leucine codons, and the isoacceptors exhibit sequence variability and structure heterogeneity in the anticodon region due to degeneracy of the leucine codons (3,4); and (iii) leucyl-tRNA synthetase (LeuRS), a class Ia aaRS, contains a second active site known as the editing site to clear the mischarged non-cognate aa-tRNA (5–10). Translocation of the tRNA 3′-terminus between this editing site and the aminoacylation site is considered to be crucial for the proofreading function (8–10). In the case of the isoleucine system, discrete tRNA determinants were found to be specific for the editing reaction (11). Thus far, nucleotides specifically involved in editing have not been identified in tRNALeu.
Due to the characteristics mentioned above, tRNALeu recognition elements have been widely studied in prokaryotic and eukaryotic systems, including the bacterium Escherichia coli (12–17), archaeon Haloferax volcanii (18), yeast Saccharomyces cerevisiae cytoplasmic system (19), bean Phaseolus vulgaris cytoplasmic system (20), and human cytoplasmic and mitochondrial systems (21–24). Although there are some minor variations in recognition systems across different organisms, the discriminator base and the tertiary structure are the major recognition elements (1,19,33). S. cerevisiae cytoplasmic tRNALeu is a remarkable exception in that it adopts a more sequence-specific recognition system based on nucleotides 73, 35 and 37, while the other systems depend more upon the tertiary structure of the tRNA molecule (19).
Leucyl-tRNA synthetase from Aquifex aeolicus (AaLeuRS) was reported to possess many evolutionary remnant characteristics. It has a unique αβ heterodimeric structure that mimics primitive aaRS enzymes with separate active sites and tRNA-binding domains (25). AaLeuRS aminoacylates a cognate minihelix believed to be the tRNA ancestor (26), and its editing domain can function as an isolated domain (27). To investigate the tRNA–synthetase interactions, we performed enzymatic and chemical probing on tRNALeu complexed with LeuRS as well as kinetic analysis on tRNALeu variants. We found that all the recognition elements previously identified in other organisms play an important role in the AaLeuRS/tRNALeu system [except for nucleotide 35 found to be an identity element in the yeast system (19)], suggesting a common origin for this recognition mode. In addition, we found that some recognition elements of tRNALeu are specific for aminoacylation while others are required for the editing reaction. This study shows that the tRNA scaffold can provide distinct determinants for the aminoacylation and editing reactions and highlights the complexity of the tRNA–synthetase interactions during the different steps from the initial binding to the final release of the product.
MATERIALS AND METHODS
Materials
l-leucine, dithiothreitol (DTT), NTP, 5′-GMP, inorganic pyrophosphatase and iodine were purchased from Sigma (USA); [α-32P]ATP, [γ-32P]ATP, NTP[α-35S]s and [14C]-l-leucine (300 Ci/mol) were obtained from Amersham Biosciences (England); and GF/C filters were purchased from Whatman Company (Germany). T4 DNA ligase and restriction endonucleases were obtained from Sangon Company (Shanghai Branch, Canada). Phosphodiesterase was purchased from Worthington Biochemical Co. (USA), and T4 polynucleotide kinase and bacterial alkaline phosphatase were from New England Biolabs (Canada). RNase T1, RNase T2 and RNase V1 were from MBI Fermentas (Lithuania), Invitrogen (USA) and Pierce (USA), respectively. T7 RNA polymerase was purified from an overproducing strain in our laboratory (28), and tRNA nucleotidyl transferase (CCase) was also purified from an overproducing strain.
Preparation of protein and tRNA
AaLeuRS and its β-subunit were purified according to published methods (25). Purified A. aeolicus tRNALeuGAG and tRNALeuCAA overproduced in E. coli were gifts from Dr J. Cavarelli. The 5′-end labeling of tRNAs with [γ-32P]ATP was performed by the action of T4 polynucleotide kinase on tRNAs previously dephosphorylated with alkaline phosphatase (29). The 3′-end labeling of the isolated tRNA was achieved by [α-32P]ATP exchange in the presence of CCase (G. Keith, personal communication). Labeled tRNA was purified by electrophoresis on a 10% polyacrylamide/8M urea gel, followed by passive elution in 10 mM Tris–HCl (pH 7.5) containing 0.3 M NaCl and 0.5 mM EDTA, and precipitation by ethanol after phenol/chloroform extraction. The samples were then dissolved in water and renatured by heating for 2 min at 80°C and cooling down for 10 min at room temperature. Transcription with phage T7 RNA polymerase was performed according to previous protocols (26).
Nuclease footprinting
Incubation of protein–tRNA complexes with RNase T1, RNase T2 and RNase V1 was performed in 100 mM Tris–HCl (pH 7.8), 30 mM KCl and 12 mM MgCl2 for 10 min at 20°C. Each reaction was performed in a 15 μl reaction volume containing 10 pmol total tRNA (as carrier), 5′-end- or 3′-end-[32P]-labeled tRNA (100 000 Cerenkov counts), 0 or 5 μM (for tRNALeuGAG) or 10 μM (for tRNALeuCAA) AaLeuRS, and RNase (0.5 U for T1 and T2 and 0.01 U for V1). Prior to enzymatic digestion, tRNA and AaLeuRS were incubated on ice for 20 min. Reactions were stopped by the addition of 15 μl of ‘Stop Mix’ (0.6 M sodium acetate, 3 mM EDTA, and 0.1 μg/μl total tRNA), followed by phenol/chloroform extraction and ethanol precipitation. An alkaline ladder was obtained by the incubation of the 5′-end- or 3′-end-labeled tRNA (100 000 Cerenkov counts) and 1 μg of total tRNA in 100 mM NaHCO3 (pH 9.0) at 80°C for 10 min. A guanine ladder was obtained by RNase T1 digestion of denatured tRNA as follows: 1 μg of total tRNA containing labeled tRNA (100 000 Cerenkov counts) was denatured by heating at 80°C for 5 min, followed by rapid cooling at room temperature and digestion in 7 M urea, 1 mM EDTA and 20 mM sodium citrate (pH 4.5) in the presence of 2 U of RNase T1 for 10 min at 80°C. The radioactive tRNAs were washed twice with 80% ethanol, dried and quantified (Cerenkov); dissolved in loading buffer (95% v/v formamide, 20 mM EDTA, and 0.1% w/v dyes); and heated for 2 min at 80°C. The radioactive bands were separated on a 10% polyacrylamide/8M urea gel and analyzed using a FUJIX Bio-Imaging Analyzer BAS 2000 (Japan).
