Abstract
Mutagenic abasic (AP) sites are generated directly by DNA-damaging agents or by DNA glycosylases acting in base excision repair. AP sites are corrected via incision by AP endonucleases, removal of deoxyribose 5-phosphate, repair synthesis, and ligation. Mammalian DNA polymerase β (Polβ) carries out most base excision repair synthesis and also can excise deoxyribose 5-phosphate after AP endonuclease incision. Yeast two-hybrid analysis now indicates protein–protein contact between Polβ and human AP endonuclease (Ape protein). In vitro, binding of Ape protein to uncleaved AP sites loads Polβ into a ternary complex with Ape and the AP-DNA. After incision by Ape, only Polβ exhibits stable DNA binding. Kinetic experiments indicated that Ape accelerates the excision of 5′-terminal deoxyribose 5-phosphate by Polβ. Thus, the two central players of the base excision repair pathway are coordinated in sequential reactions.
DNA is subject to spontaneous hydrolytic degradation and assault by metabolic byproducts, such as oxygen radicals and alkylating agents, as well as numerous environmental agents (1, 2). Genetic instability can result from this damage and is counteracted by various pathways of DNA repair. Nucleotide excision repair acts on a wide variety of DNA lesions and is mediated in mammalian cells by some 25 proteins acting in an orchestrated fashion (3, 4). Base excision repair (5) handles a more limited set of altered bases through the action of DNA glycosylases, such as the enzyme that releases uracil from DNA (6, 7). DNA glycosylases produce AP sites, which also arise from spontaneous loss of normal bases, destabilization by base damage, or base elimination by free radical attack (1, 2). Abasic (AP) sites threaten genetic stability because they block replication and are mutagenic (8).
Both direct and glycosylase-generated AP sites are substrates for AP endonucleases, which initiate repair by incising immediately 5′ to the AP site (5). The major AP endonuclease of human cells, Ape protein (also called Ref1, Hap1, or Apex), is a member of a large family of nucleases related to exonuclease III of Escherichia coli (5, 9). Ape protein has broad specificity for AP sites (10) and is abundantly present in the nucleus of human cells (11, 12). The next step of base excision repair is the removal of 5′-terminal deoxyribose 5-phosphate (dRp), the product of Ape, followed by DNA repair synthesis to fill in the small gap, and ligation to complete the repair (1). In mammalian cells, most of the repair synthesis is carried out by DNA polymerase β (Polβ) (13, 14). A distinct dRp excision activity has been recovered in small amounts from calf thymus (15). However, a recent report demonstrated that Xenopus DNA Polβ has an intrinsic activity that releases 5′-dRp by β-elimination (16); this mechanism has been confirmed for human Polβ (17). This finding suggested that one protein can catalyze consecutive steps (dRp removal and repair synthesis) in the base excision pathway. Both the nucleotide excision (3, 4) and DNA mismatch repair (1) pathways act by coordinating protein activities. We wished to determine whether base excision repair also might proceed in an orchestrated fashion mediated by specific protein–protein interactions.
MATERIALS AND METHODS
Two-Hybrid Methods.
The APE cDNA (12) was amplified by PCR (18) and inserted in-frame into the vector pAS1 (19) to generate pAS1–Ape, which encodes a Gal4DB–Ape fusion protein. Similarly, the Polβ cDNA (14) was amplified and inserted in-frame in the vector pACT2 to generate pACT-Polβ, encoding a Gal4AD–Polβ fusion protein. The structures of the fusion constructs were confirmed by DNA sequencing. Plasmid pAS1–Ape was transformed into yeast strain Y190 (MATa gal4 gal80 his3 trp1–901 ade2–101 ura3–52 leu2, 3–112 URA3::GAL-lacZ LYS2::GAL(UAS)-HIS3 cyhr) (20). Plasmids pACT-Polβ or pSE1111 then were introduced and the cells plated on synthetic medium lacking tryptophan, leucine, and histidine, and containing 25 mM 3-amino-1,2,4-triazole (19). After incubation at 30°C for 5 days, colonies were transferred to a nitrocellulose membrane by blotting and stained for β-galactosidase activity (19). For cell extracts to assay β-galactosidase activity, 20-ml cultures were grown ≥3 days in selective medium as above. The cells were collected, lysed by shaking with glass beads (500 μm diameter), and the extracts assayed using the fluorogenic substrate 4-methylumbelliferyl β-d-galactoside (21). The positive control was a yeast strain carrying the E. coli lacZ gene on a plasmid (R. Brennan and R. H. Schiestl, personal communication).
