Abstract
The 3′ end of mammalian introns is marked by the branchpoint binding protein, SF1, and the U2AF65-U2AF35 heterodimer bound at an adjacent sequence. Baker's yeast has equivalent proteins, branchpoint binding protein (BBP) (SF1) and Mud2p (U2AF65), but lacks an obvious U2AF35 homolog, leaving open the question of whether another protein substitutes during spliceosome assembly. Gel filtration, affinity selection and mass spectrometry were used to show that rather than a U2AF65/U2AF35-like heterodimer, Mud2p forms a complex with BBP without a third (U2AF35-like) factor. Using mutants of MUD2 and BBP, we show that the BBP–Mud2p complex bridges partner-specific Prp39p, Mer1p, Clf1p and Smy2p two-hybrid interactions. In addition to inhibiting Mud2p association, the bbpΔ56 mutation impairs splicing, enhances pre-mRNA release from the nucleus, and similar to a mud2::KAN knockout, suppresses a lethal sub2::KAN mutation. Unexpectedly, rather than exacerbating bbpΔ56, the mud2::KAN mutation partially suppresses a pre-mRNA accumulation defect observed with bbpΔ56. We propose that a BBP–Mud2p heterodimer binds as a unit to the branchpoint in vivo and serves as a target for the Sub2p-DExD/H-box ATPase and for other splicing factors during spliceosome assembly. In addition, our results suggest the possibility that the Mud2p may enhance the turnover of pre-mRNA with impaired BBP-branchpoint association.
INTRODUCTION
Pre-mRNA splice site consensus sequences are sampled multiple times during spliceosome assembly (1). For example, early in assembly the yeast 5′ splice site is bound by the U1 snRNP particle through protein and snRNA-based contacts. U1 snRNP also interacts with the branchpoint region through association with the branchpoint binding protein (BBP), a homolog of the mammalian SF1 protein (2,3). Subsequent recruitment of the U2 snRNP particle displaces BBP and its binding partner Mud2p (the homolog of mammalian U2AF65) from the branchpoint region to permit base pairing between the U2 snRNA and the pre-mRNA branchpoint consensus (4). The displacement of BBP and Mud2p from the branchpoint region correlates with a weakening of the U1 snRNP association with the splicing apparatus, an apparent requirement for subsequent binding of U6 snRNA at the 5′ splice site. Although U2 snRNA basepairs with the pre-mRNA substrate, initial U2 snRNP contact is likely protein-mediated and, in mammals, may be accomplished in part through U2AF65 interaction with the SAP155 subunit of the U2 snRNP particle (also called SF3b155) (5). Two-hybrid interaction between Mud2p and the Saccharomyces cerevisiae SAP155 homolog, Hsh155p, suggest that this interaction is conserved in yeast, implicating Mud2p function after the U1-snRNP-dependent step of commitment complex formation (6). While U1 displacement appears mediated by Prp28p, stable U2 snRNP addition requires participation of the Prp5p and Sub2 DExD/H-box proteins. Other DExD/H-box proteins promote subsequent steps in spliceosome assembly (Brr2p, Prp2, Prp16p) and product release (Prp22p and Prp43p) (7,8).
U2AF65 is recovered from mammalian cells as a heterodimer with U2AF35, a protein that crosslinks to the 3′ splice site during spliceosome assembly (9–11). Homologs of U2AF35 are broadly represented in nature and have been shown to function in splicing in man, flies, nematodes and fission yeast (11–14). Since no U2AF35 homolog is obvious in budding yeast, Mud2p may have diverged to the degree that it acts independently or simply no longer provides an anchor for 3′ splice site recognition factors. Alternatively, a structurally distinct but functionally related protein may bind Mud2p to serve this purpose or otherwise facilitate co-transcriptional assembly of the yeast mRNP (15).
BBP interacts with at least two essential U1 snRNP proteins, Prp39p and Prp40p, in addition to Mud2p in a step that commits the pre-mRNA to the splicing pathway (3,16). While mammalian U2AF65 binds a critical polypyrimidine stretch upstream of the 3′ splice site and is required for splicing, yeast splicing does not require either a polypyrimidine element or Mud2p (4,17,18). The yeast branchpoint motif is more rigorously conserved than the mammalian sequence, however, and BBP is necessary for yeast cell viability. The loss of either BBP or Mud2p increases pre-mRNA abundance in the cytoplasm (19,20), thereby linking both proteins to the nuclear retention of newly transcribed RNA (21). While BBP and Mud2p bind one another in vitro, the degree to which these proteins act in a concerted or independent function during spliceosome assembly or nuclear pre-mRNA retention in vivo is less clear.
Here, we report the results of experiments designed to investigate the composition and function of cellular Mud2p and BBP complexes. Like U2AF65, Mud2p is found to exist as a heterodimer. However, rather than interacting with a U2AF35 counterpart, Mud2p, is recovered bound to BBP. The phylogenetically conserved BBP–Mud2p interaction [(22,23) and references within] does not require RNA association and can be isolated essentially free of other splicing factors at modest salt concentration. Evidence is provided to show that four proteins interacting with both BBP and Mud2p in the two-hybrid assay bind one protein of this pair directly and the other through a BBP–Mud2p bridge that requires the first 56 amino acids of BBP. Deletion of this sequence from BBP enhances pre-mRNA export from the nucleus and results in pre-mRNA instability, an effect most notable with RNAs containing branchpoint motif mutations. Surprisingly, rather than enhancing the bbpΔ56 defect, we find that deletion of MUD2 increases pre-mRNA recovery in the bbpΔ56 background. Mutations in either gene suppress a null allele of SUB2, supporting the view that the BBP–Mud2p heterodimer is an intracellular target of the Sub2p DExD/H-box ATPase that acts in splicing and mRNA export. Together, these data support the existence of a simple BBP–Mud2p commitment factor bridging multiple contacts in spliceosome assembly and raises the intriguing possibility that Mud2p may enhance the turnover of transcripts with aberrant branchpoint–BBP interaction.