Iodine footprinting
The four phosphorothioate-containing tRNA transcripts were prepared according to a previously described procedure (26). Each phosphorothioate analogue (NTPαS) was separately incorporated into tRNAs at 5% (0.2 mM) of the corresponding NTP to ensure an average incorporation of one phosphorothioate per tRNA molecule during the in vitro transcription. The tRNA transcripts were quantified by absorbance at 260 nm and separated by 12% polyacrylamide gel electrophoresis (PAGE). The transcripts were visualized by UV shadowing and eluted from the gel slices, as in the case of end-labeled tRNAs described in the previous paragraph. After ethanol precipitation, the tRNA transcripts were dissolved in water, heated to 60°C, and cooled slowly to 25°C to refold the tRNA. Then, 5 µg of each transcript was incubated in 50 mM Tris–HCl (pH 8.1) and 7% formamide with 1 U of bacterial alkaline phosphatase at 65°C for 15 min. This step was repeated once. After phenol/chloroform extraction and ethanol precipitation, the dephosphorylated tRNAs were labeled with 20 µCi [γ-32P]ATP by the action of 20 U T4 polynucleotide kinase in 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT and 10 U RNase inhibitor. The labeled tRNAs were then separated by denaturing 10% PAGE. Gel slices containing full-length transcripts were excised using the autoradiogram as a guide, soaked in elution buffer overnight at 4°C, and the tRNA was recovered by ethanol precipitation. For RNA footprinting, 40 000 Cerenkov counts of each labeled tRNA transcript was mixed with 1 μM of unlabeled tRNA and 5 μM LeuRS in a 10 μl volume reaction containing 10 mM Hepes–KOH (pH 7.4), 10 mM NaCl and 10 mM MgCl2. The mixture was incubated at room temperature for 3 min; then 1 μl of 10 mM iodine was added to start the cleavage reaction. Control experiments contained water instead of the enzyme. After 1 min at room temperature, the reaction was stopped by adding 40 μl of 0.4 M Na acetate (pH 6.0). Following phenol/chloroform extraction and ethanol precipitation in the presence of glycogen, the reaction products were separated on denaturing 12% polyacrylamide sequencing gels and visualized by autoradiography. Exposed films were scanned and relative band intensities were analyzed.
Enzymatic assays
The aminoacylation activity of AaLeuRS was determined at 50°C in a reaction mixture containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 4 mM ATP, 0.5 mM DTT, 20 μM [14C]-l-leucine and 5 nM enzyme. The kinetic constants for tRNALeu were determined using various concentrations (from 0.125 μM to 20 μM) of the relevant tRNALeu substrates. Mis-aminoacylation assays were carried out as described previously (17), except that 10 μM [3H]-l-isoleucine (30 Ci/mmol) and 1 μM enzyme were used.
The hydrolytic editing activities of AaLeuRS toward mischarged [3H]Ile-tRNALeu and mutated derivatives were measured in reaction mixtures containing 100 mM Tris–HCl (pH 7.5), 30 mM KCl, 12 mM MgCl2, 0.5 mM DTT and 1 μM [3H]Ile-tRNALeu (270 μCi/μmol) at 37°C. The reaction was initiated by adding 3 nM AaLeuRS. At various time intervals, aliquots were quenched and precipitated with 5% trichloroacetic acid as described previously (27). The spontaneous hydrolysis in the absence of the enzyme (less than 10%) were subtracted and determined as the average of three independent experiments.
The total editing assay based on the measurement of ATP hydrolysis was performed as reported previously (11) and was performed at 65°C in 100 μl reaction mixtures containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 5 mM DTT, 1 U/ml RNase inhibitor, 2 U/ml pyrophosphatase, 3 mM [γ-32P]ATP (3–5 cpm/pmol), 50 mM isoleucine, 5 μM tRNALeu transcripts and 1 μM AaLeuRS. Aliquots (10 μl) of the editing reaction were removed and mixed with 350 μl of quenching liquid containing 6% activated charcoal, 7% HClO4 and 10 mM tetrasodium pyrophosphate. After centrifugation, the amount of inorganic phosphate [32P] in 50 μl of supernatant was quantified by scintillation counting. The final rate of ATP hydrolysis was determined as the average of three independent experiments.
RESULTS
The heterodimeric AaLeuRS possesses several evolutionary remnant characteristics (25–27), including the ability to charge a minihelixLeu, a property that is not shared with the more ‘modern’ E. coli or human cytoplasmic LeuRS (16,26,30). These data suggest that the ancient AaLeuRS may have retained some primitive tRNA recognition mechanism that we intend to elucidate in the present study. First, we performed probing investigations by nuclease and iodine cleavage in order to locate the enzyme interacting areas on the tRNA. To this end, we deduced a set of tRNA residues to be mutated and analyzed for their aminoacylation properties as well as their editing properties.
Nuclease probing and footprinting experiments on AatRNALeuGAG and AatRNALeuCAA complexed with AaLeuRS
The tRNA probing was performed with purified overexpressed AatRNALeuCAA and AatRNALeuGAG radiolabeled at their 5′-ends or 3′-ends. The structures of the two tRNAs were probed using the RNases T1, T2 and V1. RNase T2 preferentially cleaves after unpaired residues, and RNase T1 cuts after unpaired G residues. RNase V1 acts on double-stranded sequences or higher-order structures. The background hydrolysis by water or traces of contaminating metal cations was distinguished from probe-induced digestion by control experiments performed without probes. The two AatRNAs showed a similar pattern of cleavage (Figures 1 and 2, − lanes). The acceptor arm and the anticodon stem-loop exhibited strong cleavages. On the other hand, only faint cuts were observed in the variable arm and D-loop.
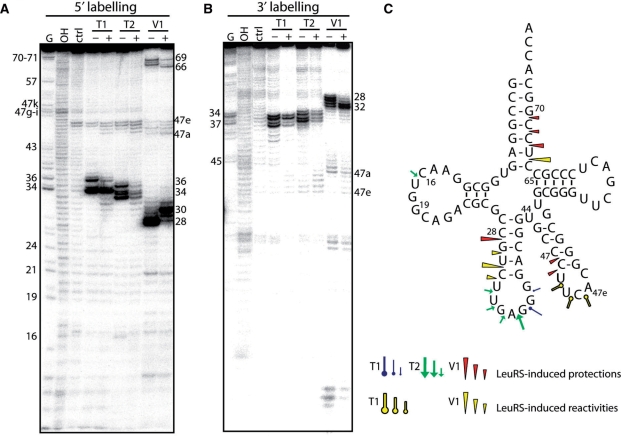
Figure 1.