Electrophoretic Mobility-Shift Assay (EMSA).
Oligonucleotides containing either a uracil or a tetrahydrofuran residue (23-F) were 32P-5′-end labeled and annealed to unlabeled complementary DNA as previously described (22). AP site DNA (18-AP and 51-AP) was generated by incubating uracil-containing substrates with 1 unit of uracil DNA glycosylase (GIBCO/BRL) for 5 min at 37°C. For a typical binding reaction, 0.05 pmol (5 nM) of duplex DNA substrate was mixed (where indicated) with Ape protein purified from HeLa cells (11) (20 ng, ≈55 nM) or human Polβ (23) (20 ng, ≈55 nM) in 50 mM Hepes⋅KOH, pH 7.5, 100 mM KCl, 10% glycerol, and either 10 mM MgCl2 (Mg) or 4 mM EDTA (EDTA). The binding reaction (final volume 10 μl) was incubated at 0°C for 5 min and resolved on a 8% nondenaturing polyacrylamide gel (22) followed by autoradiography. In “supershift” experiments, the proteins and DNA were incubated as described above, and the appropriate rabbit polyclonal antiserum, generated against purified Ape from HeLa cells (11, 12) or recombinant human Polβ (S. Linn, personal communication), was then added, and the incubation continued for 20 min at 0°C. The mixtures then were resolved on nondenaturing gels as detailed above.
dRp Substrate and Excision Assay.
An oligonucleotide containing a dUMP residue at position 22 was 3′-labeled with dideoxy-[α-32P]ATP using terminal deoxynucleotidyltransferase and annealed to the complementary oligonucleotide (13). For each experiment, the double-stranded substrate (typically 10 pmol in 30 μl) was treated at 37°C for 30 min with 60 units of purified E. coli uracil-DNA glycosylase (kindly provided by D. Mousbaugh, Oregon State University) to create an AP site at position 22 in the uracil-containing strand (13). For each dRp excision reaction, 0.5 pmol of the newly formed AP site-containing substrate was cleaved to completion (≥95%) by incubation for 10 min at 37°C in an 8-μl reaction with 6 ng of purified endonuclease IV (24) in 50 mM Hepes⋅KOH, pH 7.5/50 mM NaCl/2 mM DTT/2.5% glycerol/100 μg/ml BSA. Purified Polβ (9 fmol) and 0, 8, 80, or 800 fmol of Ape protein were added, and the mixtures (final volume 10 μl) were incubated at 22°C. At the indicated times, samples were removed from the reactions and treated with NaBH4 (final concentration 340 mM) to stabilize the AP sites (16). The samples were ethanol-precipitated, resuspended in H2O, mixed with an equal volume of formamide-containing loading buffer, and analyzed on a denaturating 20% polyacrylamide gel (18).