MATERIALS AND METHODS
Yeast strains and assays
Yeast strains BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), BY4741 (MATA his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), mud2::KAN (MATα, his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mud2::KAN), smy2::KAN (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 smy2::KAN), BBP–TAP (MATA his3Δ1 leu2Δ0 met15Δ0 ura3Δ0, MSL5-TAP) and Mud2-TAP (MATA his3Δ1 leu2Δ0 met15Δ0 ura3Δ0, MUD2-TAP) were obtained from Open Biosystems (Huntsville, AL, USA). The bbpΔ56 mutation was introduced by inverse PCR (5′TAGTGGATCCCTAACTAGGGAACAAATA3′ and 5′TAGGGATCCCATCAACAATTTCCTTTTATTGTG3′) on plasmid pRS414-BBP (obtained from Michael Rosbash). The deletion derivative was then subcloned into YIpLac 211 (24), linearized with BglII and introduced into the genome by two-step replacement into wild-type yeast to generate the bbpΔ56 strain. FOA selected colonies were scored for excision of the wild-type BBP allele by PCR. The double mutant strains were created using standard yeast genetic techniques (25). The wild-type MUD2 and BBP genes with approximately 300 bp of flanking sequence were subcloned by PCR into YCplac22 (24) for complementation studies. The structures of all novel constructs were confirmed by DNA sequencing prior to use. Plasmids containing the RPS17A-based Acc (splicing), Nde/Acc (export) and SD5 (no intron) lacZ reporters genes were previously described in detail (26,27). The lacZ reporter strains were induced with 2% galactose in complete medium at 30°C for 4 h prior to β-galactosidase measurements. Each assay was replicated 4–10 times and pair wise comparisons of the normalized enzyme units were made using a two-tailed t-test of significance.
Yeast two hybrid studies were conducted with full-length PCR products (primer sequences available upon request) fused to the GAL4 DNA binding (DB) and activation domains (AD) on vectors pACT and pAS2, respectively, in yeast strain pJ69-4A (28). The yeast transformants were scored for transactivation of the endogenous reported genes by colony growth at 30°C on medium lacking adenine or on medium lacking histidine and containing 20 mM 3-aminotriazole. Autostimulation of the chromosomal reporter genes was scored in the recombinant pAS2 and pACT transformants in the absence of the binding partner.
RNA analysis
RNA was recovered by breaking yeast on a vortex mixer with sterile glass beads followed by multiple extractions using phenol:chloroform:isolamyl alcohol (25:24:1) and ethanol precipitation. Primer extension was performed on 12–24 μg of total RNA using a 32P end-labeled RPS17A exon II primer RB1 (5′CGCTTGACGGTCTTGGTTC3′) and Superscript reverse transcriptase (Invitrogen) at 37°C for 1 h (29). The cDNA products were resolved on a denaturing 7 M urea 5% polyacrylamide gel. Northern blot analysis was performed with the RPS17A and ADE3 hybridization probes on RNA resolved by electrophoresis on a 1% agarose formaldehyde gel and transferred to a charged nylon membrane (30,31). Band intensities were visualized Typhoon 9600 phosphorimager and quantified with Image Quant software GE Healthcare (Piscataway, NJ, USA).
Protein analysis
Conditions for metabolically labeling yeast proteins with Trans 35S-label (ICN) were previously described in detail (32). In short, yeast were labeled for 4 h at 30°C and then broken with glass beads in buffer A [10 mM HEPES (pH 7.9), 10 mM KCl, 200 mM NaCl, 10% glycerol, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzamide, 0.5% NP-40] and the lysate cleared by centrifugation at 14 000g. Sequential protein A agarose and calmodulin agarose affinity (TAP) selection was done as previously modified (32). Samples recovery was estimated using with Image Quant software GE Healthcare (Piscataway, NJ, USA). Large-scale TAP selection was performed similarly with 10 l of yeast broken under liquid nitrogen in buffer A with a Spex Certiprep 5850 freezer mill (6). The recovered proteins were digested with trypsin, concentrated with a C18 column, and resolved by two-dimensional liquid chromatography followed by mass analysis with a Deca Quadruopole ion trap mass spectrometer (ThermoElectron, Waltham, MA, USA). RNase digestions were performed using RNase A at 20 μg/ml Sigma (St. Louis, MO, USA) or Ambion RNase cocktail (Ambion, Applied Biosystems, Austin TX, USA) at a 1/25 dilution for 30 min at room temperature prior to TAP selection. The mass intensity lists were screened against the non-redundant National Center for Biotechnology Information database using MASCOT software (Matrix Science, Boston, MA, USA) filtered with the default cutoff score of 20. Western blot confirmation of the TAP-selected protein was done using an antibody directed against the residual calmodulin-binding protein (CBP) (anti-CBP; Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA). The whole TAP epitope was assayed by western blot using a horseradish peroxidase anti-horseradish peroxidase antibody, PAP (Sigma) as described (33).
Size fractionation was performed by resolving 50 μl of calmodulin-agarose selected BBP–TAP or Mud2–TAP complex on a Shimadzu model LC2010C HPLC fitted with the 2 ml injection loop and tandem 15 cm Chromegapore MSE P columns of 2000A and 300A pore diameter. Chromatography was conducted in 200 mM NaCl, 50 mM Tris-HCL, pH 8.0, 0.1% NP40 at 0.5 ml/min. Proteins from alternate fractions were concentrated by TCA precipitation and analyzed by western blot using the anti-horse radish peroxidase antibody (Sigma).
