Abstract
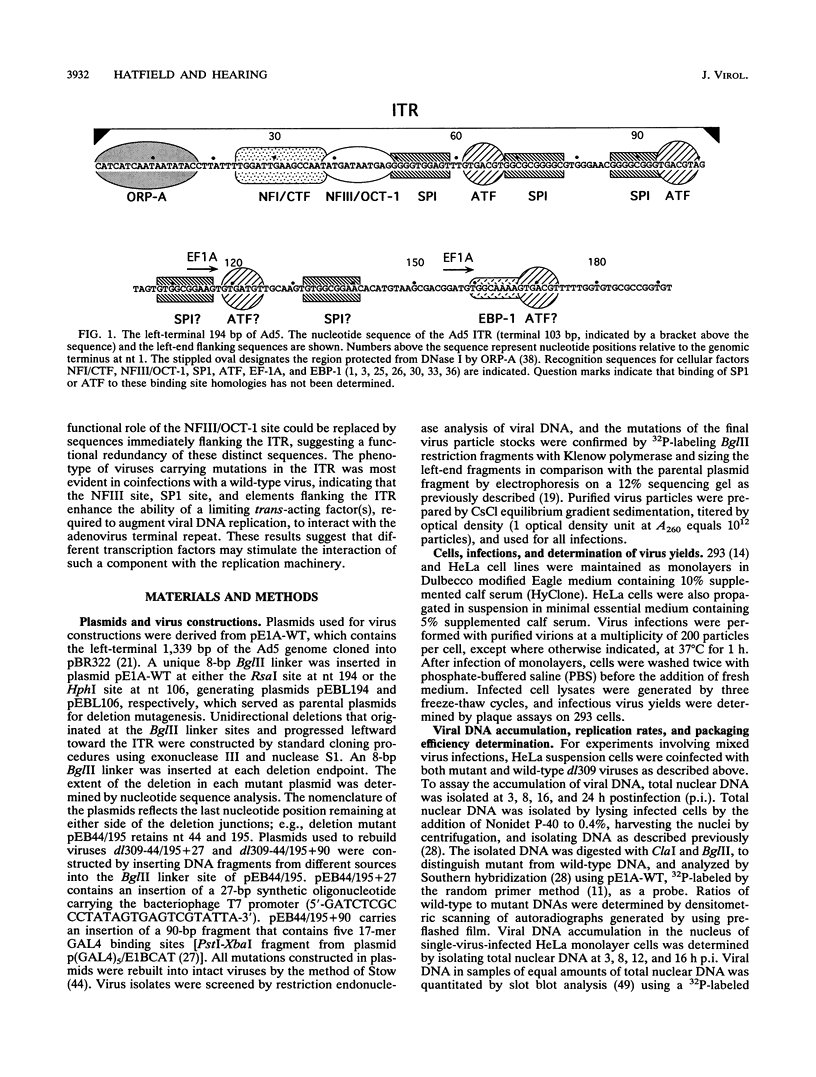
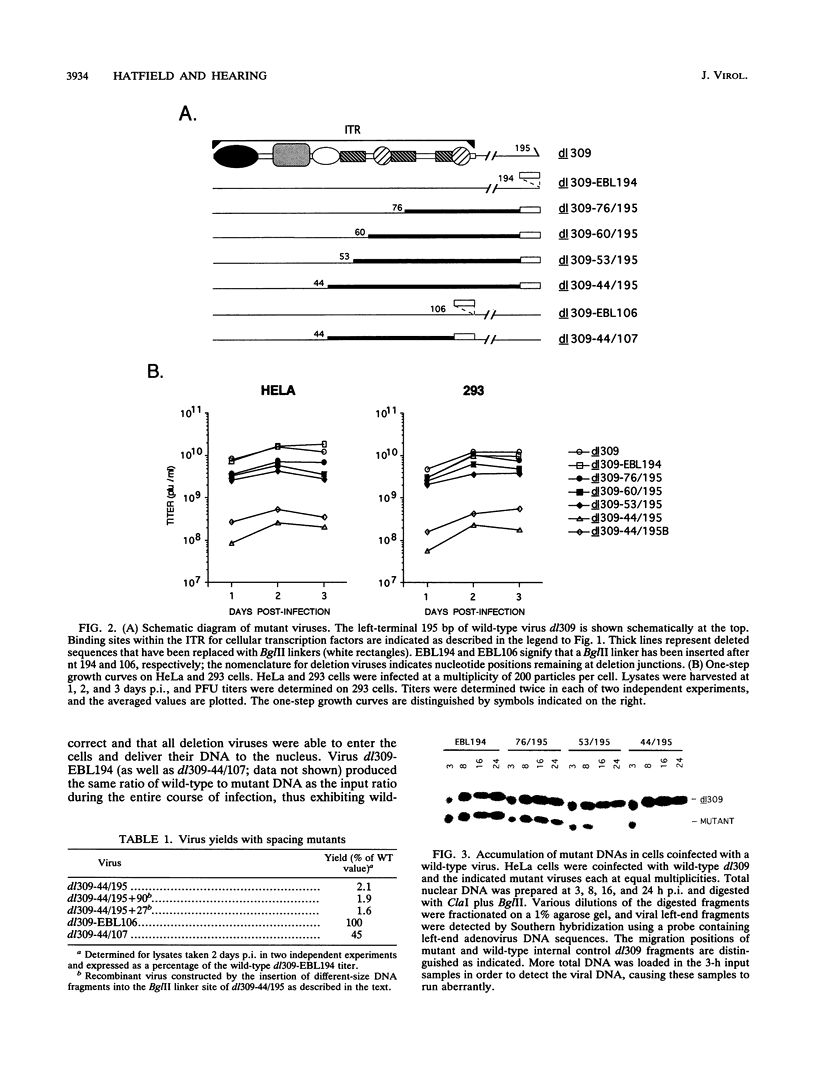
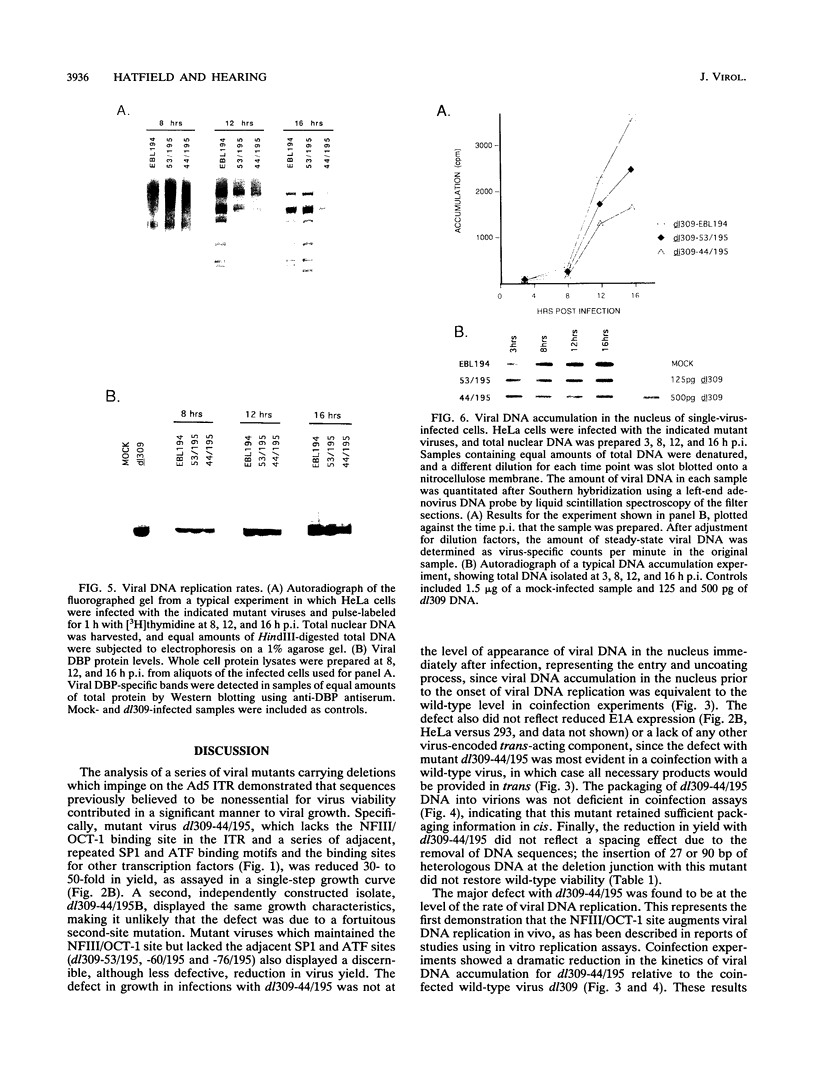
The inverted terminal repeat (ITR) of adenovirus type 5 (Ad5) is 103 bp in length and contains the origin of DNA replication. Cellular transcription factors NFI/CTF and NFIII/OCT-1 bind to sites within the ITR and participate in the initiation of viral DNA replication in vitro. The ITR also contains multiple copies of two conserved sequence motifs that bind the cellular transcription factors SP1 and ATF. We have analyzed a series of viruses that carry deletions at the left terminus of Ad5. A virus carrying a deletion of the NFIII/OCT-1, SP1, and ATF sites within the ITR (mutant dl309-44/107) was wild type for virus growth. However, the deletion of these elements in addition to sequences immediately flanking the ITR (mutant dl309-44/195) resulted in a virus that grew poorly. The analysis of growth parameters of these and other mutants demonstrate that the NFIII/OCT-1 and adjacent SP1 sites augment the accumulation of viral DNA following infection. The function of these elements was most evident in coinfections with a wild-type virus, suggesting that these sites enhance the ability of a limiting trans-acting factor(s), that stimulates viral DNA replication, to interact with the ITR. The results of these analyses indicate functional redundancy between different transcription elements at the left terminus of the Ad5 genome and demonstrate that the NFIII/OCT-1 site and adjacent SP1 site, previously thought to be nonessential for adenovirus growth, play a role in viral DNA replication in vivo.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett P., Clark L., Hay R. T. A cellular protein binds to a conserved sequence in the adenovirus type 2 enhancer. Nucleic Acids Res. 1987 Mar 25;15(6):2719–2735. doi: 10.1093/nar/15.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher J., Robinson E. C., Hay R. T. Interactions between the adenovirus type 2 DNA polymerase and the DNA binding domain of nuclear factor I. New Biol. 1990 Dec;2(12):1083–1090. [PubMed] [Google Scholar]
- Bruder J. T., Hearing P. Nuclear factor EF-1A binds to the adenovirus E1A core enhancer element and to other transcriptional control regions. Mol Cell Biol. 1989 Nov;9(11):5143–5153. doi: 10.1128/mcb.9.11.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Chen M., Mermod N., Horwitz M. S. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J Biol Chem. 1990 Oct 25;265(30):18634–18642. [PubMed] [Google Scholar]
