Abstract
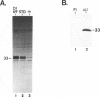
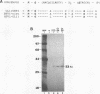
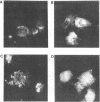
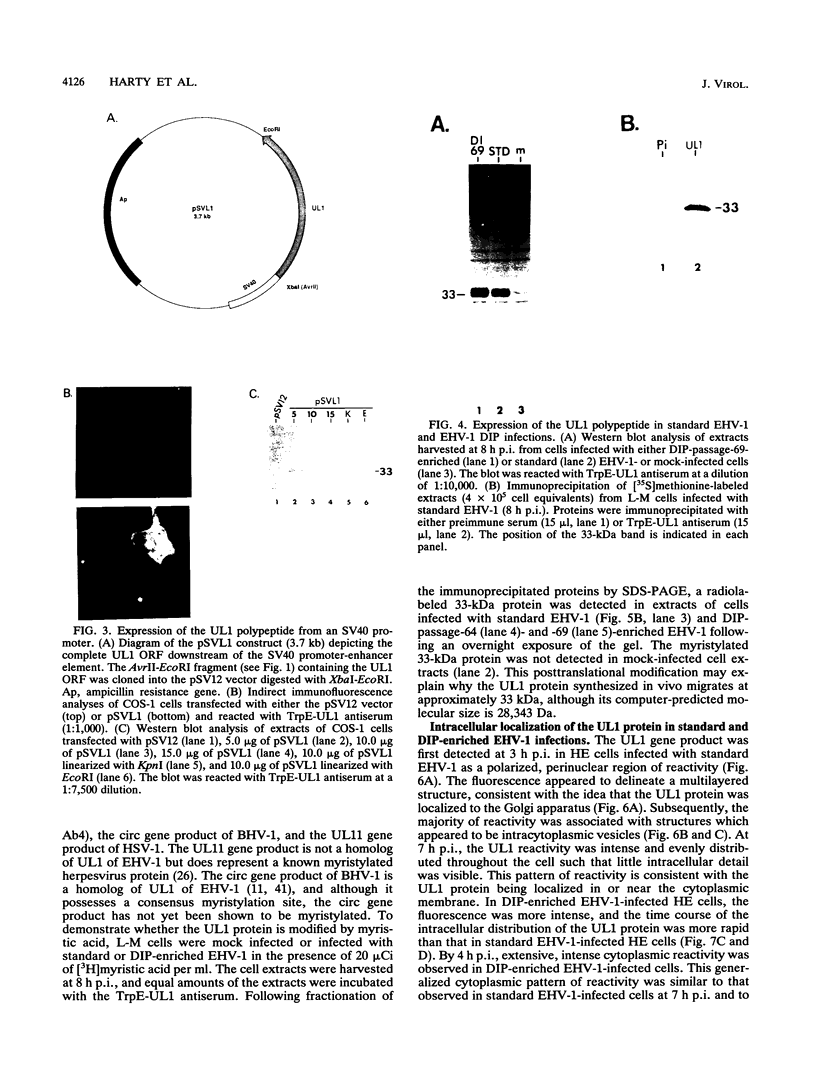
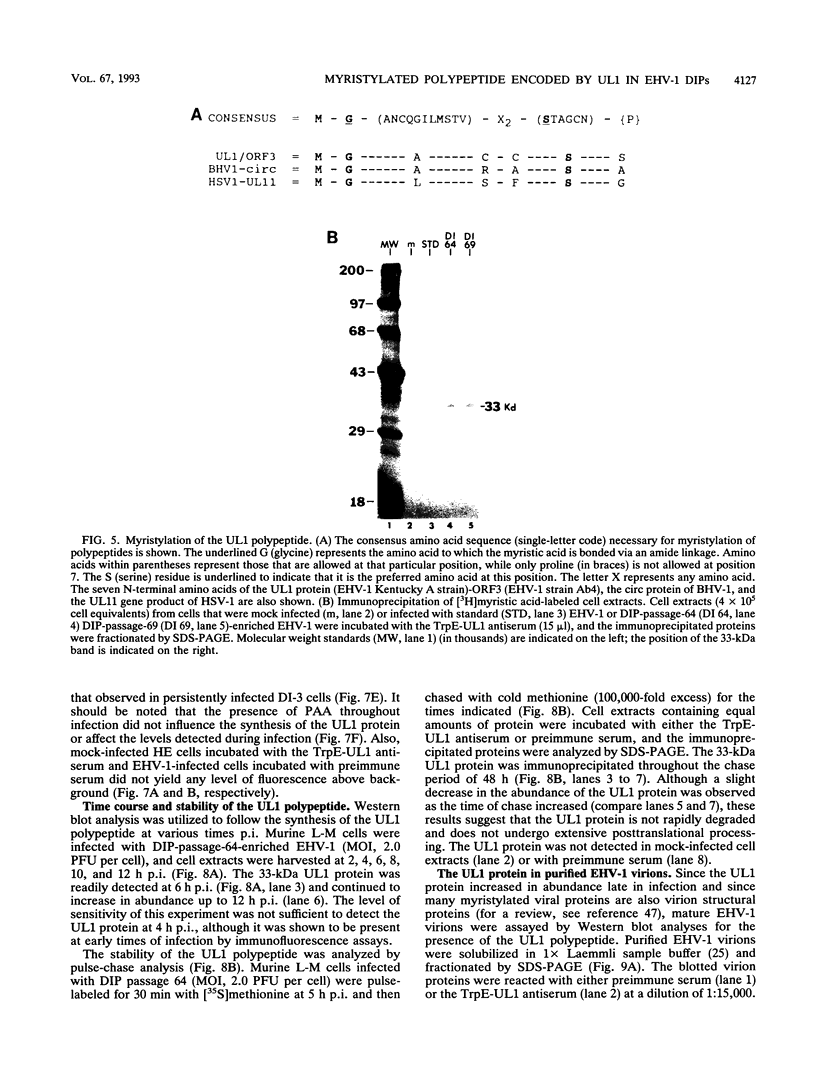
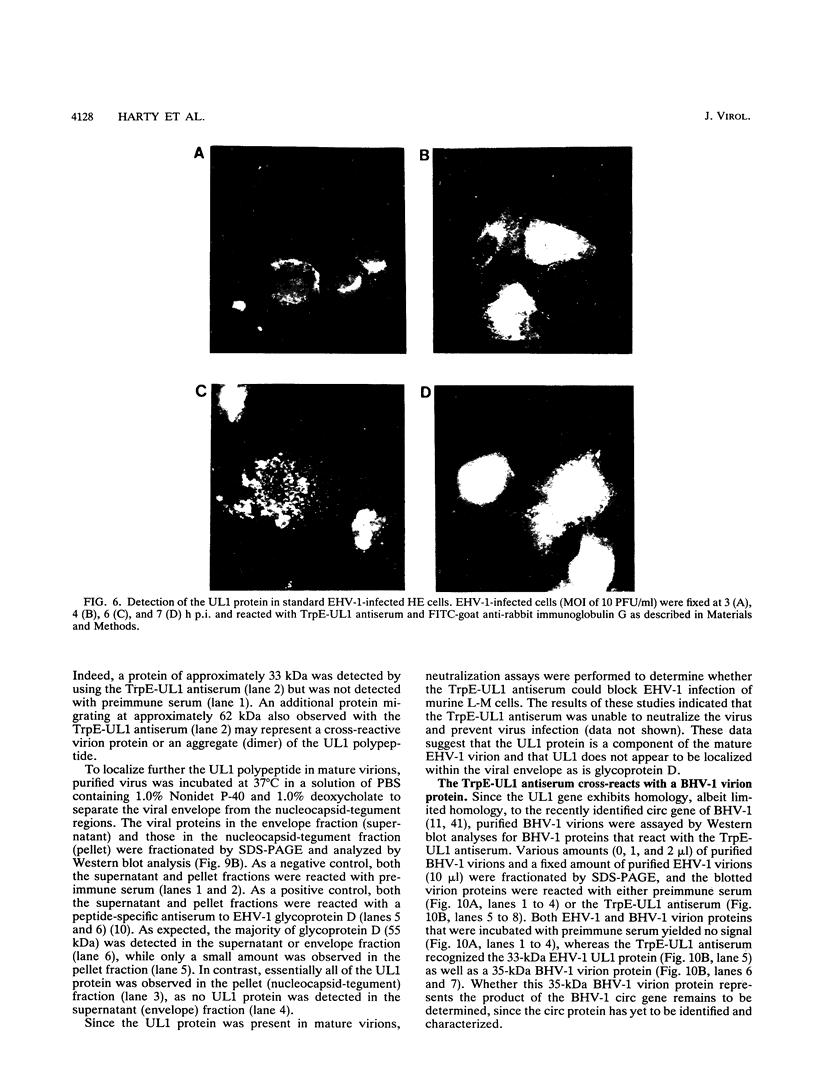
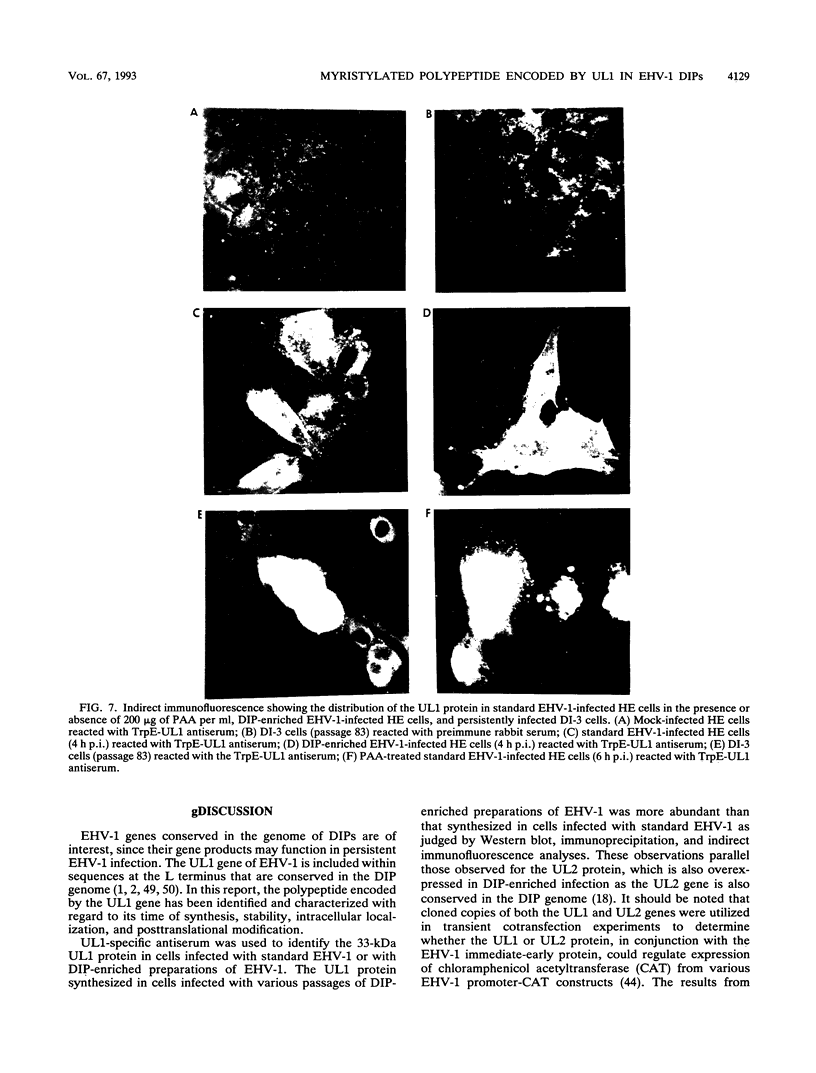
Equine herpesvirus 1 (EHV-1, Kentucky A strain) preparations enriched for defective interfering particles (DIPs) can readily establish persistent infection. The UL1 gene, which is conserved in the genome of DIPs that mediate persistent infection, maps between nucleotides 1418 and 2192 (258 amino acids) from the L (long) terminus. UL1 has no homology with any known gene encoded by herpes simplex virus type 1 but has limited homology to open reading frame 2 of varicella-zoster virus and the "circ" gene of bovine herpesvirus type 1. Previous work showed that the EHV-1 UL1 gene belongs to the early kinetic class and is transcribed as a 1.2-kb polyadenylated mRNA (R. N. Harty, R. R. Yalamanchili, and D. J. O'Callaghan, Virology 183:830-833, 1991). In this report, the UL1 protein was identified and characterized as a 33-kDa polypeptide in EHV-1-infected cells by using rabbit polyclonal antiserum raised against a TrpE-UL1 fusion protein (amino acids 7 to 258 of UL1) synthesized in Escherichia coli. Results from Western blot (immunoblot), immunoprecipitation, indirect immunofluorescence, and biochemical analyses indicated that the UL1 polypeptide (i) is more abundant in cells infected with DIP-enriched virus than in cells infected with standard EHV-1, (ii) is synthesized as early as 3 h postinfection (p.i.) in infection with standard virus or in infection with DIP-enriched virus preparations and increases in abundance up to 12 h