Abstract
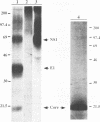
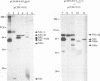
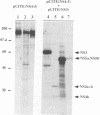
Hepatitis C virus (HCV) possesses a positive-sense RNA genome which encodes a large polyprotein of 3,010 amino acids. Previous data and sequence analysis have indicated that this polyprotein is processed by cellular proteases and possibly by a virally encoded serine protease localized in the N-terminal domain of nonstructural protein NS3. To characterize the molecular aspects of HCV protein biogenesis and to clearly identify the protein products derived from the HCV genome, we have examined HCV polyprotein expression by using the vaccinia virus T7 transient expression system in transfected cells and by cell-free translation studies. HCV proteins were identified by immunoprecipitation with region-specific antisera. Here we show that the amino-terminal region of the HCV polyprotein is processed in vitro by cellular proteases releasing three structural proteins: p21 (core), gp37 (E1), and gp61 (E2). Processing of the nonstructural region of HCV was evident in transfected cells. Two proteins of 24 and 68 kDa were immunoprecipitated with anti-NS2 and NS3 antisera, respectively. Antiserum against NS4 recognized three proteins of 6, 26, and 31 kDa, while antisera specific for NS5 immunoprecipitated two polypeptides of 56 and 65 kDa, indicating that each of these two genes encodes at least two different proteins. When the NS3 protease domain was inactivated by replacing the proposed catalytic Ser-1165 with Ala, processing at several sites was abolished. When Ser-1164 was mutated to Ala, no effect on the processing was observed. Cleavage activities at three of the four sites affected by NS3 were shown to occur in trans, while processing at the carboxy terminus of NS3 could not be mediated in trans. These results provide a detailed description of the protein products obtained from the processing of the HCV polyprotein. Furthermore, the data obtained implicate NS3 as a serine protease and demonstrate that a catalytically active NS3 is necessary for cleavage of the nonstructural region of HCV.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cahour A., Falgout B., Lai C. J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992 Mar;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990 Jul;177(1):159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S., Anderson D. K., Retzel E. Comparisons of the pestivirus bovine viral diarrhoea virus with members of the flaviviridae. J Gen Virol. 1988 Oct;69(Pt 10):2637–2643. doi: 10.1099/0022-1317-69-10-2637. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Moennig V., Horzinek M. C. Recent advances in pestivirus research. J Gen Virol. 1989 Feb;70(Pt 2):253–266. doi: 10.1099/0022-1317-70-2-253. [DOI] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C. J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989 May;63(5):1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Watanabe Y., Takeuchi K., Suzuki T., Katayama T., Takebe Y., Saito I., Miyamura T. Expression of processed core protein of hepatitis C virus in mammalian cells. J Virol. 1991 Jun;65(6):3015–3021. doi: 10.1128/jvi.65.6.3015-3021.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Lai C. J. Cleavage of dengue virus NS1-NS2A requires an octapeptide sequence at the C terminus of NS1. J Virol. 1990 Sep;64(9):4573–4577. doi: 10.1128/jvi.64.9.4573-4577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara M., Tsukiyama-Kohara K., Maki N., Asano K., Yamaguchi K., Miki K., Tanaka S., Hattori N., Matsuura Y., Saito I. Expression and characterization of glycoprotein gp35 of hepatitis C virus using recombinant vaccinia virus. J Gen Virol. 1992 Sep;73(Pt 9):2313–2318. doi: 10.1099/0022-1317-73-9-2313. [DOI] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Mason P. W. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989 Apr;169(2):354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Shin S. U., Morrison S. L. Production and properties of chimeric antibody molecules. Methods Enzymol. 1989;178:459–476. doi: 10.1016/0076-6879(89)78034-4. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Soe L. H., Shieh C. K., Baker S. C., Chang M. F., Lai M. M. Sequence and translation of the murine coronavirus 5'-end genomic RNA reveals the N-terminal structure of the putative RNA polymerase. J Virol. 1987 Dec;61(12):3968–3976. doi: 10.1128/jvi.61.12.3968-3976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler K. R., Rosser D. S., Berk A. J. Analysis of adenovirus transforming proteins from early regions 1A and 1B with antisera to inducible fusion antigens produced in Escherichia coli. J Virol. 1984 Jan;49(1):132–141. doi: 10.1128/jvi.49.1.132-141.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Kubo Y., Boonmar S., Watanabe Y., Katayama T., Choo Q. L., Kuo G., Houghton M., Saito I., Miyamura T. The putative nucleocapsid and envelope protein genes of hepatitis C virus determined by comparison of the nucleotide sequences of two isolates derived from an experimentally infected chimpanzee and healthy human carriers. J Gen Virol. 1990 Dec;71(Pt 12):3027–3033. doi: 10.1099/0022-1317-71-12-3027. [DOI] [PubMed] [Google Scholar]
- Weiland E., Stark R., Haas B., Rümenapf T., Meyers G., Thiel H. J. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J Virol. 1990 Aug;64(8):3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskerchen M., Collett M. S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991 Sep;184(1):341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- Zhang L., Mohan P. M., Padmanabhan R. Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992 Dec;66(12):7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]