RESULTS
Mud2p and BBP bind one another in vivo
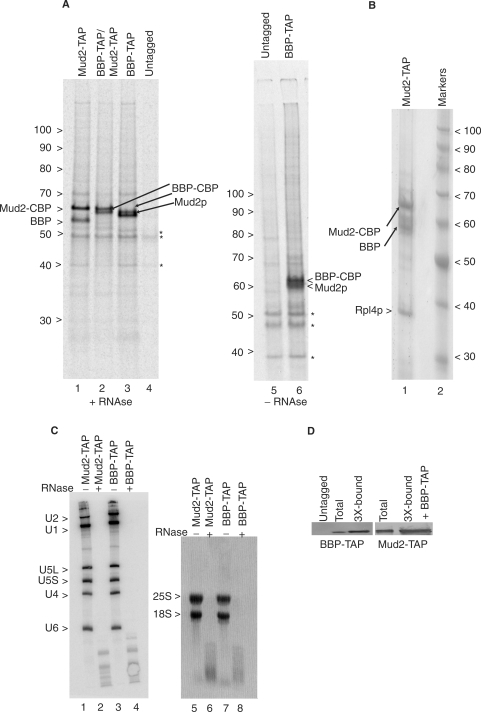
To screen for proteins that interact with Mud2p, we metabolically labeled yeast with 35S-amino acids for 4 h and then used tandem affinity purification [TAP (33)] to identify co-purifying factors (Figure 1A, lane 1). In contrast to snRNP or spliceosome preparations previously assayed in this way (32–35), the pattern of 35S-labeled proteins recovered with Mud2-TAP at 200 mM NaCl is quite simple, with the 65 kDa fusion protein derivative, Mud2-CBP, and a single additional band that migrates as a 57 kDa protein. Mud2-CBP consists of the native Mud2 protein joined to ∼5 kDa of CBP sequence that persists after TAP (33). Confirmation of the Mud2-CBP assignment was made by western blot using an anti-CBP antibody (see Materials and methods section). A few minor bands are also seen in this preparation, several which appear as background in the untagged control sample (lane 4 and see Figure 3D). RNase A is included at levels sufficient to degrade all detectable full-length rRNA and splicing-relevant snRNA (Figure 1C) although the pattern of recovered proteins appears equivalent without RNase addition (e.g. compare Figure 1A lanes 3 and 6).
Figure 1.
Tandem affinity purification of BBP–TAP and Mud2–TAP. (A) Autoradiogram of a 7% polyacrylamide gel of metabolically labeled proteins selected by sequential IgG agarose and calmodulin agarose chromatography from yeast that express the indicated TAP-tagged gene constructs as genomic integrants. The untagged lane shows background proteins (asterisks). Samples in lanes 1–4 were pre-treated with RNase A prior to TAP selection while samples 5–6 were not. The numbers at the left indicate the positions of unlabeled protein molecular weight markers. Bands corresponding to the untagged Mud2p and BBP proteins and the proteins with the residual CBP tags are indicated. (B) Coomassie blue stain of unlabeled Mud2-TAP and co-purifying proteins. Lane 2 shows the migration of protein molecular weight markers. (C) Yeast used in panel a treated (+) or not (−) with RNase A hybridized with probes specific for the spliceosomal snRNAs (lanes 1–4) or stained with ethidium bromide for the 25S and 18S ribosomal RNAs (lanes 5–8). (D) Single-step recovery of BBP–TAP and Mud2–TAP by TAP by calmodulin agarose chromatography. To avoid band distortion due to sample overloading, the unfractionated (total) protein lanes contain 1/3 the equivalent amount of sample. Untagged = calmodulin agarose recovered material recovered from an untagged extract.
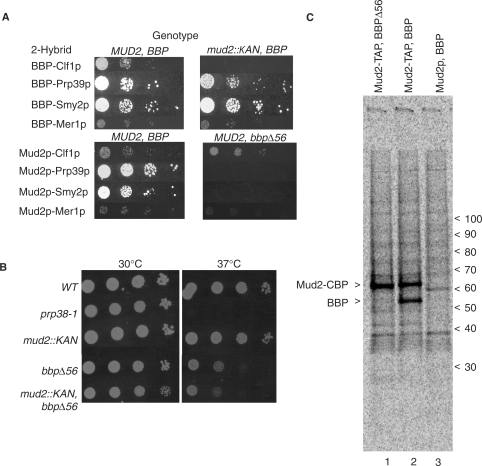
Figure 3.
Bridged interactions of the BBP–Mud2p heterodimer. (A) Two hybrid interactions conducted in yeast wild-type for both genes (MUD2, BBP), deleted for MUD2 (mud2::KAN, BBP), or containing a non-lethal mutation within BBP (MUD2, bbpΔ56). Reporter gene activity was scored after 3 days at 30°C. (B) Direct growth assay in the absence of 2-hybrid plasmids of yeast with wild-type MUD2 and BBP alleles (WT), deleted for MUD2 (mud2::KAN, BBP), containing a non-lethal mutation within BBP (MUD2, bbpΔ56) or both mutations (mud2::KAN, bbpΔ56) at 23 and 37°C. (C) Tandem affinity selection of metabolically labeled proteins with Mud2–TAP from yeast that express the bbpΔ56 allele (lane 1) and wild-type BBP allele (lane 2). Lane 3 shows background proteins isolated from an untagged strain. The positions of unlabeled protein markers are shown on the right.
In a parallel culture, yeast that express a TAP-tagged BBP derivative were similarly scored for the pattern of recovered proteins. Here also two prominent proteins, 63 and 60 kDa in apparent molecular weight were recovered (Figure 1A, lane 3). Western analysis shows that the 63 kDa band is BBP–CBP, which migrates more slowly than predicted for this 55 kDa fusion protein (data not shown). The 57 kDa and 60 kDa proteins that co-purify with Mud2–TAP and BBP–TAP, respectively, are roughly the predicted sizes for native BBP (50 kDa) and Mud2p (60 kDa). In agreement with this assignment, when Mud2p and BBP are both expressed as TAP-tagged derivatives, the pattern of recovered proteins is equivalent except that the bands corresponding to the untagged BBP and Mud2p are now absent (lane 2).
Although the background is somewhat higher, a similar protein pattern is seen when the isolation is scaled up and unlabeled Mud2–TAP or BBP–TAP complexes are visualized with Coomassie blue (Figure 1B; data not shown). Mass spectrometry was used to confirm the p60-Mud2p assignment and identifies BBP and ribosomal protein Rpl4p in the preparation (see Materials and methods section). While not ruling out the possibility of functional association, we note that the Rpl4p band appears to be of lower abundance than Mud2p or BBP in the preparative (Coomassie blue-stained) sample and that it co-migrates with a background band in the radiolabeled preparations, suggesting that Rp14p may be a contaminant of the preparation. Lower scoring mass analysis signatures were sometimes observed for splicing factors Prp40, Prp8, Prp28, Brr2p, Prp6 and Isy1 as well as for the heat shock protein, Ssb2, and for the Ded1p and Dbp2p DExD/H-box factors. Since none of these proteins correspond to prominent 35S-labeled bands or Coomassie stained proteins, the associations likely reflect weak or transient interactions with the BBP–Mud2p complex or the presence of a small amount of a more elaborate (multi-subunit) complex.
