Abstract
Programmed ribosomal frameshifting (PRF) is a process by which ribosomes produce two different polypeptides from the same mRNA. In this study, we propose three different kinetic models of +1 PRF, incorporating the effects of the ribosomal E-, P- and A-sites toward promoting efficient +1 frameshifting in Escherichia coli. Specifically, the timing of E-site tRNA dissociation is discussed within the context of the kinetic proofreading mechanism of aminoacylated tRNA (aa-tRNA) selection. Mathematical modeling using previously determined kinetic rate constants reveals that destabilization of deacylated tRNA in the E-site, rearrangement of peptidyl-tRNA in the P-site, and availability of cognate aa-tRNA corresponding to the A-site act synergistically to promote efficient +1 PRF. The effect of E-site codon:anticodon interactions on +1 PRF was also experimentally examined with a dual fluorescence reporter construct. The combination of predictive modeling and empirical testing allowed the rate constant for P-site tRNA slippage (ks) to be estimated as ks ≈1.9 s−1 for the release factor 2 (RF2) frameshifting sequence. These analyses suggest that P-site tRNA slippage is the driving force for +1 ribosomal frameshifting while the presence of a ‘hungry codon’ in the A-site and destabilization in the E-site further enhance +1 PRF in E. coli.
INTRODUCTION
Programmed ribosomal frameshifting (PRF) is a coded shift in reading frame during translation of an mRNA transcript. Consequently, one transcript may yield two different protein products, an inframe product and a frameshifted product. PRF has been observed to occur in various organisms including prokaryotes and eukaryotes. In +1 PRF, the ribosome skips over one nucleotide toward 3′ direction. +1 PRF has been observed in Escherichia coli in the translation of prfB to produce release factor 2 (RF2) (1). In Saccharomyces cerevisiae two retrotransposable elements, Ty1 and Ty3 (2,3), and three genes, ABP140 (4), EST3 (5) and OAZ1 (6) use +1 PRF. The expression of mammalian antizyme has also been shown to involve +1 PRF (7).
Several features have been shown to facilitate +1 PRF: (i) low levels of aminoacylated-tRNA (aa-tRNA) corresponding to the in-frame A-site codon, i.e. hungry codons (8); (ii) the ability of P-site tRNA to form near-cognate interactions with the shifted frame codon, i.e. slippery sequence (9); and (iii) the presence of a stimulatory signal, such as a Shine–Dalgarno (SD)-like sequence upstream of the frameshifting site (10) or an RNA secondary structure downstream of the frameshifting site (3). Both (i) and (iii) may promote a pause in translation elongation, which allows more time for a recoding event to occur, suggesting that +1 PRF is kinetically driven (11).
Several mechanistic models have been proposed to explain +1 PRF (11–13). The kinetic model of Baranov et al. (13) illustrated the dependence of frameshift efficiency on the stability of the P-site interaction and the concentration of incoming aa-tRNA available for the zero and +1 frames. This kinetic model is consistent with observations from several frameshifting studies. For example, the codon: anticodon interaction in the +1 frame of the P-site has been shown to affect the amount of frameshifted products (9). Overexpression of the cognate P-site tRNAs has also been shown to dramatically reduce +1 PRF in yeast and vice versa (2,14,15).
Recent experimental observations suggest that the E-site plays a crucial role in the efficiency of +1 PRF in E. coli (16). In that study, premature release of E-site tRNA from the ribosome correlated with high levels of frameshifting products. A mutagenesis study of 23S rRNA has also illustrated the correlation between E-site tRNA binding and the maintenance of reading frame (17). A recently published study shows that RF2 programmed frameshifting is inversely correlated with E-site stability in E. coli (18). To date, no published kinetic model of +1 PRF has explained the effect of E-site tRNA release on +1 PRF.
In the present study, we propose a new mathematical model for +1 PRF in E. coli, which incorporates the effects of E-, P- and A-site interactions in promoting high levels of frameshifting. Previously published theories of +1 PRF usually focus on a single aspect of +1 PRF [e.g. A-site tRNA abundance, stability of P-site tRNA–ribosome interaction and etc. (8,9,14,15)]. Here, we present a model synthesizing previously observed effects of all three ribosomal tRNA-binding sites on +1 PRF efficiency in E. coli. Of particular note, this is the first model combining the concepts of kinetic proofreading of aa-tRNA selection (19) with the allosteric model (20) to describe +1 PRF. The proposed mathematical model suggests that the rate of P-site tRNA slippage is the most significant parameter in the +1 PRF event, while the abundance of cognate aa-tRNA and the rate of E-site tRNA release act synergistically to promote highly efficient +1 PRF.
Kinetic model
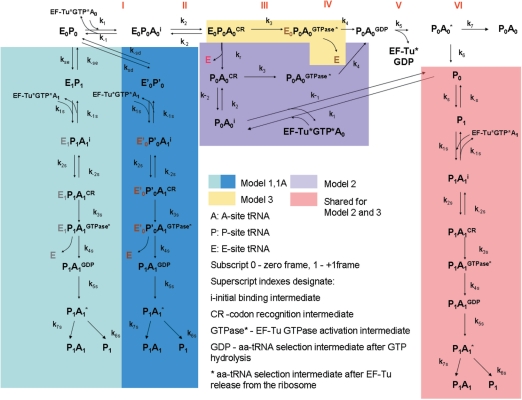
An elegant series of biochemical studies have contributed to a very detailed kinetic model of A-site tRNA selection (19). In this model, fast initial binding of the ternary complex EF-Tu:aa-tRNA:GTP is followed by codon recognition. Codon recognition triggers EF-Tu GTPase activation, which leads to the GTP hydrolysis and dissociation of EF-Tu from the ribosome. Factor dissociation is followed by the spontaneous accommodation of the acceptor end of the aa-tRNA into the A-site or the rejection of the aa-tRNA by proofreading. This concept is illustrated along the top of Figure 1.