- Cheng L., Kelly T. J. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell. 1989 Nov 3;59(3):541–551. doi: 10.1016/0092-8674(89)90037-8. [DOI] [PubMed] [Google Scholar]
- Cleat P. H., Hay R. T. Co-operative interactions between NFI and the adenovirus DNA binding protein at the adenovirus origin of replication. EMBO J. 1989 Jun;8(6):1841–1848. doi: 10.1002/j.1460-2075.1989.tb03579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenjaerts F. E., De Vries E., Pruijn G. J., Van Driel W., Bloemers S. M., Van der Lugt N. M., Van der Vliet P. C. Enhancement of DNA replication by transcription factors NFI and NFIII/Oct-1 depends critically on the positions of their binding sites in the adenovirus origin of replication. Biochim Biophys Acta. 1991 Aug 27;1090(1):61–69. doi: 10.1016/0167-4781(91)90037-m. [DOI] [PubMed] [Google Scholar]
- Cutt J. R., Shenk T., Hearing P. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J Virol. 1987 Feb;61(2):543–552. doi: 10.1128/jvi.61.2.543-552.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Gräble M., Hearing P. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J Virol. 1990 May;64(5):2047–2056. doi: 10.1128/jvi.64.5.2047-2056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräble M., Hearing P. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J Virol. 1992 Feb;66(2):723–731. doi: 10.1128/jvi.66.2.723-731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. X., Bailey S., Bodley J. W., Anderson D. Characterization of the small RNA of the bacteriophage phi 29 DNA packaging machine. Nucleic Acids Res. 1987 Sep 11;15(17):7081–7090. doi: 10.1093/nar/15.17.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. X., Erickson S., Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science. 1987 May 8;236(4802):690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Hatfield L., Hearing P. Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology. 1991 Sep;184(1):265–276. doi: 10.1016/0042-6822(91)90843-z. [DOI] [PubMed] [Google Scholar]
- Hay R. T., McDougall I. M. Viable viruses with deletions in the left inverted terminal repeat define the adenovirus origin of DNA replication. J Gen Virol. 1986 Feb;67(Pt 2):321–332. doi: 10.1099/0022-1317-67-2-321. [DOI] [PubMed] [Google Scholar]