Nuclease probing of AatRNALeuGAG in the free form or in complex with AaLeuRS. tRNALeuGAG was labeled at its 5′-end (A) or 3′-end (B). Probing was conducted in the presence (+) or absence (−) of LeuRS. OH and T1 are ladders of the tRNA under the denaturing condition; Ctrl is the control without any probe. The probes comprised RNase T1, RNase T2 and RNase V1. Numbers refer to tRNA nucleotide positions. (C) Cloverleaf structure of tRNA summarizing the reactivity changes observed in the tRNA following AaLeuRS binding. The symbols and color codes for the probes are indicated in the figure. Three intensities of cuts/modifications for each probe are shown (strong, medium and moderate).
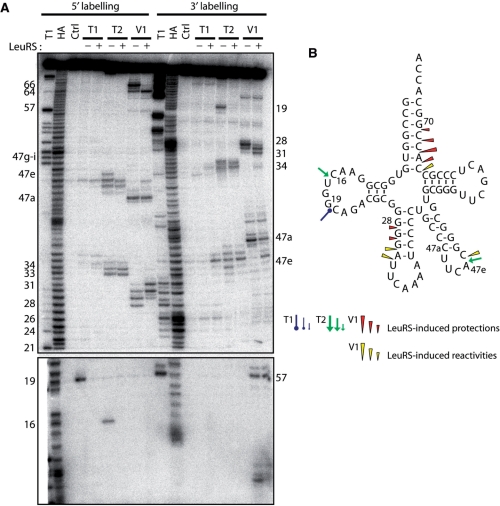
Figure 2.
Nuclease probing of AatRNALeuCAA in the free form or in complex with AaLeuRS. (A) tRNALeuCAA was labeled at its 5′-end or 3′-end. Probing was done in the presence (+) or absence (−) of LeuRS. OH and T1 are ladders of the tRNA under the denaturing condition; Ctrl is the control without any probe. The probes comprised RNase T1, RNase T2 and RNase V1. Numbers refer to tRNA nucleotide positions. (B) Cloverleaf structure of tRNA summarizing the reactivity changes observed in the tRNA following AaLeuRS binding. The symbols and color codes for the probes are indicated in the figure. Three intensities of cuts/modifications for each probe are shown (strong, medium and moderate).
Probing AatRNALeuGAG complexed with AaLeuRS (Figure 1, + lanes) revealed significant reactivity changes in the anticodon stem-loop (residues 28–36) and acceptor arm (residues 66–69). Protections were also observed in the less reactive D-loop and variable arm regions. The reactivity changes mostly comprised cleavage protections (C28, U67, C68, C69 and U32∼G37); however, in some cases, reactivity increases were observed in the anticodon stem (G29, U30 and C31), acceptor arm (C66), and variable arm (U47b-d), indicating that the tRNA structure had been remodelled following synthetase binding.
Probing of AatRNALeuCAA under the same conditions led to comparable results at the level of the acceptor and anticodon arms (Figure 2, + lanes). However, no protection of the anticodon loop by LeuRS could be detected (see RNase T2 cleavages, Figure 2), suggesting that the complex of LeuRS with AatRNALeuCAA is distinct from that with AatRNALeuGAG. This difference implies alternative interpretations. The two tRNAs may bind to the enzyme in different ways, suggesting the existence of a distinct set of interacting residues and probably identity elements. The tRNAs might also have been probed in two different states which could alternatively be the ground state, the aminoacylation or the editing state or the so-called exit state (8,10). For further investigation, we selected AatRNALeuGAG because it displayed protections that might reveal interactions with the anticodon arm. These putative contacts were especially interesting to analyze because it is generally assumed that bacterial LeuRSs do not recognize the anticodon of their tRNAs (13,16).
Iodine probing and footprinting experiments on AatRNALeuGAG complexed with AaLeuRS
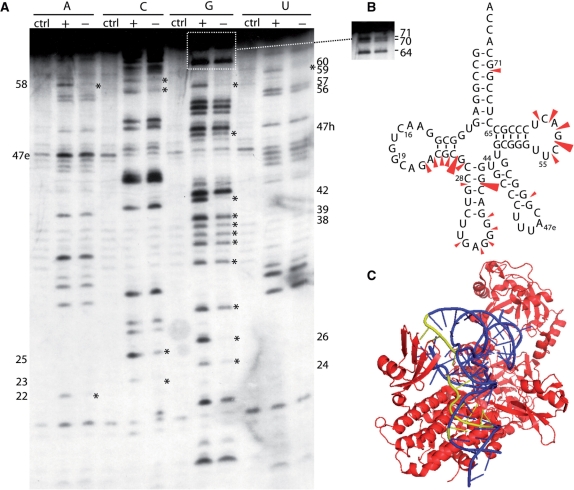
Iodine-based footprinting depends on the reactivity of the internucleotide phosphorothioate linkage with iodine, leading to the cleavage of the RNA transcript at the insertion site of the phosphorothioate nucleotide. Compared with RNases, iodine is a small probe, allowing it to access residues buried in the tertiary structure of the tRNA. The resulting cleavages affect nearly all the tRNA residues, providing more data than that provided by RNase cleavage. In addition, iodine can access nucleotides that are usually inaccessible to RNases due to hindrance from the tRNA–synthetase complex. Four transcripts of the more nuclease-reactive AatRNALeuGAG were individually synthesized; their aminoacylation by AaLeuRS was comparable to that of the native transcript, as determined by the Vmax and Km values (see Supplementary Data S1). The AaLeuRS footprinting on tRNALeuGAG detected by iodine was largely comparable to that detected by RNases (Figure 3). However, the protections detected in the anticodon loop were weaker than those observed with RNases T1 and T2. This might be due to the small size of the iodine molecule which can more easily access the tRNA for cleavage. Additional strong protections were observed in the D-arm (A22, C23, G24, C25 and G26) and T-loop (C56, G57, A58, C59 and U60). A strong cleavage was also found at position 42 in the anticodon stem. Together with the results of RNase reactivity, the above results strongly support the existence of an interaction between the enzyme and the tRNA in the anticodon arm.