The BBP–Mud2p complex is predominantly a heterodimer
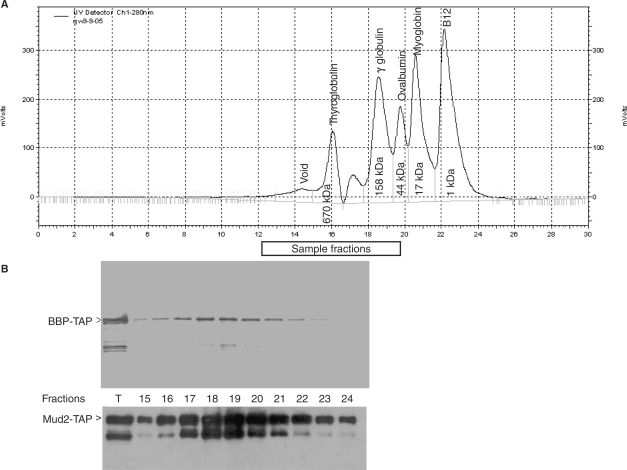
The cellular levels of BBP and Mud2p appear equivalent (36) and the Coomassie blue stained BBP and Mud2p bands are of similar intensity, suggesting a 1:1 stoichiometry in the recovered sample. In support of this, after normalization for the greater sulfur content of Mud2p, the 35-S ratio of the metabolically labeled proteins was calculated to be approximately 1.15 when Mud2-TAP is used as bait and 0.87 when BBP–TAP is used as bait. While supportive of equimolar recovery, these experiments do not distinguish between a BBP–Mud2p heterodimer and higher order complexes. To address this, we resolved the calmodulin agarose purified Mud2–BBP complex by tandem HPLC with 2000Å and 300Å sepharose columns. Complex recovery is generally quite efficient, typically 50–75% (Figure 1D). In contrast to the sharp focus of the molecular weight standards (Figure 2A), the BBP–TAP selected complex resolves as a broad peak with the greatest recovery between fractions 18–20 (Figure 2B, upper panel, note that here the protein is visualized with an antibody against the TAP tag, consequently only a single protein is visible). An equivalent distribution is seen when Mud-TAP is used for selection (Figure 2B, lower panel). The highest signals for BBP–TAP and Mud2–TAP correspond to fractions overlapping the γ globulin (158 kDa) and ovalbumin (44 kDa) protein standards. As we previously found no evidence for homodimerization of either Mud2p or BBP (6), this pattern indicates the presence of significant amounts of a BBP–Mud2p heterodimer, which has a predicted mass of ∼130 kDa (including the full TAP epitope). Given the spread of signal and the somewhat greater recovery of the TAP tagged partner indicated by the metabolic labeling experiment, monomeric BBP–TAP (73 kDa) and Mud2–TAP (80 kDa) are likely present as well. While the existence of higher order structures (e.g. tetrameric) cannot be ruled out, such complexes are not abundant under these isolation conditions.
Figure 2.
BBP–Mud2p purifies as a heterodimer. (A) HPLC resolution of molecular weight standards by sequential 2000Å and 300Å sepharose columns. (B) Western blots of the BBP–TAP and Mud2–TAP selected on calmodulin agarose and resolved by HPLC. The blots were developed with an antibody against the TAP epitope. T, total protein applied to the tandem size exclusion columns. The sample numbers correspond to the same column fractions indicated in (A).
Bridging interactions contribute to the previously reported two hybrid interactions of BBP and Mud2p
Four proteins, splicing factors Clf1p, Prp39p and Mer1p, and the largely uncharacterized Smy2 protein have been reported to interact with both BBP and Mud2p by the two-hybrid assay (3,6,37–39). These interactions presumably reflect protein-based contacts made by BBP and Mud2p during spliceosome assembly or in other functional contexts (see Discussion section). Since BBP and Mud2p bind one another we thought it is possible that indirect (i.e. bridging) interactions might contribute to the observed two hybrid pattern. As a first step in addressing this issue, we sought confirmation of the reported interactions using full-length protein constructs. Although differences exist in the extent of host cell reporter gene transactivation, each two-hybrid pair shows detectable activity in the wild-type yeast (Figure 3A). For most constructs, the two hybrid interactions are reciprocal when the GAL4 DNA-binding and transactivation domains are swapped. Autostimulation by CLF1 and PRP39 in the DNA binding cassette prevented this determination in these two cases.
Since MUD2 is not essential, the BBP associations can be assayed in the mud2::KAN null background to learn which, if any, is Mud2p-dependent. The Prp39p, Mer1p and Smy2p signals all persist in the mud2::KAN mutant, showing that none of these proteins require Mud2p for BBP association (Figure 3A). In contrast, the Clf1p-BBP signal is lost in the mud2::KAN mutant, strongly suggesting that binding of this essential splicing factor with BBP is indirect and bridged through Mud2p.
The two-hybrid results presented earlier, support direct association of BBP with Prp39p, Mer1p and Smy2p but do not rule out independent interactions of these proteins with Mud2p. As BBP is required for cell viability, we cannot assay for Mud2p-based interactions in its absence. We reasoned, however, that mutation of the Mud2p interaction domain of BBP would not be a lethal and would allow us to score for bridged two-hybrid associations. Previous studies localized the Mud2p binding site to the N-terminal half of BBP [see (19), (40) and references within]. We created a BBP derivative, bbpΔ56, from which only the most N-terminal region of the proposed interaction domain was deleted and tested this derivative for biological activity. The bbpΔ56 allele complements the lethality of bbp::KAN although the mutant grows more slowly at both 30°C and 37°C than wild-type yeast or the mud2::KAN mutant (Figure 3B). While BBP is readily affinity purified with Mud2-TAP, no band unique to the bbpΔ56 sample can be recovered with Mud2-TAP (Figure 3C compare lane 1 with lanes 2 and 3). Thus, unlike the full length protein, BbpΔ56p does not stably bind Mud2p.