- Hearing P., Samulski R. J., Wishart W. L., Shenk T. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J Virol. 1987 Aug;61(8):2555–2558. doi: 10.1128/jvi.61.8.2555-2558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell. 1986 Apr 25;45(2):229–236. doi: 10.1016/0092-8674(86)90387-9. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hen R., Borrelli E., Sassone-Corsi P., Chambon P. An enhancer element is located 340 base pairs upstream from the adenovirus-2 E1A capsite. Nucleic Acids Res. 1983 Dec 20;11(24):8747–8760. doi: 10.1093/nar/11.24.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. M., Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989 Jun;63(6):2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Leza M. A., Hearing P. Cellular transcription factor binds to adenovirus early region promoters and to a cyclic AMP response element. J Virol. 1988 Aug;62(8):3003–3013. doi: 10.1128/jvi.62.8.3003-3013.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Mensa-Wilmot K., Carroll K., McMacken R. Transcriptional activation of bacteriophage lambda DNA replication in vitro: regulatory role of histone-like protein HU of Escherichia coli. EMBO J. 1989 Aug;8(8):2393–2402. doi: 10.1002/j.1460-2075.1989.tb08369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles V. J., Cortes P., Stone N., Reinberg D. The adenovirus inverted terminal repeat functions as an enhancer in a cell-free system. J Biol Chem. 1989 Jun 25;264(18):10763–10772. [PubMed] [Google Scholar]
- Mul Y. M., Van der Vliet P. C. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 1992 Feb;11(2):751–760. doi: 10.1002/j.1460-2075.1992.tb05108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul Y. M., Verrijzer C. P., van der Vliet P. C. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol. 1990 Nov;64(11):5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. A., Fletcher C., Burrow C. R., Heintz N., Roeder R. G., Kelly T. J. Transcription factor OTF-1 is functionally identical to the DNA replication factor NF-III. Science. 1988 Sep 2;241(4870):1210–1213. doi: 10.1126/science.3413485. [DOI] [PubMed] [Google Scholar]
- Ooyama S., Imai T., Hanaka S., Handa H. Transcription in the reverse orientation at either terminus of the adenovirus type 5 genome. EMBO J. 1989 Mar;8(3):863–868. doi: 10.1002/j.1460-2075.1989.tb03447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne T. F., Berk A. J. Far upstream initiation sites for adenovirus early region 1A transcription are utilized after the onset of viral DNA replication. J Virol. 1983 Feb;45(2):594–599. doi: 10.1128/jvi.45.2.594-599.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn J. M., van der Vliet P. C., Dathan N. A., Mattaj I. W. Anti-OTF-1 antibodies inhibit NFIII stimulation of in vitro adenovirus DNA replication. Nucleic Acids Res. 1989 Mar 11;17(5):1845–1863. doi: 10.1093/nar/17.5.