Abstract
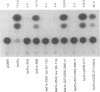
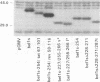
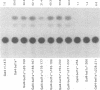
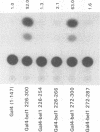
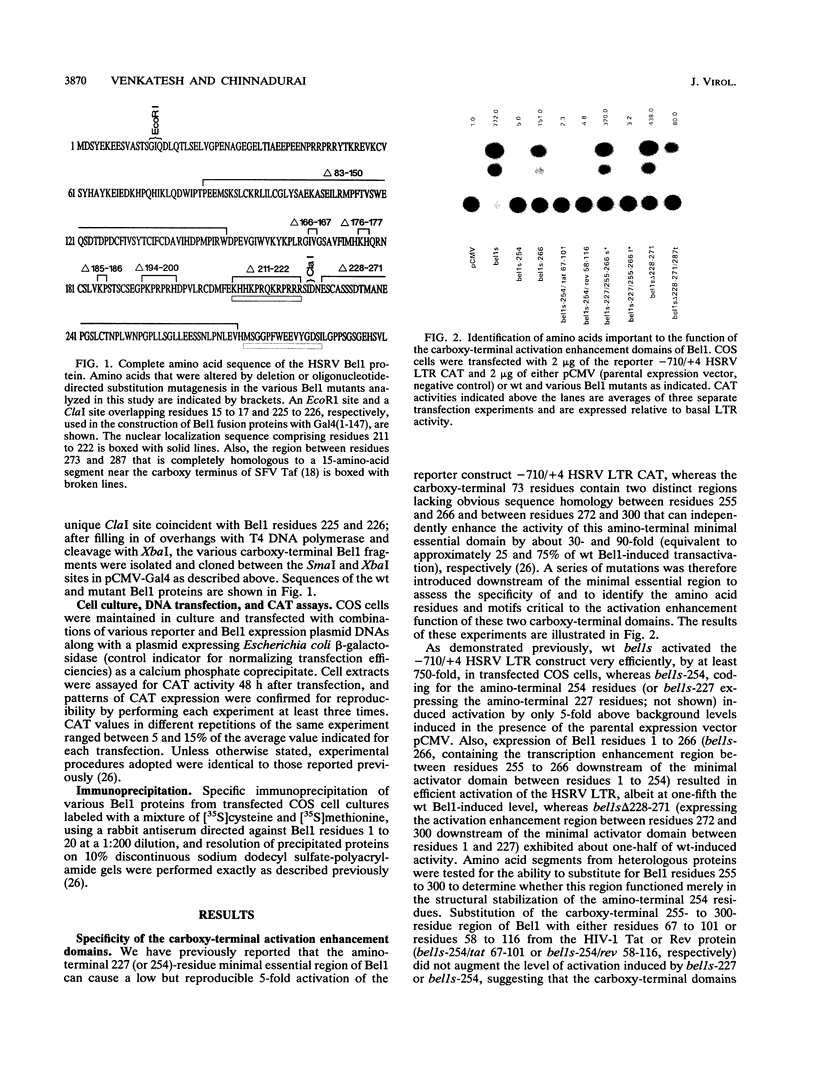
The bel1 gene of human spumaretrovirus (HSRV) encodes a 300-amino-acid nuclear protein termed Bel1 that is a potent activator of transcription from the cognate long terminal repeat (LTR). Bel1 can also efficiently activate the human immunodeficiency virus type 1 (HIV-1) LTR. We have previously shown that the amino-terminal 227-residue region (minimal activator region) of Bel1 can activate the HSRV LTR at low levels and that two distinct domains within the carboxy-terminal 73 residues, from residues 255 to 266 and 272 to 300, that bear little sequence homology can independently enhance the activity of the minimal activator domain (L. K. Venkatesh, C. Yang, P. A. Theodorakis, and G. Chinnadurai, J. Virol. 67:161-169, 1993). We now report on the further characterization of these two transcriptional enhancement regions. Mutational analysis of the region comprising residues 255 to 266 indicates that a cluster of leucine residues is critical to the function of this region. Also, residues 273 to 287, which are identical in sequence to a 15-amino-acid segment near the carboxy terminus of the simian foamy virus transcriptional activator Taf, can independently enhance the activity of the minimal activator region. To delineate the region(s) of Bel1 that could function autonomously as an activator domain, we tested the activity of chimeric proteins comprising either wild-type or functionally defective forms of Bel1 fused to the DNA binding domain, Gal4(1-147), of the yeast transcriptional activator Gal4 on a synthetic promoter comprising Gal4 DNA binding sites linked to the adenovirus E1B TATA box (minimal promoter). Gal4-Bel1 was found to activate basal transcription from the E1B TATA box at least 35-fold, and the region responsible for this activation function was localized to the carboxy-terminal 73 amino acids. When the transcriptional enhancement regions were tested for autonomous activator function as Gal4(1-147) chimeras, residues 272 to 300, but not 255 to 266, were found to activate transcription efficiently when targeted to the E1B TATA motif and also to HSRV and HIV-1 LTRs. The highly conserved region between amino acids 273 and 287 alone was found to activate transcription efficiently when targeted to the HSRV LTR but not to the E1B TATA box or the HIV-1 LTR. Thus, our results demonstrate that the carboxy-terminal 29-amino-acid region (residues 272 to 300) contributes to Bel1 transactivation by functioning as an autonomous activator of TATA motif-directed transcription in a manner similar to that of other modular transcriptional activators.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bothe K., Aguzzi A., Lassmann H., Rethwilm A., Horak I. Progressive encephalopathy and myopathy in transgenic mice expressing human foamy virus genes. Science. 1991 Aug 2;253(5019):555–557. doi: 10.1126/science.1650034. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990 May 24;345(6273):361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Human immunodeficiency virus as a prototypic complex retrovirus. J Virol. 1991 Mar;65(3):1053–1056. doi: 10.1128/jvi.65.3.1053-1056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügel R. M., Rethwilm A., Maurer B., Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987 Jul;6(7):2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Sun J. D., Garrett E. D., Cullen B. R. Functional organization of the Bel-1 trans activator of human foamy virus. J Virol. 1993 Apr;67(4):1896–1904. doi: 10.1128/jvi.67.4.1896-1904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J., Chinnadurai G. Synergistic activation of the human immunodeficiency virus type 1 promoter by the viral Tat protein and cellular transcription factor Sp1. J Virol. 