Figure 3.
Iodine probing of AatRNALeuGAG containing phosphorothioate nucleotides. (A) AatRNALeuGAG was labeled at its 5′-end and probed in the presence (+) or absence (−) of AaLeuRS; Ctrl is the control without iodine. (B) Cloverleaf structure of AatRNA summarizing the protections observed in the tRNA following AaLeuRS binding. Three intensities of protections are shown. (C) View of the T. thermophilus LeuRS/tRNALeu complex (2BYT) (10). The enzyme is shown in red and the tRNA is in blue. The tRNA nucleotides protected from iodine cleavage in AatRNA are highlighted in yellow.
AaLeuRS footprints explored by tRNA mutagenesis
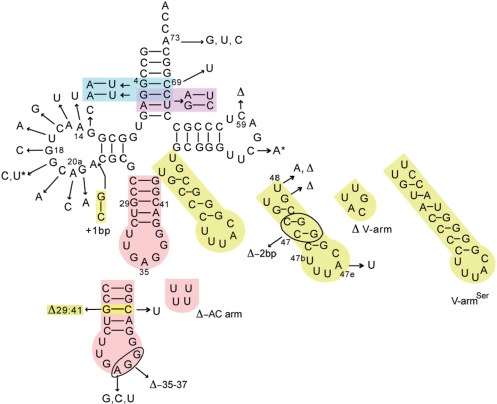
The footprinting of AaLeuRS on AatRNALeu revealed several protection areas in which identity elements may be present. In order to identify these nucleotides, transcripts of wild-type AatRNALeuGAG and a total of 34 mutants were designed, constructed and transcribed in vitro by using T7 RNA polymerase (Figure 4). The three-dimensional structure of mutant tRNA molecules was stable at 50°C in melting point assays (data not shown). The kinetic parameters of AaLeuRS for these transcripts were assayed and are shown in Table 1.
Figure 4.
Summary of AatRNALeu constructs in this study. The AatRNA variants are derived from AatRNALeuGAG. The arrows indicate the mutation locations; bp, base pair; Δ, deletions. Colors are used for the multiple mutants, and the asterisks indicate the double mutation 19–56.
Table 1.
Kinetic constants of AaLeuRS for AatRNALeuGAG and derived mutants in the aminoacylation reaction
| tRNA domain | Mutants of AatRNA | kcat (s−1) | Km (μM) | kcat/Km (μM−1s−1) | kcat/Km (relative) |
|---|---|---|---|---|---|
| native | 1.28 ± 0.34 | 1.10 ± 0.26 | 1.16 | 1.0 | |
| Acceptor-arm | A73G | 0.06 ± 0.015 | 7.40 ± 2.6 | 0.008 | 0.007 |
| A73U | 0.19 ± 0.05 | 7.65 ± 1.4 | 0.025 | 0.022 | |
| A73C | 0.15 ± 0.06 | 6.29 ± 2.1 | 0.024 | 0.021 | |
| A5:U68&G6:C67 | 1.46 ± 0.41 | 1.60 ± 0.45 | 0.91 | 0.78 | |
| A4:U69&A5:U68 | 1.67 ± 0.33 | 0.83 ± 0.21 | 2.01 | 1.73 | |
| C69U | 0.098 ± 0.014 | 3.29 ± 0.45 | 0.03 | 0.026 | |
| D-arm | G13C | 0.56 ± 0.08 | 0.32 ± 0.056 | 1.75 | 1.51 |
| A14U | 0.53 ± 0.12 | 18.00 ± 2.7 | 0.029 | 0.025 | |
| A15U | 1.13 ± 0.15 | 1.21 ± 0.18 | 0.93 | 0.8 | |
| C16G | 1.34 ± 0.31 | 1.05 ± 0.12 | 1.28 | 1.10 | |
| U17A | 1.03 ± 0.26 | 1.18 ± 0.3 | 0.87 | 0.75 | |
| G18C | 0.56 ± 0.062 | 5.20 ± 0.87 | 0.11 | 0.095 | |
| G19C | nm | nm | nm | nm | |
| C20A | 1.21 ± 0.17 | 1.50 ± 0.25 | 0.81 | 0.70 | |
| A20aC | 0.94 ± 0.14 | 2.19 ± 0.38 | 0.43 | 0.37 | |
| G21A | 0.59 ± 0.077 | 1.37 ± 0.22 | 0.43 | 0.37 | |
| G13:C23 | 1.061 ± 0.32 | 3.76 ± 0.63 | 0.28 | 0.24 | |
| G19U:C56A | 0.94 ± 0.13 | 1.49 ± 0.14 | 0.63 | 0.54 | |
| Anticodon-arm | A35U | 1.52 ± 0.22 | 1.60 ± 0.21 | 0.95 | 0.82 |
| A35C | 1.58 ± 0.37 | 0.94 ± 0.16 | 1.68 | 1.45 | |
| A35G | 1.34 ± 0.24 | 0.97 ± 0.21 | 1.38 | 1.19 | |
| Δ35-37 | 1.52 ± 0.32 | 0.77 ± 0.11 | 1.97 | 1.70 | |
| ΔG29:C41 | 1.01 ± 0.21 | 0.89 ± 0.092 | 1.13 | 0.97 | |
| C41U | 0.84 ± 0.12 | 0.85 ± 0.23 | 0.99 | 0.85 | |
| ΔAC | 1.43 ± 0.19 | 2.77 ± 0.31 | 0.52 | 0.45 | |
| Variable-arm | V-armSer | 0.11 ± 0.026 | 4.03 ± 0.55 | 0.027 | 0.023 |
| V-arm-2bp | 0.75 ± 0.15 | 1.42 ± 0.26 | 0.53 | 0.46 | |
| ΔV-arm | 0.38 ± 0.05 | 4.20 ± 0.57 | 0.09 | 0.078 | |
| U48A | 0.64 ± 0.082 | 3.64 ± 0.53 | 0.18 | 0.16 | |
| ΔG47k | 0.48 ± 0.05 | 4.02 ± 0.61 | 0.12 | 0.10 | |
| ΔU48 | nm | nm | nm | nm | |
| A47eU | 1.41 ± 0.15 | 0.85 ± 0.076 | 1.66 | 1.43 | |
| TψC-arm | ΔC59 | nm | nm | nm | nm |
| A58G | 1.57 ± 0.41 | 9.16 ± 1.86 | 0.17 | 0.15 |
All kinetic data were reproduced at least 3 times; nm: not measurable.