p.i., (iii) appears to be associated with the rough endoplasmic reticulum-Golgi apparatus early in infection (3 to 4 h p.i.), while a diffuse cytoplasmic pattern of fluorescence is observed late in infection (7 to 8 h p.i.), (iv) is modified by myristic acid as it contains a consensus N-terminal myristylation site and is readily labeled with [3H]myristic acid, and (v) is associated with mature EHV-1 virions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumann R. P., Dauenhauer S. A., Caughman G. B., Staczek J., O'Callaghan D. J. Structure and genetic complexity of the genomes of herpesvirus defective-interfering particles associated with oncogenic transformation and persistent infection. J Virol. 1984 Apr;50(1):13–21. doi: 10.1128/jvi.50.1.13-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann R. P., Staczek J., O'Callaghan D. J. Cloning and fine mapping the DNA of equine herpesvirus type one defective interfering particles. Virology. 1986 Sep;153(2):188–200. doi: 10.1016/0042-6822(86)90022-x. [DOI] [PubMed] [Google Scholar]
- Buss J. E., Der C. J., Solski P. A. The six amino-terminal amino acids of p60src are sufficient to cause myristylation of p21v-ras. Mol Cell Biol. 1988 Sep;8(9):3960–3963. doi: 10.1128/mcb.8.9.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughman G. B., Robertson A. T., Gray W. L., Sullivan D. C., O'Callaghan D. J. Characterization of equine herpesvirus type 1 immediate early proteins. Virology. 1988 Apr;163(2):563–571. doi: 10.1016/0042-6822(88)90297-8. [DOI] [PubMed] [Google Scholar]