Since BbpΔ56p impairs or blocks Mud2p binding, proteins which interact with Mud2p through a BBP-dependent bridge were anticipated to show reduced or abolished two-hybrid signals in the bbpΔ56 background. As seen in Figure 3A, Mud2p continues to interact with Clf1p under these conditions while the Prp39p, Smy2p and Mer1p two-hybrid interactions are lost. Based on this, we propose that Clf1p most likely binds Mud2p directly and that Prp39p, Mer1p and Smy2p interact with Mud2p through a BBP-dependent bridge.
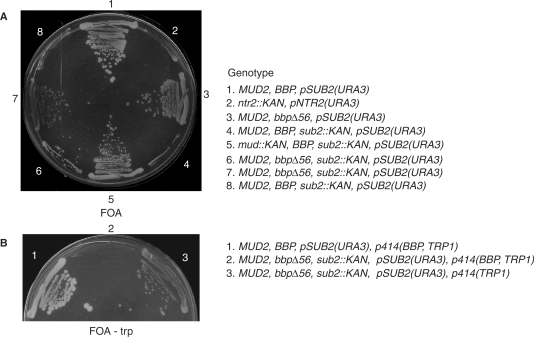
Previously, Guthrie and colleagues (41) showed that deletion of MUD2 suppresses the lethality (or near lethality) of a sub2 loss-of-function allele, suggesting that the dissociation of Mud2p from pre-mRNA is an important function of Sub2p. Our results show that Mud2p and BBP bind one another in vivo, raising the possibility that the BBP–Mud2p heterodimer is the target of Sub2p activity. To investigate this further we tested whether the bbpΔ56 mutation likewise compensates for the loss of SUB2. Here a URA3-marked plasmid copy of SUB2 [i.e. pSUB2(URA3)], was introduced into yeast bearing a chromosomal sub2::KAN null allele in a wild-type background or in the presence of either the mud2::KAN or the bbpΔ56 mutation (Figure 4A). When selected for plasmid loss on five floroorotic acid (FOA) medium, no visible colonies form after 3 days at 30°C with the sub2::KAN mutant in an otherwise wild-type background (streaks 4 and 8) confirming the vital requirement for Sub2p function. Similar results are seen with a control strain in which a lethal ntr2::KAN mutation is complemented by a URA3-linked plasmid that expresses a functional NTR2 allele (streak 2). In contrast, the mud2::KAN, sub2::KAN double mutant (streak 5) and the bbpΔ56, sub2::KAN double mutant (streaks, 6 and 7) are viable in the absence of the pSUB2(URA3) plasmid. Importantly, re-introduction of a plasmid-based wild-type copy of BBP on a TRP1-marked plasmid in the bbpΔ56, sub2::KAN double mutant background restores the essentiality of pSUB2(URA3) (i.e. p414-BBP, TRP1; Figure 4B, streak 2). As expected, this same strain transformed an empty vector (i.e. p414-TRP; streak 3) or a wild-type yeast strain transformed with the p414-BBP, TRP1 plasmid (streak 1) continue to form colonies after pSUB2(URA3) loss. The ability of both mud2::KAN and bbpΔ56 to suppress sub2::KAN lethality bolsters the view that BBP and Mud2p functionally interact in vivo and strongly suggests that the BBP–Mud2p heterodimer is a substrate for Sub2p.
Figure 4.
Mutation of the Mud2p interaction domain of BBP suppresses sub2::KAN lethality. (A) Yeast being the wild-type or the indicated mutant alleles of the MUD2, BBP, SUB2 transformed with a URA3-linked plasmid copy of SUB2 [pSUB2(URA3)] plated at 30°C on FOA medium to select for plasmid loss. The fully wild-type strain (steak 1) and a mutant in the essential NTR2 gene (ntr2::KAN) complimented by a URA3-linked wild-type plasmid (pNTR2(URA3)) are included as positive and negative controls for growth, respectively. Streaks 6 and 7 are different meiotic isolates bearing the same bbpΔ56 mutation. (B) Re-introduction of a functional BBP allele (or an empty vector, streak 3) on a second plasmid in wild-type yeast (steak 1) or the bbpΔ56 background (streak 2). Each strain also is transformed with the pSUB2(URA3) plasmid. Growth is for 3 days at 30°C FOA medium without tryptophan.
Deletion of MUD2 suppresses pre-mRNA loss in the bbpΔ56 background
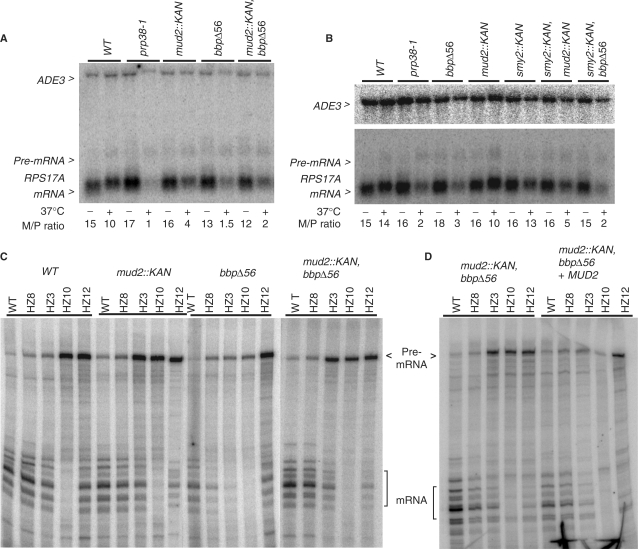
Since BBP and Mud2p are believed to act cooperatively in branchpoint binding, yeast with the mud2::KAN or bbpΔ56 mutations might be expected to show similar growth characteristics if the only consequence of bbpΔ56 was the loss of Mud2p interaction. However, the bbpΔ56 mutant and the mud2::KAN, bbpΔ56 double mutant grow more slowly than the mud2::KAN strain, suggesting that some additional function of BBP is impaired by the bbpΔ56 lesion (Figure 3B). When pre-mRNA is assayed at 23°C, the mRNA to pre-mRNA ratio (a measure of splicing efficiency) of the RPS17A transcript appears almost equivalent in the wild-type, single and double mutant backgrounds (Figure 5A; the intronless ADE3 mRNA is presented as a sample loading control). At 37°C, splicing is more impaired in the mud2::KAN mutant when compared to the wild-type strain although it remains better than what is observed after inactivation of the essential U4/U6.U5 tri-snRNP protein, Prp38-1p (30). The bbpΔ56 mutant has a somewhat stronger splicing defect than that seen with mud2p::KAN mutant at 37°C, consistent with the lower vitality of this strain. We see no exacerbation of this splicing defect in the mud2::KAN, bbpΔ56 double mutant background (Figure 5A and see below), however. While there is some experimental variability in splicing efficiency with temperature shift, we see no clear splicing defect with the smy2::KAN knockout mutant and no reproducible exacerbation of the mud2::KAN or bbpΔ56 splicing defects by this mutation (Figure 5B).