RESULTS
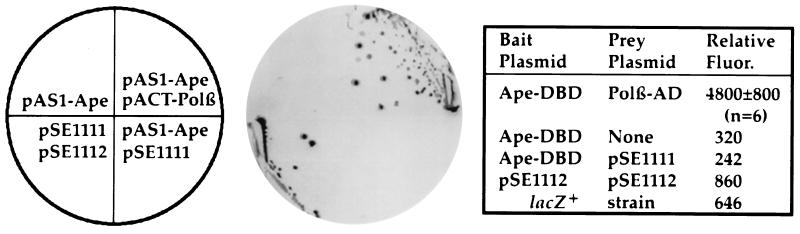
We initially tested for interaction between Ape and Polβ using the yeast two-hybrid system (25). Fusions of Ape to the yeast Gal4 DNA-binding domain (Gal4DB–Ape) and of Polβ to the Gal4 transactivation domain (Gal4AD–Polβ) were coexpressed in Saccharomyces cerevisiae bearing appropriate Gal4-regulated selection and reporter genes (20). Activation of these genes by reconstitution of Gal4 activity was observed in cells coexpressing the Gal4DB–Ape and the Gal4AD–Polβ fusions (Fig. 1). No reporter gene activation was observed when the Gal4DB–Ape fusion was expressed alone or with a different protein fused to the Gal4 transactivation domain (Gal4AD–Snf4 encoded by pSE1111, ref. 19; Fig. 1). Direct assay of β-galactosidase showed strong activation of the lacZ reporter gene in six independent transformants coexpressing GalDB–Ape and Gal4AD–Polβ (Fig. 1). Two-hybrid analysis gave no indication that Ape interacts with another component of the base excision repair pathway, human alkylpurine-DNA glycosylase (26). Additional control experiments found no evidence for nonspecific interactions of Polβ with other proteins (yeast Snf1 (19) or Epstein–Barr virus LMP1 (27) (data not shown). Thus, Ape and Polβ interact specifically in vivo.
Figure 1.
Interaction of Ape and Polβ in the yeast two-hybrid system. (Left) Schematic of plasmids present in indicator strain Y190 (20). Plasmid pAS1–Ape encodes the Gal4DB–Ape fusion; pACT–Polβ encodes the Gal4AD–Polβ fusion; pSE1111 encodes a Gal4AD-Snf4 fusion; pSE1112 encodes a Gal4DB-Snf1 fusion (19). (Center) Indicator plate for β-galactosidase expression from a GAL promoter-lacZ fusion and His+ selection from a GAL promoter-HIS3 fusion. (Right) Quantitation of β-galactosidase expression by a fluorescent assay. Ape-DBD is the Gal4DB–Ape fusion, Polβ-AD the Gal4AD–Polβ fusion. Six independent cotransformants of pAS1-Ape and pACT-Polβ were assayed for β-galactosidase activity; single isolates of the other transformants were assayed. An S. cerevisiae lacZ+ strain (obtained from R. Brennan and R. H. Schiestl, Harvard School of Public Health, Boston) was assayed as a positive control.
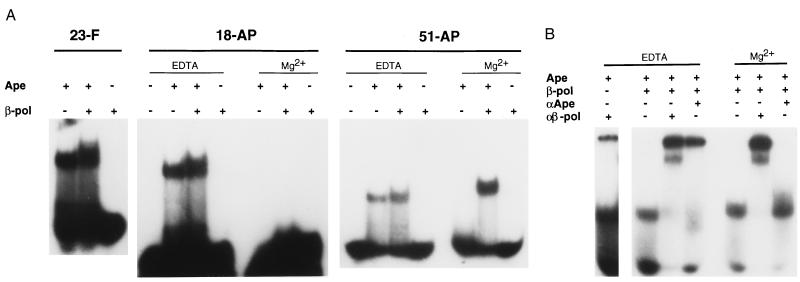
We next sought in vitro evidence for the interaction of Ape and Polβ. Because these two proteins act in consecutive steps of base excision repair, we tested whether they might interact on a DNA substrate. We recently have shown that Ape protein can form stable complexes with uncleaved AP sites in DNA (22). Such complexes are detectable by filter binding or EMSA and are enhanced in the presence of the metal chelator EDTA (22). In the experiments shown here, Ape⋅AP–DNA complexes were seen by EMSA for 18-bp, 23-bp, and 51-bp duplex oligonucleotides containing AP sites (Fig. 2A, upper bands; lower bands correspond to the free DNA). The subsequent addition of purified Polβ produced complexes of slightly slower mobility in EMSA than those found with Ape alone (Fig. 2A). When Mg2+ was added to support Ape incision activity (22), no complex of Ape alone or with Polβ was detected with the 18-bp substrate (Fig. 2A), probably due to denaturation of the short (9- and 8-nucleotide) strands after cleavage. However, incubations with the 51-mer generated a substantial amount of complex in Mg2+ when both Ape and Polβ were present. This result shows that, as expected, Ape alone does not remain bound to an incised AP site, although Polβ can bind such a site strongly. This experiment did not resolve whether Ape remained bound after cleavage in the presence of Polβ, or whether both proteins are present in the complexes formed under noncleavage conditions.