- Caughman G. B., Staczek J., O'Callaghan D. J. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate early/early/late regulation of viral gene expression. Virology. 1985 Aug;145(1):49–61. doi: 10.1016/0042-6822(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Chow M., Newman J. F., Filman D., Hogle J. M., Rowlands D. J., Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987 Jun 11;327(6122):482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- Daley G. Q., Van Etten R. A., Jackson P. K., Bernards A., Baltimore D. Nonmyristoylated Abl proteins transform a factor-dependent hematopoietic cell line. Mol Cell Biol. 1992 Apr;12(4):1864–1871. doi: 10.1128/mcb.12.4.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauenhauer S. A., Robinson R. A., O'Callaghan D. J. Chronic production of defective-interfering particles by hamster embryo cultures of herpesvirus persistently infected and oncogenically transformed cells. J Gen Virol. 1982 May;60(Pt 1):1–14. doi: 10.1099/0022-1317-60-1-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., McGeoch D. J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986 Apr;67(Pt 4):597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- Flowers C. C., O'Callaghan D. J. Equine herpesvirus 1 glycoprotein D: mapping of the transcript and a neutralization epitope. J Virol. 1992 Nov;66(11):6451–6460. doi: 10.1128/jvi.66.11.6451-6460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego C., Gupta S. K., Winitz S., Eisfelder B. J., Johnson G. L. Myristoylation of the G alpha i2 polypeptide, a G protein alpha subunit, is required for its signaling and transformation functions. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9695–9699. doi: 10.1073/pnas.89.20.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. L., Baumann R. P., Robertson A. T., Caughman G. B., O'Callaghan D. J., Staczek J. Regulation of equine herpesvirus type 1 gene expression: characterization of immediate early, early, and late transcription. Virology. 