To explore the function of the protections detected in the acceptor arm, two mutants carrying double base-pair changes and one mutant with a single base-pair change were constructed (Figure 4 and Table 1). In the case of the mutant A5:U68&G6:C67 in which both the fifth and sixth base pairs were exchanged, the kinetic parameters were not modified significantly. A slight increase in catalytic efficiency was detected in the mutant A4:U69&A5:U68, suggesting that the enhanced flexibility of the acceptor stem favored aminoacylation. However, the introduction of a single-point mutation that created a wobble base pair at position 4:69 drastically reduced the catalytic efficiency because of a loss of both catalytic and binding performances. These data suggest that the regular helical conformation of the acceptor stem is more essential than a specific sequence.
On the opposite branch of the L-shape of the tRNA molecule, the anticodon arm exhibited extensive and strong probe protections. These protections (nucleotides 28, 29 and 42) as well as the reactivity increases observed for nucleotides 30 and 31 suggested that AaLeuRS might interact with some nucleotides in the anticodon arm and modify the tRNA structure and the reactivity of the free nucleotides. The crystallographic structure of the archaeon Pyrococcus horikoshii LeuRS/tRNALeu complex structure demonstrates that the tRNA anticodon loop does not come into contact with LeuRS, while the anticodon stem interacts with the helix bundle domain—a specific domain found in class Ia aminoacyl-tRNA synthetases and usually dedicated to tRNA anticodon stem-loop recognition (8). Seven tRNA variants were synthesized in order to corroborate the reactivity signals with kinetic parameters (Figure 4 and Table 1). The central base A35, which is an identity element in the yeast system (19), was substituted by three other bases without affecting the catalytic performances. The same absence of effect was observed in a mutant with deletion of three nucleotides from the loop (nucleotides 35–37). Obviously, these data suggest that the weak protections detected in the anticodon loop do not result from crucial interactions occurring with the enzyme. Neither the deletion of the base pair G29–C41 (ΔG29 : C41) nor its change to a wobble G–U pair (C41U) exerted any effect on the catalytic properties. Nevertheless, the complete deletion of the anticodon arm (ΔAC-arm) induced a significant loss of affinity without any effect on the kcat value. Taken together, these data suggest that the anticodon loop does not contribute to the aminoacylation reaction. However, one cannot exclude the possibility that the detected protections reflect interactions that occur during other processes such as editing for instance (see later).
The variations in the RNase and iodine reactivities exhibited by the variable arm were comparable to those detected in the anticodon arm. The crystallographic structure of P. horikoshii LeuRS shows that the synthetase binds the variable arm with a leucyl-specific tRNA-binding domain located at the C-terminal end of the molecule (8). However, the β-subunit of AaLeuRS lacks approximately 90 residues at the C-terminus when compared with that of the P. horikoshii enzyme, and AatRNALeu also differs from its P. horikoshii counterpart by one additional base pair in the variable arm and 1 more nucleotide in the loop. These differences suggest that the mechanism of tRNALeu binding in A. aeolicus might differ from that in the P. horikoshii system. 7 tRNALeu variants were constructed. Removing the variable arm (ΔV-arm) or changing its orientation (U48A and ΔG47k) (10) resulted mainly in affinity decreases (Figure 4 and Table 1). A drastic modification of the orientation in the mutant ΔU48 completely abolished the aminoacylation of the molecule. The tRNA aminoacylation was relatively insensitive to the deletion of two base pairs (V-arm-2bp); however, increasing the size of the variable arm by introducing the variable arm from tRNASer (V-armSer) resulted in a considerable drop in kcat and affinity. Substitution of the nucleotide A47e for U did not induce obvious changes, suggesting that the protections seen at this level do not result from specific base contacts. Taken together, these results appear very similar to those obtained with the anticodon arm. The variable arm appears to provide binding energy for the tRNA–synthetase complex formation. It can be reduced in size, but the orientation and the maximal length must be preserved.
The tertiary structure is a complex network that defines the amino acid accepting identity of AatRNALeu
The tRNALeu identity has been extensively studied in E. coli. Together with the discriminator base A73, the tertiary structure resulting from interactions U8–A14, A15–U48, G18–U55 and G19–U56 is crucial to define the leucine identity (12–17). Remarkably, all the AatRNALeu isoacceptors possess completely conserved D-arm and T-arm. A set of 12 tRNA variants with respect to the D-arm was constructed (Figure 4 and Table 1). The substitution of residues 14, 18 and 19 drastically reduced the aminoacylation capacity of the tRNA with a major effect on the affinity. Obviously, disrupting the tertiary base-pair interactions U8–A14, G18–U55 and G19–U56 was deleterious with respect to the charging capacity, whereas changing the A15–U48 interaction was not critical. Substitutions of the other nucleotides from the loop, which are not involved in the tertiary base-pair interactions, had no obvious effects on the aminoacylation efficiency, except for the substitution of residues 20a and 21 that resulted in a 2.5-fold decrease in the catalytic efficiency. The same effect was observed following lengthening of the D-stem by insertion of the G13–C23 base pair. Clearly, the tertiary structure of tRNA is essential for its binding and aminoacylation. This is also illustrated by the compensatory double mutant G19U–C56A that restored the tertiary base pair and the aminoacylation activity simultaneously. Two tRNA variants with changes located in the T-loop were also constructed. One mutant carried a deletion of nucleotide 59 (in the variable pocket), and the other had a substitution of A58 to G, which forms a tertiary interaction with U54. Both variants showed significant defects in aminoacylation, suggesting that the structure of the T-loop is critical for tRNA recognition and aminoacylation.
The discriminator base A73 is crucial for aminoacylation
Nucleotide A73 is phylogenetically conserved in the tRNALeu of various organisms. Substitution of A73 with G73, U73 and C73 led to a 143-, 45- and 48-fold decrease in leucylation, respectively (Figure 4 and Table 1). This result indicated that the discriminator base A73 in tRNALeu is crucial for its recognition by AaLeuRS, as in all other species.