Figure 1.
The three kinetic models for +1 PRF in E. coli. Steps I–VI illustrate the non-frameshifting translation elongation process: I. initial binding; II codon recognition; III. GTPase activation; IV. GTP hydrolysis; V. EF-Tu dissociation; VI. accommodation. In Model 1, both E-site and P-site tRNAs slip into the +1 frame and follow the +1 frame aa-tRNA selection to produce frameshifted proteins (P1A1). In Model 1A, both E-site and P-site tRNAs are destabilized by stimulatory signals and follow the +1 frame aa-tRNA selection to produce frameshifted proteins (P1A1). Models 2 and 3 differ in the timing of the E-site tRNA dissociation step (in Model 2, E-site tRNA dissociation occurs during the codon recognition step while in Model 3, E-site tRNA dissociates after codon recognition). Both Models 2 and 3 result in the formation of ribosomes with only P-site tRNA (P0), which can slip to the +1 frame to form P1 and result in the formation of frameshifted proteins (P1A1).
Other recent studies suggested that events at the ribosomal E-site are involved in coordinating this process, specifically that E-site tRNA dissociation occurs prior to GTP hydrolysis (21). Functional studies suggest that +1 PRF efficiency is linked to the E-site occupation and the identity of the E-site tRNA (16–18). Following the allosteric model of the elongation cycle, the E-site is occupied at the start of each cycle prior to aa-tRNA selection, and A-site tRNA binding promotes release of the E-site tRNA, followed in turn by peptidyl transfer and translocation. During translocation the deacylated tRNA is shifted from the P-site to the E-site. Thus there are two events that affect the E-site occupancy: aa-tRNA selection and translocation. The previously observed effects of tRNA abundance and amino acid starvation on +1 PRF efficiency strongly suggest that +1 PRF occurs during A-site tRNA selection (2). Importantly, recent X-ray crystal structures show that the E-site tRNA can form 1–3 base-pairing interactions with the mRNA (22,23). Thus E-site tRNA destabilization may make ribosomes more prone to frameshifting by reducing the extent of tRNA–mRNA interactions.
Because the exact timing of dissociation is unknown, three different models of +1 PRF in E. coli that differ in the timing of E-site tRNA release (Figure 1) were constructed in the present study. In Model 1, simultaneous slippage of E- and P-site tRNAs is hypothesized to occur before aa-tRNA selection (shaded in light blue). In this model, the rate constant of the simultaneous slippage (kse) is determined by the stability of the E0P0 complex (ribosomes with E- and P-site occupied). The stability of this complex depends on the identity of the P-site tRNA (13,14), and to some extent on the E-site tRNA (24,25). The 3′ slippage results in the formation of E1P1 complex, in which both E-site and P-site tRNAs have been shifted by one base. Thereafter, the ribosome can follow the normal elongation cycle to produce frameshifted proteins (P1A1). Similarly, in Model 1A (shaded in dark blue in Figure 1), stimulatory signals may destabilize E0P0, yielding an unstable complex E′0P′0. As this destabilization occurs, the codon at the A site is shifted, leaving both zero frame aa-tRNA (A0) and +1 frame aa-tRNA (A1) as near-cognate ternary complexes. The binding of A1 to the A site will trigger the release of the E-site tRNA. This step is then followed by the slippage of the P-site tRNA to base pair with the +1 frame. Frameshifted products (P1A1) would then be produced by following the remaining steps of aa-tRNA selection by ribosomes.
Slippage could also occur during aa-tRNA selection. To accommodate for the unclear timing of E-site tRNA release, two additional models are proposed. In Model 2, E-site tRNA dissociation occurs during the codon recognition step. E-site empty ribosomes formed at this step can either continue with the subsequent steps of aa-tRNA selection or undergo the reverse reaction to yield initial binding complex P0A0 i. P0A0 i can again undergo the aa-tRNA selection or release the aa-tRNA to form ribosomes with only P-site tRNA occupied (P0). Depending upon the slippage constant (ks), tRNA in the P0 state can slip to base pair with the +1 frame and form the P1 state. P1 can then go through the +1 frame aa-tRNA selection and produce the frameshifted proteins (P1A1). Alternatively, the E-site tRNA might dissociate after codon recognition (Model 3). In this model, E-site empty ribosomes (P0) can be formed consequent to aa-tRNA rejection during the accommodation step. Importantly, because the initial binding of aa-tRNA is fast and non-specific, Model 2 would result in the formation of a significantly larger fraction of the ribosomes in P0 states as compared to Model 3.
MATERIALS AND METHODS
Computation of the kinetic model
All three models were mathematically described by systems of ordinary differential equations (see text in Supplementary Data). Assuming steady state, the expressions of intermediate concentrations in terms of initial reactant (E0P0) were solved by Matlab 7.2 (Mathworks Inc., USA). By applying the empirically determined rate constants and assumed ranges of rate constants of P-site tRNA slippage, and rate constants of E-site tRNA release (Tables S1 and S2 in Supplementary Data) with different aa-tRNA concentrations (Table S3 in Supplementary Data), the amount of non-frameshifted proteins (P0A0) and frameshifted proteins (P1A1) were calculated. The frameshift efficiency (FS%) in the model is defined as the ratio of P1A1 to total proteins (P0A0 + P1A1) multiplied by 100%.