Figure 5.
Splicing inhibition by the mud2::KAN and bbΔ56 mutations. (A) Northern analysis of RNA extracted from wild-type yeast (WT), the ts splicing mutant, prp38-1, and yeast with the bbpΔ56 and mud2::KAN single mutations and with the combined bbpΔ56 plus mud2::KAN double mutant background. RNA harvested from yeast grown continuously at 23°C (−) and after a 2 h shift to 37°C (+) was hybridized with radiolabeled probes to detect the intronless ADE3 mRNA and the RPS17A pre-mRNA and mRNA. The relative abundance of mRNA and pre-mRNA (M/P ratio) is presented below the image. (B) Northern analysis as in panel A with added samples smy2::KAN and smy2::KAN combined with bbpΔ56 or mud2::KAN. (C) Primer extension analysis of RNA isolated from the indicated yeast backgrounds that express RPS17A reporter gene constructs with the wild-type intron (WT), branchpoint mutants (HZ8, HZ3, HZ10) and 5′ splice site mutant (HZ12). (D) Primer extension of RNA from the bbpΔ56, mud2::KAN double mutant before (left) and after (right) add back of MUD2.
We next investigated the impact of the mud2::KAN and bbpΔ56 deletions on RPS17A reporter gene transcripts with splice site consensus mutations (26). For wild yeast, the pre-mRNA to mRNA ratio increases only modestly when the branchpoint consensus, UACUAAC, is changed to UAUUAAC (HZ8) or UCCUAAC (HZ3) while splicing is essentially abolished when the branchpoint nucleotide is changed in UACUACC (HZ10) [Figure 5C and (26)]. A 5′ splice site mutant, GUAUGU ->GUAUAU (HZ12), also shows a decrease in splicing efficiency in wild-type yeast. In the mud2::KAN background, these splicing defects are modestly exacerbated with the greatest impact seen with HZ3 where the pre-mRNA/mRNA ratio increases ∼2-fold compared to the wild-type strain.
Reporter transcript splicing in the bbpΔ56 background showed two distinctive features. First, although this lesion is outside the RNA binding KH domain core (approximately amino acids 149–133), splicing is much more sensitive to mutations at the branchpoint consensus when compared with wild-type yeast or the mud2::KAN mutant; splicing of the 5′ splice site mutant appears largely unaffected. Second, even as mRNA levels drop, pre-mRNA levels do not accumulate to the degree observed with wild-type or mud2::KAN yeast. This characteristic is especially obvious with the HZ3 and HZ10 substrate mutations and is consistent with the observation that diminished BBP activity results in leakage of unprocessed pre-mRNA into the cytoplasm where it is sensitive to turnover by nonsense-mediated decay (42). Surprisingly, the pre-mRNA accumulation defect of bbpΔ56 is largely suppressed in the bbpΔ56, mud2::KAN double mutant background. This is most easily seen with the HZ3 substrate where pre-mRNA levels are 2- to 3-fold higher than with bbpΔ56 alone. Although a more subtle change; spliced mRNA also appears to increase (here by ∼35% when normalized to background bands) in the bbpΔ56, mud2::KAN double mutant. Importantly, transformation with a MUD2-containing plasmid reverses this effect, resulting in lowered pre-mRNA levels mostly clearly seen with the HZ3 and HZ10 branchpoint motif mutants (Figure 5D).
Since pre-mRNA is degraded by nonsense-mediated decay in the cytoplasm (42), a reduction in nuclear export of pre-mRNA in the bbpΔ56, mud2::KAN background might contribute to the increased pre-mRNA recovery. To learn if this is the case, we used two previously described splice-sensitive reporters in which the lacZ gene is inserted into exon 2 in frame with either the upstream exon (to monitor splicing) or with the intron (to monitor translation of pre-mRNA exported from the nucleus) (27). Unlike the reporter genes described before (Figure 5), the intron of this export reporter contains no pre-mature translational termination codons and remains stable in the cytoplasm (27). To better control genetic variability, β-galactosidase measurements were made in the bbpΔ56, mud2::KAN double mutant before and after reintroduction of either MUD2 or BBP (see Materials and methods section). The β-galactosidase values are adjusted for variation in gene induction by normalization with an intronless reporter cassette (19,20).
Consistent with the earlier studies (19,20), deletion of MUD2 enhances nuclear pre-mRNA export about 2-fold and results in reduced expression from the inherently inefficiently spliced mRNA reporter (Figure 6). The bbpΔ56 mutant shows a greater reduction in spliced mRNA expression and a corresponding increase in signal from the unprocessed pre-mRNA. Rather than decreasing, β-galactosidase levels increase with export reporter in the bbpΔ56, mud2::KAN double mutant background compared to either single mutant. Based on this, we conclude that the increase in pre-mRNA abundance noted before for the bbpΔ56, mud2::KAN double mutant (Figure 5C and D) cannot be due to a simple reduction in the amount of pre-mRNA exported to the cytoplasm. We note that the signal from the inefficiently spliced reporter also appears to increase slightly in the bbpΔ56, mud2::KAN double mutant when compared with the bbpΔ56 alone (P = 0.0008). While other models are conceivable, these results are consistent with the deletion of MUD2 increasing the synthesis or stability of pre-mRNA in the bbpΔ56 background (see Discussion section). Finally, while normal levels of splicing and export are seen with the smy2::KAN mutant or with the smy2::KAN, bbp Δ56 double mutant, when smy2::KAN is combined with mud2::KAN expression from the export reporter increases significantly (P = 0.0004). This genetic interaction suggests a common intracellular function of Smy2p and Mud2p.
Figure 6.