Figure 2.
Loading of Polβ onto AP sites by Ape protein. (A) Binding to AP sites in different sequence contexts. Purified human Ape protein (0.55 pmol) or Polβ (0.55 pmol) were incubated with the indicated 5′-labeled AP substrates in reactions containing either 4 mM EDTA or 10 mM MgCl2 (Mg2+), and the uncomplexed DNA (lowest bands) and protein DNA complexes (upper bands) resolved by electrophoresis in a nondenaturing gel (22). Binding substrates were 23-F, a 23-bp duplex DNA containing a tetrahydrofuran residue (10); 18-AP, an 18-bp oligonucleotide containing an AP site generated by uracil excision (10); and 51-AP, a 51-bp duplex oligonucleotide containing an AP site generated by uracil excision (13). (B) Presence of Ape and Polβ in complexes. Binding reactions with EDTA or Mg2+ were carried out with Ape or Polβ and the 51-AP substrate as described above, then Ape-specific (αApe) or Polβ-specific (αβ-pol) antisera were added. The complexes containing Ape or Polβ (middle bands) were resolved from the antibody-supershifted complexes (top bands) by electrophoresis in nondenaturing gels. In no case was significant material retained in the wells of the gels.
These questions were addressed using Ape-specific and Polβ-specific antisera in EMSAs. In EDTA, Ape and Polβ together formed complexes (middle bands in Fig. 2B) that were super-shifted quantitatively when antibody against either protein was included in the incubation. Antibody binding formed complexes that were substantially more retarded in the gel than those formed by Ape and Polβ alone (upper “super-shifted” bands in Fig. 2B). We noted that a greater proportion of the AP substrate was bound in protein-DNA complexes in the presence of antibodies. Perhaps antibody binding stabilizes the complexes of Ape and Polβ with the DNA. A minor fraction of the Ape–DNA complex was further shifted by the anti-Polβ antiserum (Fig. 2B, Left), which indicates a weak crossreactivity. In the presence of Mg2+, super-shifting was observed only with the anti-Polβ antibodies, even though both Polβ and Ape were present in the binding reaction (Fig. 2B); the anti-Ape antibodies evidently do not crossreact with Polβ in this assay. Thus, only Polβ remains stably bound to the nicked DNA substrate in complexes resolved by EMSA. This result does not rule out a looser association of Ape with Polβ on the incised DNA.