Plasmids and bacterial strains
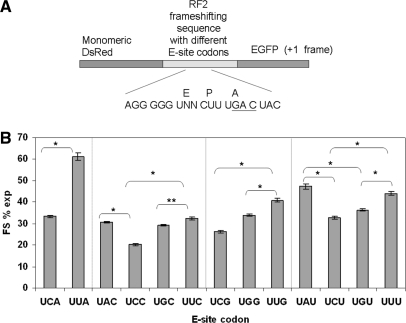
Escherichia coli XL1 blue MRF’ (Stratagene) was used in all experimental studies. The gene sequence of monomeric DsRed (26) was first cloned between HindIII and SalI sites in pEGFP vector (Clontech, USA) to create pRG plasmid, which can express DsRed-EGFP fusion protein. Different linker sequences were made from complementary oligonucleotides (Integrated DNA Technology, USA) and were cloned between SalI and BamHI sites between the coding sequence of DsRed and EGFP in the pRG plasmid. The linker sequence for the control strain is tcgacttctggctctggctctggcgag, which kept both DsRed and EGFP coding sequences in frame. The linker sequences for the mutants contained mutated RF2 frameshifting sites (tcgactagggggUNNctttgactacgag) which made EGFP coding sequence in +1 frame (UNN refers to the E-site codon when +1 frameshifting is taking place and the stop codon is underlined). The control strain expressed only the DsRed-EGFP fusion protein. The mutants expressed DsRed proteins as non-frameshifted proteins (because of the stop codon in the linker sequence) and DsRed-EGFP fusion protein as frameshifted proteins (because the stop codon is bypassed by +1 frameshifting). Thirteen mutants differing only in the E-site codon (UNN) in the recoding sites were constructed. Among the 13 mutants, the first base in the E-site codon was kept intact to maintain SD-like sequence and stop codons were avoided.
Fluorescence assay
Cells with different plasmids were cultured in 200 μl Luria–Bertani (LB) medium containing 100 μg/ml ampicillin in a 96-well plate for 24 h at 37°C, 250 rpm. The fluorescence was then measured by plate reader (SpectraMax Gemini EM, Molecular Devices). The green fluorescence was measured with excitation wavelength at 485 nm and emission at 528 nm. The red fluorescence was measured with excitation wavelength at 530 nm and emission at 590 nm. From the fluorescence measurement, the experimental frameshift efficiency (FS%exp) was obtained as the ratio of green fluorescence to red fluorescence for the mutant strains (containing RF2 sequence with different E-site codons), normalized against the fluorescence ratio of the control strain.
Chi-square analysis
Chi-square is defined as:
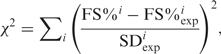 |
where i refers to different E-site codons (i = 1–13, for 13 tested E-site codons), FS% is the frameshift efficiency calculated by the model and FS%exp is the frameshift efficiency observed in the experiment. The rate constant of E-site tRNA release, kr, was assumed as kr = A′exp(−mjΔGc/RT), where A′ is the pre-exponential constant for the effect of the stimulatory signals (the same for all tested E-site codons); ΔGc is the codon:anticodon interaction in the E-site (27); mj is the modifying factor to account for other factors (e.g. tRNA:ribosome interactions, base modification, etc.) that may affect the contribution of the base pairing on kr (j = 1–6, see Table S4 in Supplementary Data); R is the gas constant (8.314 JK−1mol−1); T is the temperature (310 K). Matlab V.7.2 was used to optimize the values of ks, A′ and mj that resulted in the minimum chi-square value.
RESULTS
Mathematical model
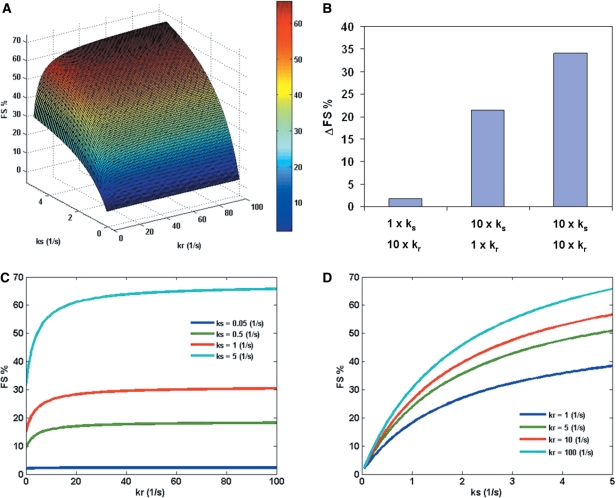
The three major variables in the model are the rate constant of P-site tRNA slippage (ks), the rate constant of E-site tRNA release (kr) and the concentration of cognate aa-tRNA for zero-frame codon in the A-site (cog.A0). To understand the synergistic effect of ks, kr and cog.A0, surface plots are used to show the effect of any two parameters on FS% while keeping the third parameter as a constant. Figure 2A shows the effect of ks and kr on FS%. An increase in FS% is observed as kr and ks are increased. Figure 2B shows an example of the synergistic effect of E-site (kr) and P-site (ks): while a 10-fold increase in kr or ks alone results in an increase in FS%, a 10-fold increase in both parameters results in a greater increase in FS% than the summation of the individual effects. Figure 2C and D show the cross-section curves of Figure 2A. This analysis suggests that the effect of kr is more significant when kr is below 10 s−1 (Figure 2C). Interestingly, the effect of kr on FS% is less important for smaller values of ks (only 1% increase in FS% with increasing kr for ks = 0.05 s−1), which suggests that the effect of the E-site tRNA release becomes prominent above a threshold value of P-site tRNA slippage (represented by ks).
Figure 2.
(A) The effect of P-site tRNA slippage (represented by ks) and E-site tRNA release (represented by kr) on FS% at fixed concentration of zero-frame cognate aa-tRNA (cog.A0 = 1%). All the other parameters are assumed to be constants (Table S1. in Supplementary Data). (B) An example of the synergistic effect of E-site (kr) and P-site (ks) (data points from Figure 2A). 1× means the parameter is the same as a randomly chosen base point (ks = 0.2 s−1, kr = 1 s−1). 10× means a 10-fold increase in the parameter. ΔFS% refers to the increase in FS% as compared to the base point. (C) The effect of kr on FS% at different values of ks (0.05–5 s−1). (D) The effect of ks on FS% at different values of kr (1–100 s−1).