The anticodon arm is required for the editing process
In this study, we showed that AaLeuRS induced protections on the anticodon stems of both AatRNALeuGAG and AatRNALeuCAA as well as the anticodon loop of AatRNALeuGAG, although the latter appeared weak when a small probe such as iodine was used. However, the mutation analysis performed on the tRNA did not reveal any interaction that was essential for tRNA aminoacylation, and deletion of the whole anticodon stem-loop resulted in only a 2.5-fold decrease in tRNA affinity (Table 1). To determine whether the protections could reflect interactions occurring during the editing process, we studied the total editing activity (the sum of pre- and post-transfer editing) by measuring the ATP consumption of the enzyme in the presence of different mutated tRNAs and isoleucine. In fact, this assay measures the ATP consumed during the repetitive and futile cycles of adenylate formation and destruction. In the absence of editing, ATP consumption is usually very low because cognate adenylates are not easily released from the enzyme and aa-tRNAs are stable in solution. The incubation time for the ATP consumption assay was chosen long enough to take into account the possible slow misactivation rates of isoleucine that could ultimately affect the ATP consumption. We also measured the post-transfer editing reaction using preformed mischarged Ile-tRNALeu. However this assay only measures the de-acylation step occurring in the CP1 editing site and does not take into account the translocation step of the mis-acylated 3′-end of the tRNA from the synthetic site to the editing site.
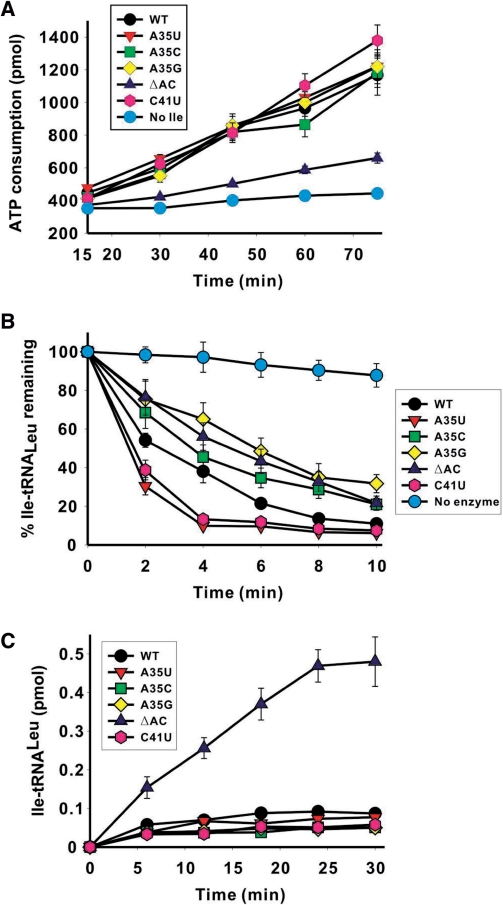
The total editing measurement revealed that in the presence of the ΔAC tRNA mutant and isoleucine, AaLeuRS did not consume ATP, indicating that the anticodon arm of AatRNALeu is required for editing. The other tRNA mutants induced ATP consumptions equivalent to that of the native tRNA (Figure 5A). These data showed that despite a modest role in tRNA aminoacylation, the anticodon arm could have a pivotal function in editing, as determined in the ATP consumption assay.
Figure 5.
Effect of AatRNALeuGAG mutations on the editing reactions of AaLeuRS. In (A)–(C), different colors represent various AatRNALeuGAG variants: AatRNALeuGAG (wild-type), black; AatRNALeuGAG (A35U), red; AatRNALeuGAG (A35C), green; AatRNALeuGAG (A35G), yellow; AatRNALeuGAG (ΔAC), blue; and AatRNALeuGAG (C41U), pink. Cyan curve shows the background rate in the ATP consumption assay in the absence of isoleucine in (A) or the spontaneous hydrolysis in the absence of the enzyme in (B). (A) ATP consumption during total editing by AaLeuRS in the presence of 50 mM isoleucine and 5 μM AatRNALeu mutant transcripts. (B) Hydrolysis of Ile-tRNALeuGAG for AatRNALeuGAG mutants by 5 nM AaLeuRS. The spontaneous hydrolysis in the absence of the enzyme (<10%) were subtracted. (C) Isoleucylation rates of 5 μM AatRNALeu mutants by 1 μM LeuRS. Data are averaged from three independent assays.
Analysis of post-transfer editing yielded a more complex picture. The de-acylation of the A35U and C41U mutants was faster and that of the mutants A35C, A35G and ΔAC was slower than the de-acylation of the native tRNA (Figure 5B). This result has several implications. First, these data suggest that the anticodon region of tRNA interacts with the enzyme during the post-transfer editing reaction. Although the effect of the substitutions was not dramatic, one cannot exclude the possibility that in vivo, the substitutions may modulate the efficiency of the post-transfer editing reaction. Second, the substitution of residue A35 or C41 induces some structural rearrangements at long distance into the editing site where the hydrolytic reaction occurs. At this level, the signal may favor or inhibit the transition state of the de-acylation reaction. On the other hand, the mutant ΔAC that is deprived of its anticodon arm loses most of its ATP consumption activity but is deacylated with only a slight decrease in efficiency. This result suggests either of two different hypotheses. (i) The mis-acylation activity of the enzyme in the presence of isoleucine and mutant ΔAC is so low that the ATP consumption can scarcely be detected. (ii) Alternatively, the anticodon arm deletion specifically impeded translocation of the mis-acylated 3′-end of the tRNA from the synthetic site to the editing site, thus inhibiting the editing reaction. This would explain that the de-acylation rate of Ile-tRNALeu, was nearly unchanged, because the de-acylation assay only monitors the last step of the editing process which is the hydrolytic step and not the translocation step. This would suggest that the interactions occurring between the enzyme and the tRNA anticodon arm are essential during the translocation step of the tRNA acceptor end. This interpretation is also consistent with the effects observed on the de-acylation rates of the mutants for A35 and C41 (see above).
In the last assay, we examined the global effect of the different mutations by measuring the mischarging level of the mutated tRNAs with isoleucine (Figure 5C). Only the ΔAC mutant appeared to exhibit a weak mischarging activity for isoleucine, which was 10-fold higher than that observed in the native tRNA. Therefore, the result reveals that AaLeuRS can activate and charge isoleucine on a tRNA deprived of the anticodon arm, and that the ΔAC deletion induces a relaxation of the aminoacylation specificity of the enzyme. Such a relaxation might be induced by a defect in the editing ability of the enzyme and, indeed, the ΔAC tRNA mutant exhibits a significant decrease in ATP consumption in the global editing assay (see above, Figure 5A).