1992 Jun;66(6):3932–3936. doi: 10.1128/jvi.66.6.3932-3936.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J., Subramanian T., Chinnadurai G. Sp1-dependent activation of a synthetic promoter by human immunodeficiency virus type 1 Tat protein. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8510–8514. doi: 10.1073/pnas.88.19.8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Garrett E. D., Cullen B. R. The Bel-1 protein of human foamy virus activates human immunodeficiency virus type 1 gene expression via a novel DNA target site. J Virol. 1992 Jun;66(6):3946–3949. doi: 10.1128/jvi.66.6.3946-3949.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Partin K. M., Löchelt M., Bannert H., Flügel R. M., Cullen B. R. Characterization of the transcriptional trans activator of human foamy retrovirus. J Virol. 1991 May;65(5):2589–2594. doi: 10.1128/jvi.65.5.2589-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagaye S., Vexiau P., Morozov V., Guénebaut-Claudet V., Tobaly-Tapiero J., Canivet M., Cathelineau G., Périès J., Emanoil-Ravier R. Human spumaretrovirus-related sequences in the DNA of leukocytes from patients with Graves disease. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10070–10074. doi: 10.1073/pnas.89.21.10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. H., Lee K. J., Kim S., Sung Y. C. Transactivation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by the human foamy virus bel1 protein requires a specific DNA sequence. J Virol. 1992 May;66(5):3236–3240. doi: 10.1128/jvi.66.5.3236-3240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Carey M., Ptashne M., Green M. R. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990 May 24;345(6273):359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- Löchelt M., Zentgraf H., Flügel R. M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991 Sep;184(1):43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Mergia A., Luciw P. A. Replication and regulation of primate foamy viruses. Virology. 1991 Oct;184(2):475–482. doi: 10.1016/0042-6822(91)90417-a. [DOI] [PubMed] [Google Scholar]
- Mergia A., Shaw K. E., Pratt-Lowe E., Barry P. A., Luciw P. A. Identification of the simian foamy virus transcriptional transactivator gene (taf). J Virol. 1991 Jun;65(6):2903–2909. doi: 10.1128/jvi.65.6.2903-2909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Muranyi W., Flügel R. M. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991 Feb;65(2):727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G. J., Rice S. A., Knipe D. M., Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988 Mar 11;239(4845):1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- Rethwilm A., Erlwein O., Baunach G., Maurer B., ter Meulen V. The transcriptional transactivator of human foamy virus maps to the bel 1 genomic region. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):941–945. doi: 10.1073/pnas.88.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh L. K., Theodorakis P. A., Chinnadurai G. Distinct cis-acting regions in U3 regulate trans-activation of the human spumaretrovirus long terminal repeat by the viral bel1 gene product. Nucleic Acids Res. 1991 Jul 11;19(13):3661–3666. doi: 10.1093/nar/19.13.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh L. K., Yang C., Theodorakis P. A., Chinnadurai G. Functional dissection of the human spumaretrovirus transactivator identifies distinct classes of dominant-negative mutants. J Virol. 1993 Jan;67(1):161–169. doi: 10.1128/jvi.67.1.161-169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. A. Foamy retroviruses. A virus in search of a disease. Nature. 1988 Jun 9;333(6173):497–498. doi: 10.1038/333497a0. [DOI] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]