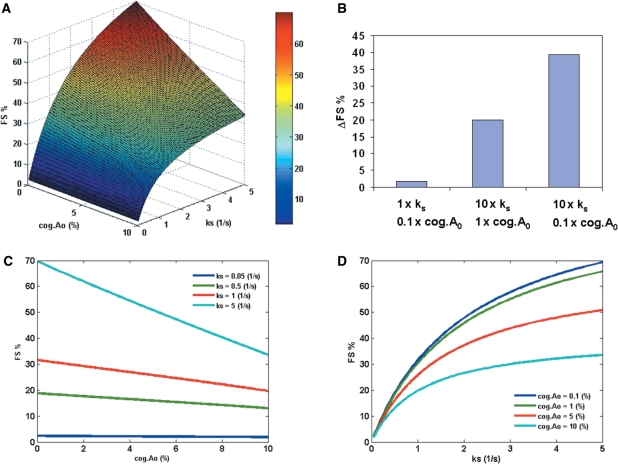
Additionally, the model reveals a synergistic effect of P-site tRNA slippage and the hungry codon (Figure 3A). The analysis suggests that FS% increases as cog.A0 decreases and as ks increases. Figure 3B shows an example of the synergistic effect of P-site (ks) and A-site (cog.A0): while a 10-fold decrease in cog.A0 or a 10-fold increase in ks results in an increase in FS%, a 10-fold change in both parameters results in a greater increase in FS% than the summation of the individual effects. Figure 3C and D show the cross-section curves of Figure 3A. Importantly, the effect of cog.A0 on FS% decreases with ks (Figure 3C). As a result, the hungry codon effect (represented by a small value of cog.A0) becomes more significant as the probability of P-site tRNA slippage increases (represented by larger ks).
Figure 3.
(A) The effect of P-site tRNA slippage (represented by ks) and the concentration of zero-frame cognate aa-tRNA (cog.A0) on FS% at fixed rate constant of E-site tRNA release (kr = 100 s−1). All the other parameters are assumed to be constants (Table S1 in Supplementary Data). (B) An example of the synergistic effect of P-site (ks) and A-site (cog.A0) (data points from Figure 3A). 1× means the parameter was the same as a randomly chosen base point (ks = 0.2 s−1, cog.A0 = 10%). 10× means a 10-fold increase in the parameter. 0.1× means a 10-fold decrease in the parameter. ΔFS% refers to the increase in FS% as compared to the base point. (C) The effect of cog.A0 on FS% at different values of ks (0.05–5 s−1). (D) The effect of ks on FS% at different values of cog.A0 (0.1–10%).
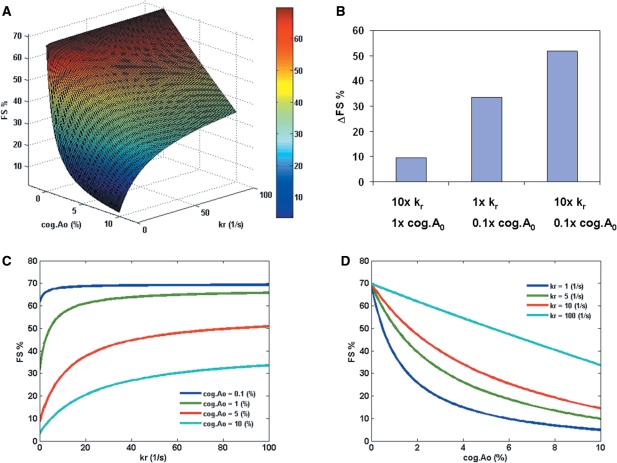
The model also shows the synergistic effect between hungry codon at the A-site and release of tRNAs from the E-site. Examination of Figures 2C and 3C shows that the effects of kr and cog.A0 become significant only for higher values of ks. Therefore, a higher value of ks (5 s−1) was chosen to study the effect of cog.A0 and kr on FS% (Figure 4A). The analysis shows that FS% increases as kr increases and as cog.A0 decreases, respectively. Figure 4B shows an example of the synergistic effect of E-site (kr) and A-site (cog.A0): while a 10-fold increase in kr or a 10-fold decrease in cog.A0 results in an increase in FS%, a 10-fold change in both parameters results in a greater increase in FS% than the summation of the individual effects. Figure 4C and D show the cross-section curves of Figure 4A. The result shows that for small cog.A0, the effect of kr is not important (Figure 4C), i.e. the effect of E-site tRNA release is less important if there is hungry codon in the A-site. Therefore, the model suggests that in the presence of a slippery P-site (high ks) with no hungry codon effect (large cog.A0), a higher rate of E-site tRNA release can still result in a higher FS%. In contrast, for lower rates of E-site tRNA release, the model predicts substantial FS% in the presence of P-site slippery sites and hungry codons (Figure 4D).
Figure 4.
(A) The effect of the concentration of zero-frame cognate aa-tRNA (cog.A0) and E-site tRNA release (represented by kr) on FS% at fixed rate constant of P-site tRNA slippage (ks = 5 s−1). All the other parameters are assumed to be constants (Table S1 in Supplementary Data). (B) An example of the synergistic effect of E-site (kr) and A-site (cog.A0) (data points from Figure 4A). 1× means the parameter was the same as a randomly chosen base point (kr = 1 s−1, cog.A0 = 10%). 10× means a 10-fold increase in the parameter. 0.1× means a 10-fold decrease in the parameter. ΔFS% refers to the increase in FS% as compared to the base point. (C) The effect of kr on FS% at different values of cog.A0 (0.1–10%). (D) The effect of cog.A0 on FS% at different values of kr (1–100 s−1).