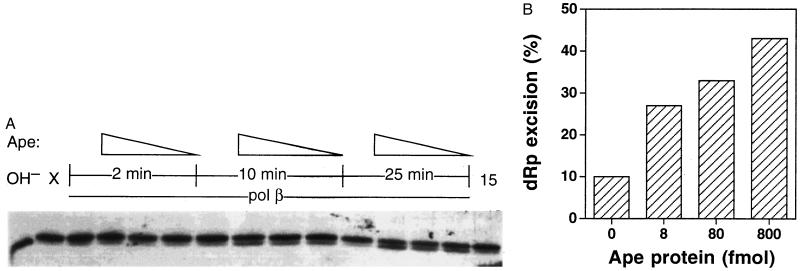
The experiments of Fig. 2 indicated that Ape acts as a loading factor for Polβ onto nonincised AP sites in DNA. The next question was whether this interaction affects the enzymatic activities of either protein. Polβ at various concentrations had no detectable effect on Ape’s AP endonuclease activity (using various substrates; refs. 10 and 11) or 3′-repair diesterase activity (11) (data not shown). However, Ape stimulated 5′-dRp excision by Polβ. The release of dRp was monitored using a 51-mer DNA substrate containing a central AP site (13) previously incised with a catalytic amount of E. coli endonuclease IV, which cleaves in the same position as Ape (5). As shown previously (16), high-resolution gel electrophoresis separates the 3′-labeled fragment bearing a 5′-dRp moiety (Fig. 3A, upper bands) from the product without dRp (Fig. 3A, lower bands). Using such a substrate, 9 fmol of Polβ alone removed only 10% of the 5′-dRp during a 25-min incubation. Addition of 8, 80, or 800 fmol of Ape protein increased the rate of dRp excision by Polβ by 2.7-, 3.3-, and 4.3-fold, respectively, for the 25-min reaction (Fig. 3B). The stimulating effect of Ape on the dRp-excision activity of Polβ also was observed for 10-min reactions (Fig. 3A) and in several independent experiments. The highest level of Ape protein alone (800 fmol) exhibited little (≤10% excision) or no detectable dRp-releasing activity over many experiments (Fig. 3A, far right lane; unpublished data). Addition of up to 18 pmol of E. coli exonuclease III, the bacterial homolog of Ape (5), did not stimulate dRp excision by Polβ (data not shown).
Figure 3.
Activation of Polβ dRp excision activity by Ape. (A) A duplex oligonucleotide containing an AP site at position 22 was cleaved with a catalytic amount of E. coli endonuclease IV, then incubated for the indicated times with purified human Polβ (9 fmol) and varying amounts of purified human Ape protein (0, 8, 80, or 800 fmol). After the incubation, the substrate bearing 5′-dRp (upper band) and the product after dRp excision (lower band) were resolved in a denaturing gel. OH−, substrate hydrolyzed with NaOH; X, the 5′-dRp substrate before incubation; 15, 5′-dRp substrate incubated 15 min with 800 fmol of Ape alone. (B) Quantitation of dRp excision. The gel in A was subjected to scanning densitometry and the ratio of the substrate (upper band in A) to product (lower band in A) used to calculate the excision of dRp in 25-min reactions.
DISCUSSION
These experiments demonstrate a previously unrecognized level of coordination in the mammalian base excision repair pathway. A specific interaction of Ape and Polβ occurs that assembles the polymerase onto an AP site in DNA. After AP site cleavage by the endonuclease, the Ape-dependent stimulation of Polβ’s dRp-excision activity then can accelerate the next step in the pathway, and perhaps the subsequent repair synthesis as well. In vivo, the relative amounts of Ape protein (350,000–7,000,000 molecules per cell) (11) and Polβ (≈50,000 molecules per cell) (28) are similar to the range used in our in vitro experiments. An excess of Ape would favor productive interaction with Polβ, such that AP sites would most likely already be associated with a molecule of Ape before the binding of Polβ.
A previous effort failed to identify the Ape–Polβ interaction using affinity chromatography (29), and we also have been unsuccessful in that approach. These negative results might result from a requirement for the Ape–Polβ interaction to occur in the context of DNA, as was the case for the EMSA approach used here. In the two-hybrid experiments, the additional proteins of Gal4 and the transcriptional apparatus also could act as a scaffold for the interaction (25).
There appear to be two branches of base excision repair in mammalian cells that differ in the DNA polymerase required (13, 30): a Polβ-dependent branch that repairs ≈90% of the AP sites and a DNA polymerase δ- or ɛ-dependent branch. Recent observations indicate that Polβ also interacts with DNA ligase III via XRCC1 protein (31, 32) and perhaps with DNA ligase I (29). Interaction of Polβ with DNase V, a bidirectional exonuclease, has been known for some time (33). Thus, Polβ may participate in multiple protein–protein interactions to coordinate steps of the base excision pathway.