Increased RNA export reporter activity in the mud2::KAN, bbΔ56 and mud2::KAN, smy2::KAN double mutant backgrounds. β-galactosidase activities from the spliced reporter and unspliced reporters are presented in the wild-type and mutant backgrounds. The β-galactosidase measurements were normalized by dividing each by that obtained with an intronless control reporter assayed in parallel and multiplying the resultant value by 1000. The bars represent the experimental standard deviations from four to ten replicate experiments. A two-tailed t-test of significance was used with pair wise comparisons.
DISCUSSION
Specific interactions between BBP and the branchpoint motif occur early in spliceosome assembly that together with the U1 snRNP particle bound to the 5′ splice site contribute to the formation of a nuclear retained structure called the commitment complex (43). In vitro, Mud2p binds BBP to enhance substrate association (23) and recent chromatin immunoprecipitation studies suggest an early-acting role for this protein during spliceosome assembly in vivo (44). In the mammalian system U2AF65 binds tightly to a smaller U2AF subunit, U2AF35 (45), which, together with DEK(46) and PUF60 (47,48) act in 3′ splice site selection. The Schizosaccharomyces pombe (fission yeast) Mud2/U2AF65 homolog also binds a smaller U2AF subunit, U2AF23, as well as stably interacting with the BBP (22). In contrast to the mammalian and fission yeast systems, our studies identify a BBP–Mud2p heterodimer as the prominent form of these commitment complex proteins in budding yeast and rule out stable association of a U2AF35-like third factor.
Previous two-hybrid and protein-interaction studies localized the Mud2p binding domain between residues 41–141 of BBP (37,40). We have refined the map of critical residues by showing that deletion of BBP residues 2–55 significantly weakens or prevents Mud2p association. The BBP–Mud2p heterodimer bridges at least four common two-hybrid interactions. Of these, the BBP-binding partners Prp39p and Mer1p have established roles in early spliceosome assembly consistent with functional associations with the BBP–Mud2p heterodimer. Prp39p is an essential U1 snRNP protein and extracts where Prp39p is inactive or absent fail to form commitment complexes (49). As such, in addition to BBP-Prp40p (3), BBP-Prp39p bolsters the cross-intron association of U1 snRNP with the 5′ splice site and branchpoint region of the pre-mRNA. Mer1p is a meiosis-specific splicing factor that interacts with the Nam8 and Bud13 proteins to promote early spliceosome assembly [(39,50,51) and references within]. The BBP–Mer1p interaction may help integrate predicted U1 snRNP-Nam8p contacts near the 5′ splice site with the U2 snRNP-Bud13p interactions at the branchpoint on inherently weak splicing substrates (6,52). Although Mud2p is not essential for cell viability, Mud2p binds the U2 snRNP SF3b subunit, Hsh155p, in addition to other U2 snRNP proteins (6,18,37) providing a possible means by which Mud2p may enhance the kinetics of spliceosome assembly (4). The Mud2p two-hybrid partner, Clf1p, also interacts genetically or biochemically with several SF3b or SF3b-associated U2 snRNP proteins [i.e. Hsh155, Rds3, Rse3p, Bud13p; see (6,53)]. And while generally thought to function with the Prp19 complex (NTC) late in splicing (54), an earlier role for Clf1p in spliceosome assembly has been suggested (32,55). Indeed, a solution structure was recently presented for the N-terminus of Clf1p interacting with the FF domain of the BBP-binding protein and U1 snRNP component, Prp40p (56). Consequently, Clf1p may act with the BBP–Mud2p complex defined here to influence U1 and U2 snRNP dynamics with the nascent pre-mRNP.
The biological relevance of the Smy2p-BBP interaction to splicing is less clear. SMY2 was originally identified as a dosage suppressor of a mutation in MYO2, a gene encoding the heavy chain of the yeast type V myosin (57). Smy2p contains a GYF motif (58) (amino acids 205–261), a domain that binds proline-rich sequences and that can be found in other splicing factors, including the U5 snRNP-associated CD2BP2 protein (59,60) and yeast Lin 1 (61,62). Although Smy2p is predicted to interact with the proline-rich domains of BBP and Prp8p (63), we detect no reduction in splicing efficiency in the smy2::KAN mutant or evidence for smy2::KAN exacerbating the mud2::KAN or bbpΔ56 splicing defects. We note, however, that Smy2p is also predicted to bind a number of cytoplasmic mRNA decay factors including Eap1p, Pat1p, Kem1p, Mot2p, Ccr4p, Pop2p (64) and that it localizes to cytoplasmic P-bodies enriched in these factors (64). While requiring additional study, the increased RNA export reporter signal in the mud2::KAN, smy2::KAN double mutant might reflect stabilization of cytoplasmic pre-mRNA in the absence of Smy2p and Mud2p. Pre-mRNA stabilization was not obvious for natural transcripts in this background (Figure 5) although this might be explained by the normally rapid degradation of pre-RNAs with premature translational termination codons.
The hypersensitivity to branchpoint mutations observed for BbpΔ56p has been with other BBP mutations (19) and for each mutant assayed impaired splicing is correlated with enhanced pre-mRNA export from the nucleus. Increased nuclear export of pre-mRNA is not limited to mutants of BBP and Mud2p, however, as mutations in a number of early acting splicing factors and other nuclear proteins also show this defect (37,65–67), indicating considerable complexity to the nuclear retained mRNP. We observe that reduced pre-mRNA recovery with branchpoint motif mutants in the bbpΔ56 background can be partially suppressed by deletion of MUD2, apparently without reducing pre-mRNA export. All else being equal, stabilizing nuclear pre-mRNA in the mud2::KAN, bbpΔ56 background might be expected to provide increased opportunity both for the precursor to be spliced or exported, both possibilities consistent with the results of our reporter gene assays (Figures 5 and 6). Nuclear pre-mRNA is subject to surveillance by the exosome (68).While the details of exosome target selection are not well understood (69), it seems reasonable that pre-mRNA assembled into spliceosomes will be less sensitive to turnover. The suppression of sub2::KAN lethality by bbpΔ56 and mud::KAN argues for the bypass of the Sub2p-dependent step, possibly enhancing spliceosome assembly. A second but not mutually exclusive possibility for the enhanced pre-mRNA recovery in the mud2::KAN, bbpΔ56 double mutant is that the presence of Mud2p may normally enhance the turnover of intron-bearing pre-mRNA in the cytoplasm. As mutations in the pre-mRNA branchpoint motif increase the level of cytoplasmic pre-mRNA (27), suppression by mud2::KAN might reasonably be most pronounced with branchpoint mutants in the bbpΔ56 background. A cytoplasmic function for Mud2p would be consistent with its localization in both the nucleus and cytoplasm (70), the association of the BBP–Mud2p heterodimer with the P-body linked Smy2p, and with the observation that the metazoan U2AF can be recovered bound to spliced mRNA and to the transcripts of intronless genes (71,72). Finally, given the functional coupling of transcription, splicing, mRNA export, translation and decay [see (73,74) and references within], it is conceivable that Mud2p acts elsewhere in the gene expression pathway to alter cellular pre-mRNA levels.