Empirical studies
To understand the importance of the release of E-site tRNA on +1 PRF, an in vivo dual fluorescence reporter system in E. coli is used to study the effect of the E-site stability on +1 PRF (see ‘Materials and methods’ section). The reporter system (Figure 5A) allows measurement of frameshift efficiency for different recoding sites by calculating the ratio of green to red fluorescence. All possible E-site codons (13 sense codon = 16 potential codons − 3 stop codons) in the RF2 frameshifting site have been tested under the condition that the SD-like sequence was kept intact. Statistical analysis was applied to all datasets according to Jacobs et al. (2004) (28). Ten replicates for the mutants and 20 replicates for the control were performed to satisfy the minimum sample requirement. The standard error for FS% for different mutants was less than 2%. The results show that the presence of an A:U pair in the second position of the E-site codon in the RF2 frameshifting site results in higher frameshifting as compared to a G:C pair in the same position (Figure 5B). Importantly, frameshift efficiency can be generally categorized into three levels based on the number of hydrogen bonds in the base pair interaction (Table 1). A:U pairs in both the second and the third positions have the lowest number of hydrogen bonds and promote the highest frameshifiting. One A:U pair and one C:G pair in the second and the third positions result in the intermediate level of frameshifting. With the highest number of hydrogen bonds, C:G pairs in both the second and the third positions result in the lowest frameshifting. An interestingly unexpected result is UGG as an E-site codon. The frameshifting efficiency for UGG in the E-site is comparable to that for one G:C and one A:U base pairs in the second and third positions in the E-site. This observation may result from factors not accounted for in the model or perhaps be a result of the reporter protein. The absence of modified nucleoside pseudouridine (Ψ) at position 38–40 in tRNATrpCCA could also be a reason for less-efficient binding of this tRNA to the E-site. It is suggested that deficiency of modified nucleosides may change tRNA structure, resulting in different ribosome:tRNA interactions (29). However, the exact reason for relatively higher FS% for UGG in the E-site is not known. These FS% data are less likely to be due to the availability of tRNA for the specific codon in the E-site, because we observed no obvious correlation between FS% and tRNA concentration for the E-site codons (Figure S1 in Supplementary Data). These experimental observations emphasize the effect of E-site stability on +1 PRF, which is consistent with the computational simulations described earlier.
Figure 5.
The effect of different E-site codon:anticodon interactions on frameshift efficiency. (A) The sequence design of the dual fluorescence reporter system. The E, P and A denote the codon in the E-, P- and A-sites when +1 frameshifting is taking place. The +1 frame A-site codon is underlined. (B) Experimentally obtained frameshift efficiency for different E-site codons. The error bars indicate the standard deviation. * indicates significant difference (P < 0.001). ** indicates significant difference (P < 0.01).
Table 1.
Three levels of +1 frameshifting efficiency for different E-site codons
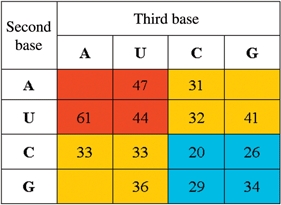 |
Red indicates highest frameshifting: A:U basepairs at both 2nd and 3rd positions of the E-site codon; yellow indicates intermediate frameshifting: One A:U and one G:C at the 2nd and 3rd position of the E-site codon; blue indicates lowest frameshifting: G:C basepairs at both 2nd and 3rd positions of the E-site codon.
Parameter estimation
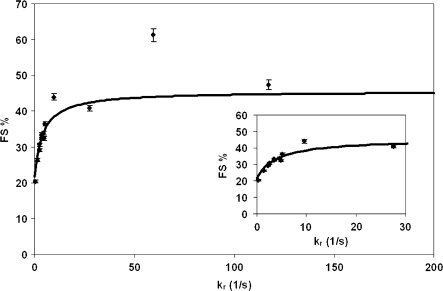
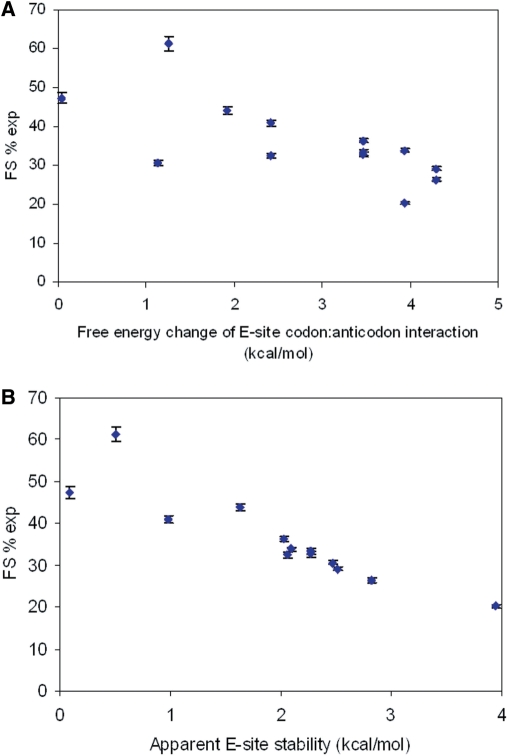
The rate constant for the P-site tRNA slippage (ks) can be estimated by combining the kinetic model and the experimental results. Changing E-site stabilities by using different E-site codons while maintaining the identity of the P-site codon enables manipulation of kr at a constant ks. kr is assumed to be a function of stimulatory signals, tRNA:mRNA (codon:anticodon) and tRNA:ribosome interactions in the E-site (see ‘Materials and methods’ section). Chi-square analyses were performed to obtain optimum values for ks and kr, which give the best fit of the model predictions and the experimental results. Figure 6 shows the data-fitting result of the model prediction (solid line) and the experimentally detected FS% (diamonds). ks was determined to be 1.9 s−1 for the RF2 frameshifting sequence and parameters for calculating kr are listed in Table S4 in Supplementary Data. Modifying factors were used to account for other factors (e.g. tRNA:ribosome interactions, base modifications) that may affect the contribution of the base pairing on kr. The modifying factor for tRNAQTA Tyr was observed to be 2.18. The value is consistent with the observation that the binding efficiency of Q34-tRNATyr to triplet programmed ribosomes is 2-fold more than G34-tRNATyr (30). Modifying factors for other tRNAs are less than one, which may suggest that for tRNAPhe, tRNALeu, tRNASer, tRNACys and tRNATrp, other interactions in the E-site could reduce the contribution of codon:anticodon interactions on kr. The correlation between FS%exp and free energy change of the codon:anticodon interactions in the E-site is shown in Figure 7A and the correlation between FS%exp and apparent E-site stability (free energy change of codon:anticodon interactions in the E-site multiplied by modifying factors) is shown in Figure 7B. Importantly, frameshifting efficiency is observed to inversely correlate with the E-site stability and this observation is more clear when codon:anticodon interactions and other interactions in the E-site are all considered.