DISCUSSION
The mechanism of tRNA recognition by the ancient AaLeuRS is similar to that by most other LeuRS enzymes and is thus highly conserved
The sequences of five tRNALeu isoacceptors from A. aeolicus are relatively well conserved. The D-arm and T-arm are completely conserved, and the variable arm exhibits only a few nucleotide variations within the loop (http://lowelab.ucsc.edu/GtRNAdb/). The acceptor stem and anticodon arm are less conserved, but they exhibit several conserved nucleotides such as G4–C69, G5–C68, G29–C41 and A35. Employing different approaches, we showed that the enzyme interacts with these conserved elements. The footprinting assay revealed that the nucleotides in the acceptor stem, anticodon arm, TΨC-arm and variable loop make contact with LeuRS. The in vitro kinetic assays showed that the conserved base A73 and the nucleotides in the D-loop, particularly those involved in the tertiary structure, are crucial to the maintenance of the aminoacylation efficiency. We showed that the conserved G–C base pairs found in the acceptor and anticodon stems do not interact in a base-specific manner, although a regular amino-acid acceptor helix is strictly required, as shown by the drastic effect of creating a G4–U69 base pair. Altogether, the results obtained with the aminoacylation system of leucine in A. aeolicus appear to be in agreement with the other leucine recognition systems. As in E. coli (17), the tertiary structure formed by the D-arm and TΨC-arm is crucial for AatRNALeu recognition by AaLeuRS. As in the human mitochondrial tRNALeu, AatRNALeu shows protections on several nucleotides in the amino-acid acceptor arm (24). Moreover, its minihelix is recognized and charged by AaLeuRS, suggesting the existence of cryptic recognition elements in the acceptor arm (26). Although the anticodon loop and variable stem-loop do not contain identity elements in AatRNALeu, nuclease protections were detected at positions 35 and 47e, as observed in S. cerevisiae (19) and H. volcanii (18), respectively, and the orientation of the variable arm is also crucial, as has been demonstrated in human cytoplasmic tRNALeu systems (21). In addition, the protections in the D-stem and anticodon stem were similar to those observed in the P. vulgaris cytoplasmic system (20) and human mitochondrial system (24).
Discrete determinants for editing and aminoacylation are found in tRNALeu
This is the second study in which the tRNA determinants for aminoacylation and editing were found to be separate. Isoleucyl-tRNA synthetase was the first enzyme to show a segregation of nucleotide determinants for the editing and aminoacylation functions of tRNA (11). Editing determinants were identified in the corner of tRNAIle of the L-shaped tRNA molecule at positions 16, 20 and 21, whereas the major determinants for aminoacylation were found in the anticodon triplet (11). Herein, we showed that the anticodon arm of AatRNALeu is essential for editing but dispensable for aminoacylation. Single substitutions of residue 35 also exerted various effects on the editing reaction, but not on the aminoacylation reaction. These results show that the anticodon arm of AatRNALeu contains specific determinants for the editing reaction, and these determinants can be distinguished from those involved in the aminoacylation step. Consequently, the interactions that occur during the editing step should be at least partially distinct from those occurring during the aminoacylation step. In the editing state, AaLeuRS would interact with the anticodon of the tRNA, whereas no such interaction would be essential during the aminoacylation state. This may explain the two anticodon loop conformations revealed by the footprinting experiments on AatRNALeuGAG and AatRNALeuCAA. The AatRNALeuGAG anticodon was protected by LeuRS, as expected from an editing state, whereas the AatRNALeuCAA anticodon was not protected, as expected from a complex in an aminoacylation state (8,10).
Interactions with the anticodon arm might be crucial for the translocation of the tRNA acceptor end from the synthetic to the editing site
Post-transfer editing catalyzed by aminoacyl-tRNA synthetases has been extensively studied. Based on structural and biochemical data, a model has been proposed wherein the flexible 3′-end of a mis-acylated tRNA is translocated from the aminoacylation active site to the hydrolytic editing site (31). Our results support the possibility that the tRNA anticodon arm may be involved in the translocation process. We illustrated that the de-acylation rate of a tRNA mutant with the deletion of its entire anticodon arm is nearly unchanged when exogenous mis-acylated tRNA is provided. However, the ATP consumption assay is severely decreased, suggesting that another step is blocked at the level of the synthetic or editing activity, as both activities are required to induce the significant ATP consumption characteristic of the editing process. We demonstrated that the synthetic activity is preserved as shown by the ability of LeuRS to mischarge isoleucine on the ΔAC tRNA mutant. However, with native tRNA and other mutated tRNAs, no detectable mis-acylation with Ile could be measured, suggesting that the enzyme efficiently edits these tRNAs. This striking difference suggests that the enzyme cannot catalyze the full post-transfer editing step in the presence of the ΔAC tRNA mutant. Therefore, as the ΔAC tRNA mutant can be efficiently deacylated in a separated assay, we suggest that after its formation in the synthetic site, the mis-acylated Ile-tRNALeu could be released into the solution without being edited in the post-transfer editing site. The release may be facilitated by the 2.5-fold decrease of the apparent affinity for AaLeuRS (Table 1). Because the post-transfer editing site is located ∼25 Å away from the active site (31) a mechanism for the translocation of the aminoacylated tRNA has been proposed (32). We suggest that the translocation step cannot be performed in the absence of interactions with the anticodon arm of the tRNA. Such interactions might be crucial to bind the tRNA in the correct conformation when the acceptor end is released from the synthetic site and is translocated into the editing site.
Taken together, our results support the existence of significant interactions between LeuRS and the tRNA anticodon arm. Except in S. cerevisiae, the tRNA anticodon was considered to be a dispensable element for the tRNALeu identity (19), and here we demonstrate that mutations of the central base A35 affect the post-transfer editing reaction. In addition, our results suggest that the deletion of the whole anticodon arm impedes the translocation of the acceptor arm from the synthetic to the editing site. The next step toward understanding further into the functional details of AaLeuRS editing will be a more extensive mutagenesis study on the tRNA anticodon arm residues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank J. Cavarelli for the tRNA gift and J. Rudinger-Thirion for advice. This work was funded by the Natural Science Foundation of China (Grant 30330180, 30570380); National Key Basic Research Foundation of China (Grant 2006CB910301); 973 project (Grant 2005CB724600) of China; Committee of Science and Technology in Shanghai (Grant 06JC14076) and Exchange Program and Programme International de Coopération Scientifique from CNRS (Grant 3606). Funding to pay the Open Access publication charges for this article was provided by Grant 2006CB910301.