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. C., Sarnow P., Duprey E., Levine A. J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983 Jul 30;128(2):480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P. J., O'Neill E. A., Wides R. J., Kelly T. J. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol Cell Biol. 1987 Feb;7(2):875–886. doi: 10.1128/mcb.7.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Hen R., Borrelli E., Leff T., Chambon P. Far upstream sequences are required for efficient transcription from the adenovirus-2 E1A transcription unit. Nucleic Acids Res. 1983 Dec 20;11(24):8735–8745. doi: 10.1093/nar/11.24.8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa M., Padmanabhan R. Comparative sequence analysis of the inverted terminal repetitions from different adenoviruses. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3831–3835. doi: 10.1073/pnas.77.7.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Baker T. A., Kornberg A. Strand separation required for initiation of replication at the chromosomal origin of E.coli is facilitated by a distant RNA--DNA hybrid. EMBO J. 1990 Jul;9(7):2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. W., Topp W. C., Engler J. A. Conserved sequences at the origin of adenovirus DNA replication. J Virol. 1982 Nov;44(2):530–537. doi: 10.1128/jvi.44.2.530-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Initiation of eukaryotic DNA replication in vitro. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J Virol. 1981 Jan;37(1):171–180. doi: 10.1128/jvi.37.1.171-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuiver M. H., van der Vliet P. C. Adenovirus DNA-binding protein forms a multimeric protein complex with double-stranded DNA and enhances binding of nuclear factor I. J Virol. 1990 Jan;64(1):379–386. doi: 10.1128/jvi.64.1.379-386.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley S. M., Hay R. T. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J. 1992 Feb;11(2):761–768. doi: 10.1002/j.1460-2075.1992.tb05109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer C. P., van Oosterhout J. A., van Weperen W. W., van der Vliet P. C. POU proteins bend DNA via the POU-specific domain. EMBO J. 1991 Oct;10(10):3007–3014. doi: 10.1002/j.1460-2075.1991.tb07851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg D. H., Ketner G. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J Virol. 1986 Mar;57(3):833–838. doi: 10.1128/jvi.57.3.833-838.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. DNA replication, the bacterial cell cycle, and cell growth. Cell. 1992 Apr 3;69(1):5–8. doi: 10.1016/0092-8674(92)90112-p. [DOI] [PubMed] [Google Scholar]