Figure 6.
Data fit of frameshift efficiency for different codon:anticodon interactions in the E-site. By using chi-square analysis, the optimum value for ks is 1.9 s−1 for RF2 frameshifting sequence. The diamonds show the experimentally obtained frameshift efficiency for different E-site codons [kr = A′exp(−mΔGc/RT), see ‘Materials and methods’ section]. The solid line indicates model predicted FS% for different kr at ks = 1.9 s−1.
Figure 7.
(A) The correlation between FS%exp and the free energy change of the E-site codon:anticodon interactions. (B) The correlation between FS%exp and the apparent E-site stability obtained by free energy change of the E-site codon:anticodon interactions multiplied by modifying factors (Table S4).
DISCUSSION
Comparison of the three models
Figure 1 presents three possible pathways for ribosomes to synthesize +1 frameshifted proteins in E. coli. We believe that all three pathways can occur in vivo but that Model 2 is the dominant pathway for +1 PRF. Model 1 involves simultaneous slippage of E- and P-site tRNAs to the +1 frame, an energetically unfavorable process less likely to occur. To test this hypothesis, the effect of kse (the rate constant that determines if the ribosome complex would get into Model 1) was studied and it was found that FS% remained at a similar level at different values of kse (Figure S2A–S2C in Supplementary Data). A recently published study observed no correlation between +1 PRF efficiency and the stability of complex of E-site tRNA base pairing with +1 frame (18). Similarly, the effect of ksd, which governs whether the ribosome complex enters Model 1A pathway, was also studied. The effect of ksd is observed to be less significant on FS% (Figure S3A–S3C in Supplementary Data). These observations suggest that Model 1 and Model 1A may contribute much less than Model 2 or 3 to the overall FS%. In Model 3, the formation of P0, the major precursor of frameshifted products, depends on aa-tRNA rejection. The aa-tRNA that reaches the accommodation step is more likely to be cognate, because it has already passed through the selective codon recognition and GTPase activation steps. As a result, this aa-tRNA is less likely to be rejected, and thus the probability for ribosomes to form P0 is less. Although in Model 2, formation of P0 also depends on dissociation of aa-tRNA, the reversible nature of the codon recognition step and higher concentration of non- and near-cognate tRNAs relative to cognate tRNA, together with same rates for forward reactions of codon recognition for both substrates, make P0 formation likely. Therefore, Model 2 would result in formation of a significantly larger fraction of the P0 state ribosomes as compared to Model 3. Thus, we propose that Model 2 is the major pathway for +1 PRF in vivo.
Role of the E-site
The function of ribosome E-site is still under debate in the literature. Some studies suggest the E-site interactions are functionally important for maintaining the reading frame (16,31,32), while others suggest the E-site tRNA binds to the ribosome in a labile manner (33,34). The results presented in this study are fundamentally helpful to explain different E-site effects suggested by different studies. In the proposed mechanism, kr represents the de-occupation of E-site tRNA. Our model results show that the effect of E-site interactions on +1 PRF is more significant when kr is smaller than 10 s−1 and the effect is less when kr is in a range of larger values (Figures 2C and 4C). We believe that different views of the E-site function can be due to the result of different experimental conditions, which produce different ranges of kr. It has been suggested that the ionic conditions, physical parameters (pH, temperature, etc.), and material preparation methods all affect the binding affinity of tRNA to the ribosome (35). Therefore, for buffer conditions or mRNA sequences for which kr is in the range of smaller values, the effect of E-site interactions on +1 frameshifting efficiency can be observed (16–18,31,32). On the other hand, for buffer conditions or mRNA sequences for which kr is in the range of larger values, the effect of E-site interactions is less important for translation elongation (33). This observation clearly demonstrates the utility of a modeling approach to help reconcile disparate observations from the literature.
A question may remain: which range of kr should be expected? The data fitting (Figure 6) shows that the range of kr is actually different for different E-site codons in vivo. For E-site codons UAU, UUA, UUG and UUU, kr are in a range of large values (≥10 s−1). For all the other tested E-site codons in the present study, the kr values are smaller (<10 s−1). Previous in vitro studies used polyU programmed ribosome to study E-site interactions (33,34), which could be the reason that smaller tRNA-binding affinities to the ribosome E-site were observed. On the other hand, weaker interactions in the ribosome E-site have been shown to reduce translational fidelity in vivo (17,18,32). In the present study, the experiments support the importance of E-site interactions in +1 PRF in E. coli (Figure 5B). The data show that an A:U base pairing in the E-site, which contains one less hydrogen bond than a G:C base pairing, results in higher frameshift efficiency. A recently published study using a monocistronic reporter system also showed that RF2 programmed frameshifting is inversely correlated with the E-site stability (18). Taken together, the experimental data in the present study and the study by Sanders et al. (18) provide independent evidence that different E-site interactions may result in different ranges of kr in vivo, illustrating the role of E-site stability on +1 PRF.