1987 May;158(1):79–87. doi: 10.1016/0042-6822(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Gray W. L., Baumann R. P., Robertson A. T., O'Callaghan D. J., Staczek J. Characterization and mapping of equine herpesvirus type 1 immediate early, early, and late transcripts. Virus Res. 1987 Sep;8(3):233–244. doi: 10.1016/0168-1702(87)90018-9. [DOI] [PubMed] [Google Scholar]
- Harty R. N., Holden V. R., O'Callaghan D. J. Transcriptional and translational analyses of the UL2 gene of equine herpesvirus 1: a homolog of UL55 of herpes simplex virus type 1 that is maintained in the genome of defective interfering particles. J Virol. 1993 Apr;67(4):2255–2265. doi: 10.1128/jvi.67.4.2255-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
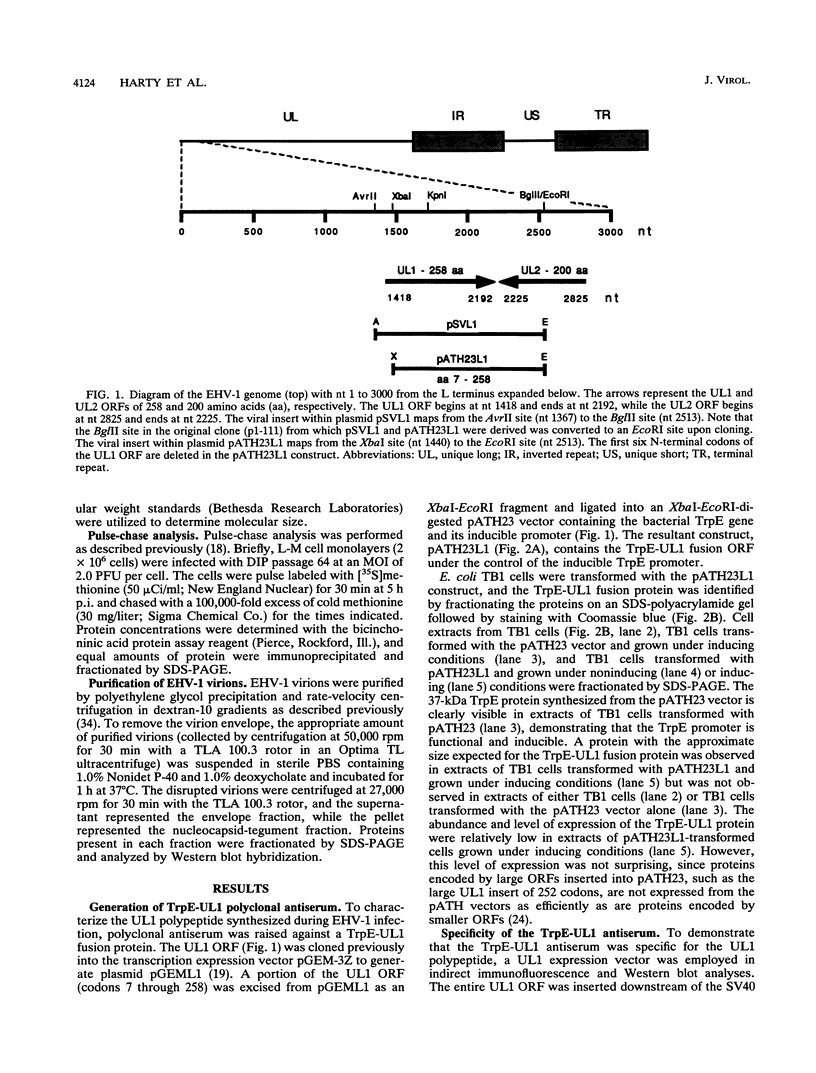
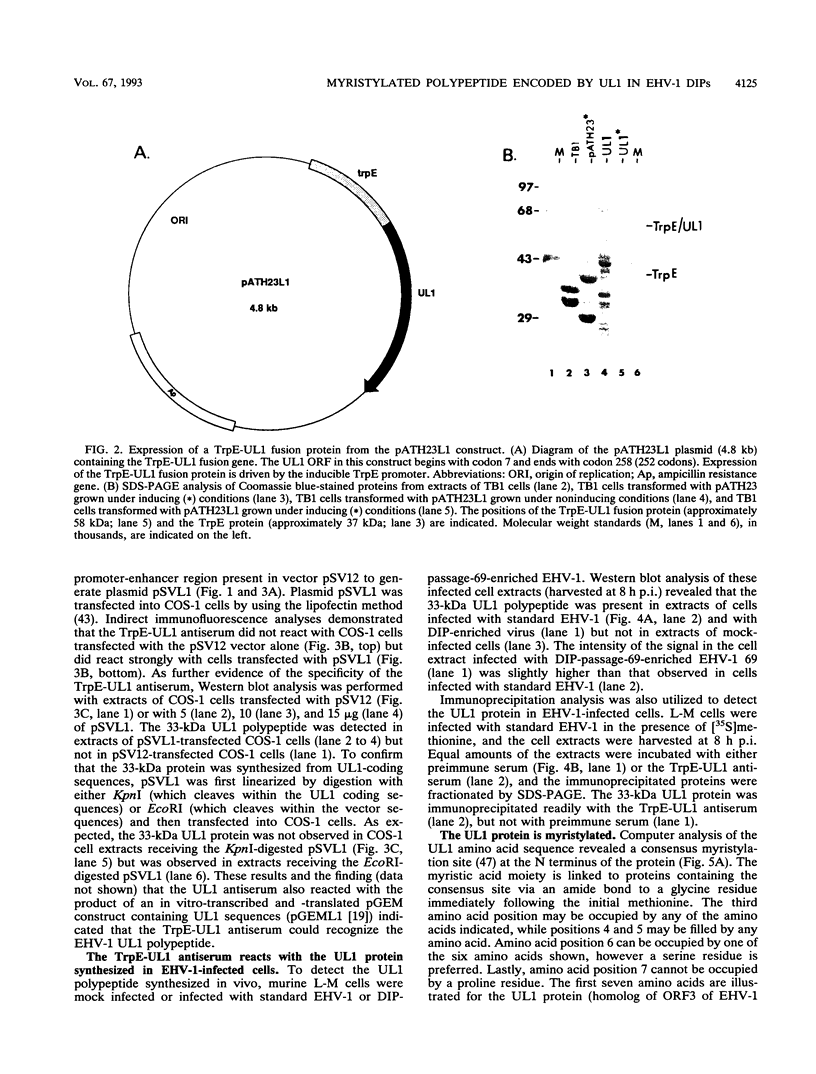
- Harty R. N., O'Callaghan D. J. Identification and expression of the UL1 gene product of equine herpesvirus 1. Virus Res. 1992 Sep 1;25(1-2):105–116. doi: 10.1016/0168-1702(92)90103-g. [DOI] [PubMed] [Google Scholar]
- Harty R. N., Yalamanchili R. R., O'Callaghan D. J. Transcriptional analysis of the UL1 gene of equine herpesvirus 1: a gene conserved in the genome of defective interfering particles. Virology. 1991 Aug;183(2):830–833. doi: 10.1016/0042-6822(91)91020-h. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology. 1979 Jan 30;92(2):495–506. doi: 10.1016/0042-6822(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Holden V. R., Yalamanchili R. R., Harty R. N., O'Callaghan D. J. ICP22 homolog of equine herpesvirus 1: expression from early and late promoters. J Virol. 