ACKNOWLEDGEMENTS
We thank Kate Zaytseva and Mingxia Zhang for technical assistance and Charles Query, Stefan Stamm and members of the Rymond lab for their helpful comments on this article. The pRS414-BBP, Acc, Nde/Acc, and SD5 plasmids were generously provided by Michael Rosbash. This work was supported by National Institutes of Health award GM42476 to BCR and infrastructure support in proteomics from the National Science Foundation award EPS-0132295. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health award GM42476 to BCR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Burge CB, Tuschl T, Sharp PA. Splicing of Precursors to mRNAs by the Spliceosome. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1999. [Google Scholar]
- 2.Kramer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 4.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, He J, Lynn B, Rymond BC. Interactions of the yeast SF3b splicing factor. Mol. Cell Biol. 2005;25:10745–10754. doi: 10.1128/MCB.25.24.10745-10754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 8.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 9.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 10.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 11.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 12.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 13.Webb CJ, Lakhe-Reddy S, Romfo CM, Wise JA. Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo. Mol. Biol. Cell. 2005;16:584–596. doi: 10.1091/mbc.E04-09-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zorio DA, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA. 1999;5:487–494. doi: 10.1017/s1355838299982225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 17.Rymond BC, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 18.Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 19.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rain JC, Legrain P. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen TH, Dower K, Libri D, Rosbash M. Early formation of mRNP. License for export or quality control? Mol. Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 22.Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S.pombe requires a stable complex of SF1-U2AF(59)-U2AF(23) EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berglund JA, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser C, Michaelis S, Mitchell A, editors. Methods in Yeast Genetics. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1994. [Google Scholar]
- 26.Jacquier A, Rodriguez JR, Rosbash M. A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985;43:423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- 27.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 28.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teem JL, Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc. Natl Acad. Sci. USA. 1983;80:4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanton S, Srinivasan A, Rymond BC. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pikielny CW, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Hobbs K, Lynn B, Rymond BC. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 2003;278:7875–7883. doi: 10.1074/jbc.M210839200. [DOI] [PubMed] [Google Scholar]
- 33.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Rymond BC. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol. Cell. Biol. 2003;23:7339–7349. doi: 10.1128/MCB.23.20.7339-7349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen N, Harris TW, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Canaran P, Chan J, Chen CK, et al. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33:D383–389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 37.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two- hybrid screens [see comments] Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 38.Chung S, McLean MR, Rymond BC. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA. 1999;5:1042–1054. doi: 10.1017/s1355838299990635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spingola M, Armisen J, Ares M., Jr Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res. 2004;32:1242–1250. doi: 10.1093/nar/gkh281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rain JC, Rafi Z, Rhani Z, Legrain P, Kramer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc. Natl Acad. Sci. USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 44.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl Acad. Sci. USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 47.Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS ONE. 2007;2:e538. doi: 10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Page-McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA. 1999;5:1548–1560. doi: 10.1017/s1355838299991938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lockhart SR, Rymond BC. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scherrer FW, Jr, Spingola M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA. 2006;12:1361–1372. doi: 10.1261/rna.2276806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 52.Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vincent K, Wang Q, Jay S, Hobbs K, Rymond BC. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics. 2003;164:895–907. doi: 10.1093/genetics/164.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CH, Yu WC, Tsao TY, Wang LY, Chen HR, Lin JY, Tsai WY, Cheng SC. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 2002;30:1029–1037. doi: 10.1093/nar/30.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung S, Zhou Z, Huddleston KA, Harrison DA, Reed R, Coleman TA, Rymond BC. Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biophys. Biochem. Acta. 2002;1576:289–297. doi: 10.1016/s0167-4781(02)00368-8. [DOI] [PubMed] [Google Scholar]
- 56.Gasch A, Wiesner S, Martin-Malpartida P, Ramirez-Espain X, Ruiz L, Macias MJ. The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains. J. Biol. Chem. 2006;281:356–364. doi: 10.1074/jbc.M508047200. [DOI] [PubMed] [Google Scholar]
- 57.Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell. Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishizawa K, Freund C, Li J, Wagner G, Reinherz EL. Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc. Natl Acad. Sci. USA. 1998;95:14897–14902. doi: 10.1073/pnas.95.25.14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kofler M, Heuer K, Zech T, Freund C. Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J. Biol. Chem. 2004;279:28292–28297. doi: 10.1074/jbc.M402008200. [DOI] [PubMed] [Google Scholar]
- 60.Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Luhrmann R. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA. 2005;11:598–608. doi: 10.1261/rna.2300805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bialkowska A, Kurlandzka A. Proteins interacting with Lin 1p, a putative link between chromosome segregation, mRNA splicing and DNA replication in Saccharomyces cerevisiae. Yeast. 2002;19:1323–1333. doi: 10.1002/yea.919. [DOI] [PubMed] [Google Scholar]
- 62.Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- 63.Kofler MM, Freund C. The GYF domain. FEBS J. 2006;273:245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- 64.Georgiev A, Sjostrom M, Wieslander A. Binding specificities of the GYF domains from two Saccharomyces cerevisiae Paralogs. Protein Eng. Des. Sel. 2007;20:443–452. doi: 10.1093/protein/gzm041. [DOI] [PubMed] [Google Scholar]
- 65.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abovich N, Legrain P, Rosbash M. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol. Cell Biol. 1990;10:6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 68.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 69.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 71.Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 72.Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 74.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]