Mechanistically, kr may be a function of mRNA:tRNA and tRNA:ribosome interactions at the E-site, stimulatory signals (SD sequence, mRNA structures, etc.), and spacing between the stimulatory signals and the E-site. The experimental results in the present study suggest that tRNA:mRNA base pairing in the E-site could be functionally important, supporting the X-ray crystal structures (22,23). Previous experimental observations also support the effect of stimulatory factors and spacing on frameshifting. For example, it has been proposed that the interaction between the SD and anti-SD sequence in E. coli prfB mRNA precludes the binding of the E-site tRNA and therefore might facilitate destabilization of the E-site tRNA (16). That study also showed that the spacing between the SD sequence and the frameshifting site is critical for high frameshift efficiency. Mutations in the SD sequence have also been shown to cause significant reductions in frameshift efficiency (10). In our model, the SD:antiSD interaction may play its role in RF2 frameshifting in E. coli in two ways. First, the presence of an SD:antiSD interaction enhances the release of E-site tRNA. As for the data fitting in this study, the rate constant for E-site tRNA release is assumed as kr = A′exp(mΔGc/RT). The presence of an SD-like sequence will result in a larger A′ and therefore result in a higher rate of E-site tRNA release, paving the way for +1 PRF in E. coli as described in Model 2. Secondly, the SD:antiSD interaction may destabilize the ribosome complex, yielding unstable complex E′0P′0, which can directly interact with +1 frame aa-tRNA as described in Model 1A.
Stimulatory elements have also been found in the Ty3 and OAZ1 + 1 PRF signals in yeast, and their effects also depended on strict spacing from the sites of frameshifting (6,36). However, there is not yet any direct experimental evidence demonstrating the effect of E-site destabilization in Ty1 and Ty3 frameshifting. The prokaryotic ribosomal structure suggests that although there is no direct contact between E-site tRNA and P-site tRNA in the ribosome, the E-site tRNA might interact indirectly with the P-site tRNA through the 16S rRNA (37–39). In agreement with these observations, our model of +1 PRF suggests that E-site tRNA dissociation might destabilize the mRNA ribosome interactions and affect the P-site tRNA slippage. Thus, ribosomes with an empty E-site may be more prone to slip.
Role of the P-site
The computational modeling shows that for small values of ks (ks = 0.05 s−1), the effects of hungry codons in the A-site, and of rates of E-site tRNA release on FS% are less significant, thus demonstrating that P-site tRNA slippage is the dominant factor for +1 PRF in E. coli. As illustrated in Figure 1, the efficiency of +1 PRF is determined by two competing reaction branches: (i) zero-frame aa-tRNA selection followed by peptidyl transfer (PT) and (ii) P-site tRNA slippage to the +1 frame, which is subsequently trapped by aa-tRNA selection and PT. The rate constant of slippage, ks depends on the stability of the P0 and P1 states. In other words, the analysis presented here indicates that the less stable P0 and more stable P1 (which gives higher ks) should result in higher FS%. This is consistent with previous experimental observations. Curran (9) showed that among 32 polynucleotides differing only in their P-site, tRNAs that form more cognate interactions with the +1 frame in the P-site had a 1000-fold increase in frameshifted proteins than tRNAs mispairing with the +1 frame. Other factors such as wobble base modification and tRNA hypomodification have been shown to weaken base-pairing and stimulate tRNA slippage in the P-site (29,40,41). It has also been suggested that features of tRNA structure outside of the anticodon contribute to the P-site stability and the ability to shift reading frames (42–44). Moreover, in yeast, a mutant form of ribosomal protein L5 (RPL5) that promoted decreased ribosomal affinity for peptidyl-tRNA also promoted increased +1 PRF at a Ty1 signal (45).
The rate constant for P-site tRNA slippage has not been previously reported in the literature. Our kinetic model combined with experiments using different E-site interactions provides an approach to estimate ks. Fitting the experimental data for RF2 frameshifting sequence (CUU U sequence in the P-site) yielded a rate constant of slippage ≈1.9 s−1. The small magnitude of ks, as compared to other rate constants in the model, is consistent with the idea that the slippage is the rate-limiting reaction in the +1 PRF mechanism.
Role of the A-site
Our model suggests that in the presence of a slippery P-site, a low availability of cognate aa-tRNA for zero-frame (cog.A0) can enhance FS% by about 2-fold (Figure 3A and C). A low concentration of cognate tRNA at the A-site (hungry codon) has been experimentally observed to promote frameshifting (46), consistent with the model. We believe that the low availability of the zero-frame cognate aa-tRNA (cog.A0) can affect +1 PRF in two ways. First, the low availability of cog.A0 slows down translation, which allows more time for the kinetically driven +1 PRF event to take place. Secondly, the low availability of cog.A0 increases the chance for the near-cognate tRNA to bind to the A-site. During the elongation cycle, both cognate and near-cognate tRNAs compete for the A-site. Since a near-cognate tRNA has more chances to be rejected after the codon recognition step or GTP hydrolysis step than the cognate tRNA, a low concentration of cognate tRNA is more likely to result in the ribosomes containing only P-site tRNA (P0), thus enhancing the probability of slippage. In support of this, studies in yeast show that mutants that only affect A-site affinities for aa-tRNAs do not affect +1 PRF efficiency (47,48).
+1 PRF in eukaryotes
The rate constants used in this study are based on data obtained using E. coli ribosomes. The finding of synergistic effects among E-, P- and A-site interactions on +1 PRF is likely to be applicable to Ty1 expression in yeast and antizyme expression in mammalian cells. However, owing to differences in aa-tRNA abundance and ribosome structures between prokaryotes and eukaryotes, eukaryotic +1 PRF signals were not tested in the present study. For Ty3 frameshifting in yeast, it is suggested that a special P-site interaction may interfere with the binding of in-frame aa-tRNA and stabilize out-of-frame decoding (6). According to our model, this observation suggests the possibility that a special tRNA interaction in the P-site may change k1s. It is also likely that the Ty3 mechanism includes another reaction pathway for P0 to directly interact with a +1 frame aa-tRNA ternary complex. We believe that a quantitative kinetic model, similar to our current model, can be built for Ty3 frameshifting in yeast to understand this unique frameshifting process better.