1992 Feb;66(2):664–673. doi: 10.1128/jvi.66.2.664-673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner T. J., Hill J. E., Myers A. M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MacLean C. A., Clark B., McGeoch D. J. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J Gen Virol. 1989 Dec;70(Pt 12):3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- McNabb D. S., Courtney R. J. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J Virol. 1992 May;66(5):2653–2663. doi: 10.1128/jvi.66.5.2653-2663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscufo N., Chow M. Myristate-protein interactions in poliovirus: interactions of VP4 threonine 28 contribute to the structural conformation of assembly intermediates and the stability of assembled virions. J Virol. 1992 Dec;66(12):6849–6857. doi: 10.1128/jvi.66.12.6849-6857.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan D. J., Hyde J. M., Gentry G. A., Randall C. C. Kinetics of viral deoxyribonucleic acid, protein, and infectious particle production and alterations in host macromolecular syntheses in equine abortion (herpes) virus-infected cells. J Virol. 1968 Aug;2(8):793–804. doi: 10.1128/jvi.2.8.793-804.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue M. L., Kemp M. C., Randall C. C., O'Callaghan D. J. Studies of the molecular anatomy of the L-M cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology. 1974 May;59(1):201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- Persing D. H., Varmus H. E., Ganem D. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J Virol. 1987 May;61(5):1672–1677. doi: 10.1128/jvi.61.5.1672-1677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Resh M. D. Specific and saturable binding of pp60v-src to plasma membranes: evidence for a myristyl-src receptor. Cell. 1989 Jul 28;58(2):281–286. doi: 10.1016/0092-8674(89)90842-8. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Tucker P. W., Dauenhauer S. A., O'Callaghan D. J. Molecular cloning of equine herpesvirus type 1 DNA: analysis of standard and defective viral genomes and viral sequences in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6684–6688. doi: 10.1073/pnas.78.11.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Dauenhauer S. A., O'Callaghan D. J. Electron microscopic study of equine herpesvirus type 1 DNA. J Virol. 1982 Apr;42(1):297–300. doi: 10.1128/jvi.42.1.297-300.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A. M., Henderson L. E., Oroszlan S., Garber E. A., Hanafusa H. Amino terminal myristylation of the protein kinase p60src, a retroviral transforming protein. Science. 1985 Jan 25;227(4685):427–429. doi: 10.1126/science.3917576. [DOI] [PubMed] [Google Scholar]
- Sefton B. M., Trowbridge I. S., Cooper J. A., Scolnick E. M. The transforming proteins of Rous sarcoma virus, Harvey sarcoma virus and Abelson virus contain tightly bound lipid. Cell. 1982 Dec;31(2 Pt 1):465–474. doi: 10.1016/0092-8674(82)90139-8. [DOI] [PubMed] [Google Scholar]
- Smith R. H., Caughman G. B., O'Callaghan D. J. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J Virol. 1992 Feb;66(2):936–945. doi: 10.1128/jvi.66.2.936-945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli C. H., Griffin B. E. Myristic acid is coupled to a structural protein of polyoma virus and SV40. Nature. 1987 Apr 9;326(6113):619–622. doi: 10.1038/326619a0. [DOI] [PubMed] [Google Scholar]