Engineered “knock-out” of either Ape or Polβ generates an embryonic-lethal phenotype without apparent tissue-specific defects (14, 34). These observations indicate a crucial role for base excision repair in animal viability. It is not known whether viable Ape-deficient cell lines can be obtained. However, Polβ-deficient mouse cells do grow in culture, where they exhibit hypersensitivity to alkylating agents (14) consistent with a critical role for Polβ in base excision repair. The possibility also should be considered that some repair defects might act by disrupting the coordinating interactions in base excision repair without altering the intrinsic activities of Polβ and Ape. Thus, the molecular basis of the Polβ–Ape interaction needs to be defined in detail.
Acknowledgments
We are indebted to Dr. Steve Elledge for supplying the two-hybrid system vectors and for much helpful advice, to Dr. Stuart Linn for providing purified Polβ and Polβ-specific antiserum, to Dr. Dale Mosbaugh for uracil-DNA glycosylase, and to Drs. R. Brennan and R. H. Schiestl for the yeast strain expressing E. coli β-galactosidase. We are grateful to Drs. James C. Wang and Graham C. Walker for helpful comments on the manuscript. The able assistance of John Davidson in constructing pAS1-Ape is gratefully acknowledged. This work was supported by an National Institutes of Health grant (GM40000) to B.D., a pilot project grant to R.A.O.B. and D.M.W. from the Kresge Center for Environmental Health, Harvard School of Public Health, a National Research Service Award fellowship (CA62845) to D.M.W., and National Institutes of Health training grants supporting R.A.O.B. (CA09078 and ES07155) and D.W. (ES07155).
ABBREVIATIONS
- AP
abasic
- Polβ
DNA polymerase β
- Ape
AP endonuclease
- dRp
deoxyribose 5-phosphate
- EMSA
electrophoretic mobility-shift assay
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 5.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 6.Demple B. Curr Biol. 1995;5:719–721. doi: 10.1016/s0960-9822(95)00143-6. [DOI] [PubMed] [Google Scholar]
- 7.Dianov G, Lindahl T. Curr Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 8.Loeb L A. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- 9.Barzilay G, Hickson I D. Bioessays. 1995;17:713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 10.Wilson D M, III, Takeshita M, Grollman A P, Demple B. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 11.Chen D S, Herman V, Demple B. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demple B, Herman V, Chen D S. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhal R K, Prasad R, Wilson S H. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 14.Sobol R W, Horton J K, Kuhn R, Gu H, Singhal R K, Prasad R, Rajewsky K, Wilson S H. Nature (London) 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 15.Price A, Lindahl T. Biochemistry. 1991;30:8631–8637. doi: 10.1021/bi00099a020. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto Y, Kim K. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 17.Piersen C E, Prasad R, Wilson S H, Lloyd R S. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 20.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 22.Wilson D M, III, Takeshita M, Demple B. Nucleic Acids Res. 1997;25:933–939. doi: 10.1093/nar/25.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randahl H, Elliott G C, Linn S. J Biol Chem. 1988;263:12228–12234. [PubMed] [Google Scholar]
- 24.Levin J D, Shapiro R, Demple B. J Biol Chem. 1991;266:22893–22898. [PubMed] [Google Scholar]
- 25.Allen J B, Walberg M W, Edwards M C, Elledge S J. Trends Biochem Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 26.Samson L, Derfler B, Boosalis M, Call K. Proc Natl Acad Sci USA. 1991;88:9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosalios G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 28.Horton J K, Srivastava D K, Zmudzka B Z, Wilson S H. Nucleic Acids Res. 1995;23:3810–3815. doi: 10.1093/nar/23.19.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasad R, Singhal R K, Srivastava D K, Molina J T, Tomkinson A E, Wilson S H. J Biol Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto Y, Kim K, Bogenhagen D F. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldecott K W, Aoufouchi S, Johnson P, Shall S. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubota Y, Nash R A, Klungland A, Schär P, Barnes D A, Lindahl T. EMBO J. 1996;15:6662–6669. [PMC free article] [PubMed] [Google Scholar]
- 33.Mosbaugh D W, Meyer R R. J Biol Chem. 1980;255:10239–10247. [PubMed] [Google Scholar]
- 34.Xanthoudakis S, Smeyne R J, Wallace J D, Curran T. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]