CONCLUSION
A detailed kinetic model for +1 PRF in E. coli has been presented and the effect of E-site stabilities on +1 PRF has been experimentally demonstrated. According to the model results, a combination of stimulatory signals leading to the release of deacylated tRNA in the E-site, tRNA slippage in the P-site, and the hungry codon effect in the A-site synergistically promote efficient +1 ribosomal frameshifting. The experimental result suggested that weaker codon:anticon interactions in the E-site correlate with higher +1 PRF efficiency in E. coli. Our mathematical analysis shows that the rate of P-site tRNA slippage is the dominant factor, while the effect of hungry codon in the A-site and E-site tRNA destabilization further enhance +1 PRF. We propose that E-site empty ribosomes, which facilitate the P-site tRNA slippage, is the driving force for +1 PRF.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was funded by New York State office of Science, Technology and Academic Research (to K.H.L.); National Institutes of Health (GM058859 to J.D.D.). We are thankful to Navneetha Santhanam, Dr Fernando Escobedo and Dr Abraham Stroock for their insightful comments and critiques of this work. We gratefully acknowledge Dr Matthew DeLisa for the DsRed gene sequence. We also acknowledge Robert Kuczenski for advice in developing the Matlab program. Funding to pay the Open Access publication charges for this article was provided by the University of Delaware.
Conflict of interest statement. None declared.
REFERENCES
- 1.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 2.Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farabaugh PJ, Zhao H, Vimaladithan A. A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: Frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asakura T, Sasaki T, Nagano F, Satoh A, Obaishi H, Nishioka H, Imamura H, Hotta K, Tanaka K, Nakanishi H, et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- 5.Morris DK, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 6.Palanimurugan R, Scheel H, Hofmann K, Dohmen RJ. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–4867. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsley D, Gallant J. On the directional specificity of ribosome frameshifting at a “hungry” codon. Proc. Natl Acad. Sci. USA. 1993;90:5469–5473. doi: 10.1073/pnas.90.12.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curran JF. Analysis of effects of tRNA:Message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harger JW, Meskauskas A, Dinman JD. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh PJ, Bjork GR. How translational accuracy influences reading frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranov PV, Gesteland RF, Atkins JF. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundararajan A, Michaud WA, Qian Q, Stahl G, Farabaugh PJ. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell. 1999;4:1005–1015. doi: 10.1016/s1097-2765(00)80229-4. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami K, Pande S, Faiola B, Moore DP, Boeke JD, Farabaugh PJ, Strathern JN, Nakamura Y, Garfinkel DJ. A rare tRNA-arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Sergiev PV, Lesnyak DV, Kiparisov SV, Burakovsky DE, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Function of the ribosomal E-site: A mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 20.Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: Features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 21.Dinos G, Kalpaxis DL, Wilson DN, Nierhaus KH. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 23.Jenner L, Rees B, Yusupov M, Yusupova G. Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep. 2007;8:846–850. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 25.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 26.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. PNAS. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klump HH. Exploring the energy landscape of the genetic code. Arch. Biochem. Biophys. 2006;453:87–92. doi: 10.1016/j.abb.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase – function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- 31.Nierhaus KH. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 32.O’Connor M, Willis NM, Bossi L, Gesteland RF, Atkins JF. Functional tRNAs with altered 3′ ends. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lill R, Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 1987;196:137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- 35.Schilling-Bartetzko S, Franceschi F, Sternbach H, Nierhaus KH. Apparent association constants of tRNAs for the ribosomal A, P, and E sites. J. Biol. Chem. 1992;267:4693–4702. [PubMed] [Google Scholar]
- 36.Li Z, Stahl G, Farabaugh PJ. Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA. 2001;7:275–284. doi: 10.1017/s135583820100190x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 38.Ramakrishnan V, Moore PB. Atomic structures at last: the ribosome in 2000. Curr. Opin. Struct. Biol. 2001;11:144–154. doi: 10.1016/s0959-440x(00)00184-6. [DOI] [PubMed] [Google Scholar]
- 39.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl Acad. Sci. USA. 1985;82:4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuchihashi Z, Brown PO. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(lys) and an AAG lysine codon. Genes Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 42.Hansen TM, Baranov PV, Ivanov IP, Gesteland RF, Atkins JF. Maintenance of the correct open reading frame by the ribosome. EMBO Rep. 2003;4:499–504. doi: 10.1038/sj.embor.embor825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz DW, Yarus M. tRNA structure and ribosomal function. II. interaction between anticodon helix and other tRNA mutations. J. Mol. Biol. 1994;235:1395–1405. doi: 10.1006/jmbi.1994.1096. [DOI] [PubMed] [Google Scholar]
- 44.Smith D, Yarus M. Transfer RNA structure and coding specificity. I. evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J. Mol. Biol. 1989;206:489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- 45.Meskauskas A, Dinman JD. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001;7:1084–1096. doi: 10.1017/S1355838201001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallant J, Lindsley D. Ribosome frameshifting at hungry codons: sequence rules, directional specificity and possible relationship to mobile element behaviour. Biochem. Soc. Trans. 1993;21:817–821. doi: 10.1042/bst0210817. [DOI] [PubMed] [Google Scholar]
- 47.Meskauskas A, Baxter JL, Carr EA, Yasenchak J, Gallagher JE, Baserga SJ, Dinman JD. Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol. Cell. Biol. 2003;23:1602–1613. doi: 10.1128/MCB.23.5.1602-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meskauskas A, Petrov AN, Dinman JD. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell Biol. 2005;25:10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.