- Telford E. A., Watson M. S., McBride K., Davison A. J. The DNA sequence of equine herpesvirus-1. Virology. 1992 Jul;189(1):304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- Towler D. A., Gordon J. I., Adams S. P., Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- Whalley J. M., Robertson G. R., Davison A. J. Analysis of the genome of equine herpesvirus type 1: arrangement of cleavage sites for restriction endonucleases EcoRI, BglII and BamHI. J Gen Virol. 1981 Dec;57(Pt 2):307–323. doi: 10.1099/0022-1317-57-2-307. [DOI] [PubMed] [Google Scholar]
- Yalamanchili R. R., O'Callaghan D. J. Organization and function of the ORIs sequence in the genome of EHV-1 DI particles. Virology. 1990 Dec;179(2):867–870. doi: 10.1016/0042-6822(90)90157-m. [DOI] [PubMed] [Google Scholar]
- Yalamanchili R. R., O'Callaghan D. J. Sequence and organization of the genomic termini of equine herpesvirus type 1. Virus Res. 1990 Feb;15(2):149–161. doi: 10.1016/0168-1702(90)90005-v. [DOI] [PubMed] [Google Scholar]
- Yalamanchili R. R., Raengsakulrach B., Baumann R. P., O'Callaghan D. J. Identification of the site of recombination in the generation of the genome of DI particles of equine herpesvirus type 1. Virology. 1990 Apr;175(2):448–455. doi: 10.1016/0042-6822(90)90429-u. [DOI] [PubMed] [Google Scholar]
- Yalamanchili R. R., Raengsakulrach B., O'Callaghan D. J. Equine herpesvirus 1 sequence near the left terminus codes for two open reading frames. Virus Res. 1991 Mar;18(2-3):109–116. doi: 10.1016/0168-1702(91)90012-k. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Holden V. R., Harty R. N., O'Callaghan D. J. Identification and transcriptional analyses of the UL3 and UL4 genes of equine herpesvirus 1, homologs of the ICP27 and glycoprotein K genes of herpes simplex virus. J Virol. 1992 Sep;66(9):5363–5372. doi: 10.1128/jvi.66.9.5363-5372.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]