Conflict of interest statement. None declared.
REFERENCES
- 1.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinberg S, Misch A, Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson JD, Hopkins NH, Roberts JW, Steitz JA, Weiner AM. Molecular Biology of the Gene, 4ed. Vol. 1. Menlo Park, CA: The Benjamin/Cummings Publishing Company; 1987. [Google Scholar]
- 4.Sprinzl M, Hartmann T, Weber J, Blank J, Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17(Suppl.):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JF, Guo NN, Li T, Wang ED, Wang YL. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 6.Chen JF, Li T, Wang ED, Wang YL. Effect of alanine-293 replacement on the activity, ATP binding, and editing of Escherichia coli leucyl-tRNA synthetase. Biochemistry. 2001;40:1144–1149. doi: 10.1021/bi0017226. [DOI] [PubMed] [Google Scholar]
- 7.Du X, Wang ED. Discrimination of tRNA(Leu) isoacceptors by the mutants of Escherichia coli leucyl-tRNA synthetase in editing. Biochemistry. 2002;41:10623–10628. doi: 10.1021/bi026000o. [DOI] [PubMed] [Google Scholar]
- 8.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 9.Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grotli M, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 10.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 11.Hale SP, Auld DS, Schmidt E, Schimmel P. Discrete determinants in transfer RNA for editing and aminoacylation. Science. 1997;276:1250–1252. doi: 10.1126/science.276.5316.1250. [DOI] [PubMed] [Google Scholar]
- 12.Normanly J, Ollick T, Abelson J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc. Natl. Acad. Sci. USA. 1992;89:5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asahara H, Himeno H, Tamura K, Nameki N, Hasegawa T, Shimizu M. Discrimination among E. coli tRNAs with a long variable arm. Nucleic Acids Symp. Ser. 1993;29:207–208. [PubMed] [Google Scholar]
- 14.Asahara H, Nameki N, Hasegawa T. In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol. 1998;283:605–618. doi: 10.1006/jmbi.1998.2111. [DOI] [PubMed] [Google Scholar]
- 15.Tocchini-Valentini G, Saks ME, Abelson J. tRNA leucine identity and recognition sets. J. Mol. Biol. 2000;298:779–793. doi: 10.1006/jmbi.2000.3694. [DOI] [PubMed] [Google Scholar]
- 16.Larkin DC, Williams AM, Martinis SA, Fox GE. Identification of essential domains for Escherichia coli tRNA(leu) aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 2002;30:2103–2113. doi: 10.1093/nar/30.10.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du X, Wang ED. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872. doi: 10.1093/nar/gkg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soma A, Uchiyama K, Sakamoto T, Maeda M, Himeno H. Unique recognition style of tRNA(Leu) by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 1999;293:1029–1038. doi: 10.1006/jmbi.1999.3219. [DOI] [PubMed] [Google Scholar]
- 19.Soma A, Kumagai R, Nishikawa K, Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNA(Leu) J. Mol. Biol. 1996;263:707–714. doi: 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- 20.Dietrich A, Romby P, Marechal-Drouard L, Guillemaut P, Giegé R. Solution conformation of several free tRNALeu species from bean, yeast and Escherichia coli and interaction of these tRNAs with bean cytoplasmic Leucyl-tRNA synthetase. A phosphate alkylation study with ethylnitrosourea. Nucleic Acids Res. 1990;18:2589–2597. doi: 10.1093/nar/18.9.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitschopf K, Achsel T, Busch K, Gross HJ. Identity elements of human tRNA(Leu): structural requirements for converting human tRNA(Ser) into a leucine acceptor in vitro. Nucleic Acids Res. 1995;23:3633–3637. doi: 10.1093/nar/23.18.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitschopf K, Gross HJ. The discriminator bases G73 in human tRNA(Ser) and A73 in tRNA(Leu) have significantly different roles in the recognition of aminoacyl-tRNA synthetases. Nucleic Acids Res. 1996;24:405–410. doi: 10.1093/nar/24.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sohm B, Frugier M, Brule H, Olszak K, Przykorska A, Florentz C. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 2003;328:995–1010. doi: 10.1016/s0022-2836(03)00373-5. [DOI] [PubMed] [Google Scholar]
- 24.Sohm B, Sissler M, Park H, King MP, Florentz C. Recognition of human mitochondrial tRNALeu(UUR) by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 2004;339:17–29. doi: 10.1016/j.jmb.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 25.Xu MG, Chen JF, Martin F, Zhao MW, Eriani G, Wang ED. Leucyl-tRNA synthetase consisting of two subunits from hyperthermophilic bacteria Aquifex aeolicus. J. Biol. Chem. 2002;277:41590–41596. doi: 10.1074/jbc.M205126200. [DOI] [PubMed] [Google Scholar]
- 26.Xu MG, Zhao MW, Wang ED. Leucyl-tRNA synthetase from the hyperthermophilic bacterium Aquifex aeolicus recognizes minihelices. J. Biol. Chem. 2004;279:32151–32158. doi: 10.1074/jbc.M403018200. [DOI] [PubMed] [Google Scholar]
- 27.Zhao MW, Zhu B, Hao R, Xu MG, Eriani G, Wang ED. Leucyl-tRNA synthetase from the ancestral bacterium Aquifex aeolicus contains relics of synthetase evolution. EMBO J. 2005;24:1430–1439. doi: 10.1038/sj.emboj.7600618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Wang E, Wang Y. A modified procedure for fast purification of T7 RNA polymerase. Protein Expr. Purif. 1999;16:355–358. doi: 10.1006/prep.1999.1083. [DOI] [PubMed] [Google Scholar]
- 29.Silberklang M, Gillum AM, RajBhandary UL. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Meth. Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- 30.Metzger AU, Heckl M, Willbold D, Breitschopf K, RajBhandary UL, Rösch P, Gross HJ. Structural studies on tRNA acceptor stem microhelices: exchange of the discriminator base A73 for G in human tRNALeu switches the acceptor specificity from leucine to serine possibly by decreasing the stability of the terminal G1-C72 base pair. Nucleic Acids Res. 1997;25:4551–4556. doi: 10.1093/nar/25.22.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 32.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 33.Hou YM. Discriminating among the discriminator bases of tRNAs. Chem. Biol. 1997;4:93–96. doi: 10.1016/s1074-5521(97)90252-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.